Abstract
The Biomolecular Interaction Network Database (BIND: http://bind.ca) archives biomolecular interaction, complex and pathway information. A web-based system is available to query, view and submit records. BIND continues to grow with the addition of individual submissions as well as interaction data from the PDB and a number of large-scale interaction and complex mapping experiments using yeast two hybrid, mass spectrometry, genetic interactions and phage display. We have developed a new graphical analysis tool that provides users with a view of the domain composition of proteins in interaction and complex records to help relate functional domains to protein interactions. An interaction network clustering tool has also been developed to help focus on regions of interest. Continued input from users has helped further mature the BIND data specification, which now includes the ability to store detailed information about genetic interactions. The BIND data specification is available as ASN.1 and XML DTD.
INTRODUCTION
The Biomolecular Interaction Network Database (BIND) is designed to capture protein function, defined at the molecular level as the set of other molecules with which a protein interacts or reacts along with the molecular outcome. The inimitable growth of the known cell map continues unabated with new data on the structure of cell signaling and metabolic networks generated by constantly improving techniques such as mass spectrometry and two-hybrid screens (1). Interaction databases such as BIND (2,3) must keep pace so that such data is manageable. In cohort, visualization and analysis tools for this data must be made available to assist in understanding this complex data.
BIND
BIND stores information about interactions, molecular complexes and pathways. Interactions occur between two biological ‘objects’, A and B, which could be protein, RNA, DNA, molecular complex, small molecule, photon (light) or gene. Molecular complexes and pathways are collections of these pairwise interactions, with some additional data. The minimum amount of information required to define an interaction is a description of A and B and a publication reference to PubMed. BIND is based on an extensive ASN.1 data specification [as previously published (4,5)] that can describe much of the detail underlying biochemical and genetic networks. XML versions of all data with accompanying DTDs are supported through the use of the NCBI programming toolkit (http://www.ncbi.nlm.nih.gov/IEB/).
The BIND specification has remained stable since version 2.0 in 2001. Initially, BIND was designed only to support physical/biochemical interactions. Stemming from collaboration with a yeast genetic mapping project (6), our current 3.0 version has a wide range of support for genetic interactions (valid when A and B are genes), where both the genetic experiment and its result can be described in detail. This demonstrates the flexibility and extensibility of our data specification approach. Apart from cumulative minor changes, the current specification version has many general external references to enable integration with private database systems that may be similar to BIND. For instance, an external reference in the BIND-Interaction object can point to an in-house interaction database. Logical collections of records are now called divisions, similar to those in GenBank (see below). Up to date UML diagrams of the 3.0 BIND specification are present in the Supplementary Material.
BIND has progressed through multiple implementation cycles, each benefiting from collaborator and community constructive feedback. As part of our continuing effort to populate BIND, we have imported data from large-scale cell mapping studies, including ones we have been a part of (6–8). Recently, all molecular interactions in PDB (9) were imported into BIND, via the validated MMDB database (10), using MMDBBIND (11). Because of currently limited server space, these 64 956 records are not available for querying through the web interface, but are freely available on our FTP site. MMDBBIND records are currently being enriched in information using a human curation process to create high-quality records that will be part of RefBIND, a curated division of BIND.
As the data in BIND expands, so too must the underlying infrastructure. The public BIND site has been currently running on a shared mid-size web server. A transition to larger, redundant servers is currently being planned in conjunction with the launch of our SeqHound service, upon which BIND depends. SeqHound is our in-house integrated database (18), similar in scope to the Entrez system (12), which contains extensive C, C++ and Perl programming APIs.
New infrastructure for data analysis and visualization tools are also being built. One such tool is Pajek (13) (see Supplementary Material), for visualizing and analyzing large networks, although it has no support for analyzing networks in the context of sequence, structure and associated annotation. We are presently developing tools to address these issues.
An interaction network clustering tool, called Molecular Complex Detection (MCODE), has been developed to help focus on regions of biological interest. MCODE detects densely interconnected regions of a molecular interaction network, which may represent molecular complexes (Bader and Hogue, submitted).
FUNCTIONAL ALIGNMENT SEARCH TOOL (FAST)
Many proteins contain a number of structural and functional modules such as SH3, SH2, kinase and DNA binding domains (14). Most of these domains mediate protein interactions with other biomolecules. A collection of interaction information, such as BIND, enables the study of the relationships between protein domain architecture and protein–protein interactions. Specifically, it is possible to classify the interactors of a protein into distinct groups based on domain composition.
As part of our research and using BIND and SeqHound as platforms, we implemented FAST as an application that displays the domain annotation for a group of functionally related proteins. In BIND, these groups of related proteins can be proteins that interact with a common partner or are found together in molecular complexes. The domain annotation is from SeqHound which contains a complete RPS-BLAST analysis of the GenBank or dataset, using the Conserved Domain Database (15) performed on our 216 Beowulf cluster.
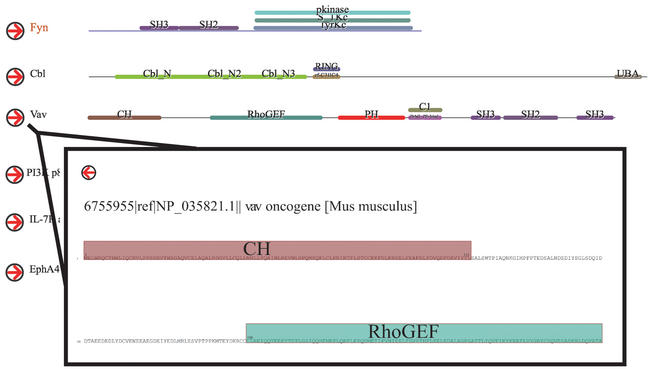
FAST has a web-based graphical interface, based on Macromedia Flash vector graphics, that displays a set of proteins and their domains. Vector graphics format was chosen as it provides improved resolution and zooming ability over bitmap images. FAST is accessible from BIND via interaction and molecular complex records. When accessed from an interaction record, the protein and its protein interactors in BIND are displayed. When accessed from a complex record, the protein subunits are displayed. Domain composition is shown as unique coloured horizontal bars above a line representing the sequence (Fig. 1). Clicking on the arrow beside each protein links a user to an expanded display where domains are shown with respect to the amino acid sequence of the protein. Users can zoom in and out to examine the boundaries of a domain of interest in more detail using the Flash control tool. A domain summary table for the protein set, containing links to information on each protein and domain, can be accessed from the FAST image page.
Figure 1.
Functional Alignment Search Tool (FAST). Domain composition for a set of proteins that interact with mouse Fyn is shown as uniquely coloured horizontal bars above a line representing the sequence. Expanded view of Vav, linked to via right-pointing red arrows, where domains are shown correctly situated on the amino acid sequence of each protein. For brevity, this figure does not show all Fyn-interacting proteins in BIND or, in the expanded view, all of the domains in Vav.
Visualization of a list of related proteins and their domains is a powerful approach to help direct future interaction studies. For example, the human and mouse variants of the protein tyrosine kinase Fyn each have nine recorded interactions in BIND (Fig. 1). The human and mouse forms of Fyn share six similar interactions, however, the mouse variant is known to interact with a second protein tyrosine kinase Vav, whereas the human Fyn currently has no recorded interaction with the human Vav homologue. Using FAST, it is easy to see that many Fyn-interacting proteins, including Vav, contain common cell-signaling modules such SH2 and SH3 domains. In combination with other tools and databases such as NCBI's CDART (17), human homologues with similar domain architectures to mouse Fyn interactors can be identified (e.g. VAV-3 and TIM). These proteins potentially interact with human Fyn.
FAST can also be used to study the topology and function of molecular complexes. A number of protein complexes were recently identified in large-scale mass-spectrometry studies (7,16). FAST can help decipher the interaction topology of these complexes by grouping proteins according to their domain composition. For example, part of the proteasome complex was identified using the protein Ygl004c as bait (BIND complex ID 11939). The domain architecture of the identified proteins reveals three distinct subgroups corresponding to three functional elements that control proteasome activity: ATPase (Rpt5, Rpt4, Rpt3, Rpt2, Rpt1), proteasome (Rpn9, Rpn7, Rpn6, Rpn5, Rpn3) and proteasome regulatory subunits (Rpn8, Rpn11).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Tony Pawson for putting forth the idea of an interaction database pre-1998, Cheryl Wolting and Ian Donaldson for past contributions, Adrian Heilbut for helping to import recent molecular complex records and colleagues in the Hogue lab and at the SLRI for helpful discussions. D.B. wrote the FAST software. This work is funded by a consortium including Genome Canada, the Canadian Institutes of Health Research (CIHR), the Ontario Research and Development Challenge Fund, IBM and MDS Proteomics.
REFERENCES
- 1.Fields S. (2001) Proteomics. Proteomics in genomeland. Science, 291, 1221–1224. [DOI] [PubMed] [Google Scholar]
- 2.Xenarios I., Salwinski,L., Duan,X.J., Higney,P., Kim,S.M. and Eisenberg,D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res., 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanzoni A., Montecchi-Palazzi,L., Quondam,M., Ausiello,G., Helmer-Citterich,M. and Cesareni,G. (2002) MINT: a Molecular INTeraction database. FEBS Lett., 513, 135–140. [DOI] [PubMed] [Google Scholar]
- 4.Bader G.D., Donaldson,I., Wolting,C., Ouellette,B.F., Pawson,T. and Hogue,C.W. (2001) BIND—the biomolecular interaction network database. Nucleic Acids Res., 29, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bader G.D. and Hogue,C.W. (2000) BIND—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics, 16, 465–477. [DOI] [PubMed] [Google Scholar]
- 6.Tong A.H., Evangelista,M., Parsons,A.B., Xu,H., Bader,G.D., Page,N., Robinson,M., Raghibizadeh,S., Hogue,C.W., Bussey,H. et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science, 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 7.Ho Y., Gruhler,A., Heilbut,A., Bader,G.D., Moore,L., Adams,S.L., Millar,A., Taylor,P., Bennett,K., Boutilier,K. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 8.Tong A.H., Drees,B., Nardelli,G., Bader,G.D., Brannetti,B., Castagnoli,L., Evangelista,M., Ferracuti,S., Nelson,B., Paoluzi,S. et al. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science, 295, 321–324. [DOI] [PubMed] [Google Scholar]
- 9.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S. et al. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Anderson,J.B., Chen,J., Geer,L.Y., He,S., Hurwitz,D.I., Liebert,C.A., Madej,T., Marchler,G.H., Marchler-Bauer,A. et al. (2002) MMDB: Entrez's 3D-structure database. Nucleic Acids Res., 30, 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salama J.J., Donaldson,I. and Hogue,C.W. (2002) Automatic annotation of BIND molecular interactions from three-dimensional structures. Biopolymers, 61, 111–120. [DOI] [PubMed] [Google Scholar]
- 12.Schuler G.D., Epstein,J.A., Ohkawa,H. and Kans,J.A. (1996) Entrez: molecular biology database and retrieval system. Methods Enzymol., 266, 141–162. [DOI] [PubMed] [Google Scholar]
- 13.Batagelj V. and Mrvar,A. (1998) Pajek–Program for large network analysis. Connections, 2, 47–57. [Google Scholar]
- 14.Pawson T. (1995) Protein modules and signalling networks. Nature, 373, 573–580. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 17.Geor L.Y., Domrachov,M., Lipman,D.J. and Bryant,S.H. (2002) CDART: Protein Homology by Domain Architecture. Genome Res., 12, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalickova K., Bader,G.D., Dumontier,M., Lieu,H.C., Betel,D., Issorlin,R. and Hogue,C.W. (2002) SeqHound: biological sequence and structure database as a platform for bioinformatics research. BMC Bioinformatics, in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.