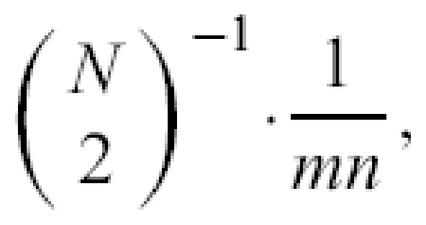Abstract
Advances in proteomics technology have enabled new proteins to be discovered at an unprecedented speed, and high throughput experimental methods have been developed to detect protein interactions and complexes en masse. Such bottom-up, data-driven approach has resulted in data that may be uninformative or potentially errorful, requiring further validation and annotation. The InterDom database focuses on providing supporting evidence for the detected protein interactions based on putative protein domain interactions. Using an integrative approach, InterDom derives potential domain interactions by combining data from multiple sources, ranging from domain fusions, protein interactions and complexes, to scientific literature. The InterDom database is available at http://InterDom.lit.org.sg.
INTRODUCTION
The study of protein interactions is essential in understanding how life's many biological processes work. Traditionally, protein interactions have been studied individually using top-down, hypothesis-driven approaches, with experiments designed to derive high quality detailed interaction information. Today, advances in proteomics technology have enabled new proteins to be discovered at an unprecedented speed, creating a need for high-throughput interaction detection methods such as two-hybrid systems (1,2) and protein chips (3) to detect protein interactions en masse. As these high throughput systems often provide only the mere detection of the physical molecular interactions, such bottom-up, data-driven approaches do not provide much functional insights about the interactions detected. Furthermore, the focus on quantity may have also resulted in a compromise on the quality of the interaction data—high throughput interaction data are now known to contain significant error rates (4).
InterDom, a database of interacting domains, is a compilation of putative protein domain–domain interactions that can be used for in silico validation and annotation of detected protein interactions and complexes. Protein domains are structural or functional units within the proteins themselves, usually evolutionarily-conserved modules of amino acid sequences. The existence of certain domains in proteins can suggest the propensity for the proteins to interact or form a stable complex to bring about certain biological functions. However, unlike protein–protein interaction detection, high-throughput experimental results for domain–domain interactions are currently unavailable. In the InterDom database, we derive putative domain–domain interactions using computational methods. We employ a recipe of different methods to infer the domain interactions from diverse data sources, and then use a probabilistic scoring system to give higher confidence to domain interactions that are derived independently by multiple methods from different data sources. We illustrate with an example how the putative domain–domain interactions in InterDom can be useful for the evidential validation of detected protein interactions.
MATERIALS AND METHODS
InterDom's main strategy is to use multiple methods and data sources to independently derive protein domain–domain interactions, and then assign higher confidence to putative domain interactions that are multiply derived. In version 1.0 of InterDom, we used four different data sources to derive putative domain–domain interactions, namely domain fusions, protein–protein interactions, protein complexes and scientific literature.
Domain fusions
The domain fusion method is based on the observation that some pairs of interacting proteins have homologs in another organism that are fused into a single protein chain. For instance, two interacting proteins in the fly genome might be found as a single longer protein in the worm genome. If such proteins, or protein domains, disparate in a first organism are fused together in a second organism, it suggests to us that they are likely to function or interact together in the first organism. The fused protein A–B is called Rosetta Stone Sequence (5).
For inferring putative interactions between protein domains, the domain fusion method looks for protein domains that are disparate in one organism but are fused together in another. By applying the domain fusion method on the proteins from the SWISS-PROT (6) database (Release 39.6), with the Pfam (7) database (Release 5.5) as a reference database of protein domains, we have deduced 1296 unique domain–domain interactions involving 683 domains.
Protein–protein interactions
Domain–domain interactions can also be derived from pairwise protein–protein interactions. Given two proteins that are known to bind to each other (e.g. in yeast-two-hybrid experiments), we can infer that the domains from the two proteins could potentially be interacting. In other words, if two proteins P1 and P2 are known to bind to each other, then we infer that domain d1,i potentially interacts with domain d2,j with a minimal probability of 1/mn, where m and n are the number of domains in proteins P1 and P2 respectively, and d1,i and d2, j are the ith and jth domains of proteins P1 and P2 respectively.
As the data source of protein–protein interactions, we have integrated the protein–protein interaction data from both the DIP (8) database (Released October 12, 2000), and the BIND (9) database (Released March 15, 2000), to form a set of 14 771 unique protein interactions for inferring domain interactions for InterDom. A total of 3503 putative domain–domain interactions were derived using this method.
Protein complexes
Interactions between proteins are not limited to binary interactions such as those detected by yeast-two-hybrids; several proteins can come together to form a multi-protein complex, and we can infer putative domain interactions from the inter-protein relations in the protein complexes. Suppose proteins P1,… , PN are known to form an N-protein complex. We can infer that the domain dr,i potentially interacts with domain ds, j with a minimal probability of
where m and n are the number of domains in proteins Pr and Ps respectively, and dr,i and ds, j are the ith and jth domains of proteins Pr and Ps respectively.
In InterDom version 1.0, we used 418 protein complexes comprising of up to 24 proteins per complex from the PDB (10) database to derive a total of 1004 putative domain–domain interactions.
Scientific literature
Despite the proliferation of sequence and structure databases, results of scientific research are still reported in scientific journals and conference proceedings in free text format. Fortunately, unique to the field of life sciences is a central repository of scientific abstracts in the MEDLINE database provided by the National Library of Medicine for public access. As such, scientific text mining is becoming an increasingly researched topic in post-genome bioinformatics (11).
In InterDom, we used the text mining approach described in (12) to automatically extract domain–domain interactions, protein–protein interactions, and protein complex information from MEDLINE abstracts as further evidential support for the domain interactions derived from the above methods. A total of 575 InterDom putative interactions have been further annotated using the interactions detected by literature mining.
False positive detection
Since InterDom uses an exhaustive approach to generate all possible domain interactions from the source data, it is important to identify potential false positives in the database. Currently, potential false positives are detected by identifying:
Potentially superfluous interactions. We used a confidence scoring system based on probabilistically-weighted odd ratios to rank domain interactions that were inferred from high throughput protein interactions and complexes. Under this scoring system, higher confidence scores are assigned to single domain interactors as well as domain interactions that were inferred from different protein interactions and complexes. Interactions that were derived solely by the domain fusion method are currently assigned a low confidence score. The confidence scores are then totaled for each inferred domain interaction, so that interactions derived from multiple data sources and methods will be assigned higher overall confidence scores. Domain interactions with low confidence scores can then be identified as likely false positives and InterDom currently uses 1.5 as the cut-off score.
Potentially problematic domains. InterDom also detects putative interactions that involve potentially problematic domains. Promiscuous domains such as SH3 can be deemed unsuitable for accounting for protein interactions, as well as rare domains that have unnecessarily high odd ratios because of their low occurrence counts in the protein data sets. In the current version, InterDom considers single-occurrence domains as well as domains with more than 50 putative interacting partners as potentially problematic domains.
RESULTS
The InterDOM system was implemented in a UNIX environment, with the InterDom data stored in a relational database in MySQL for scalability. Automated methods for searching the databases and dynamically displaying the selected tables and domain interaction graphs were built with a combination of Perl, PHP, Java and HTML.
A total of 5511 putative domain–domain interactions were inferred from the various methods described above, 3308 being identified by InterDom as potential false positives based on the above-mentioned criteria. User can choose to analyze their interaction data based on all the domain interactions derived, or only those not detected as potential false positives. About 29% (1612) of the interactions have multiple supporting evidences from the same class of data source (for example, interactions derived from more than one protein–protein interaction), but currently only 313 putative domain–domain interactions were derived from at least two disparate methods (for example, interactions inferred from domain fusion hypothesis and protein–protein interaction information).
EXAMPLE
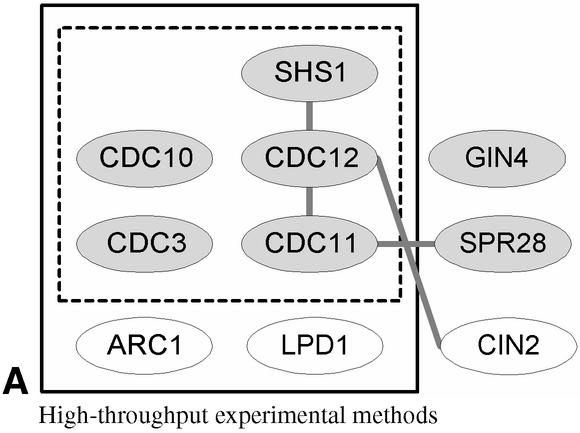
We provide an example on how InterDom can be useful for validating predicted or detected protein interactions and complexes. As an illustration, we use the septin complex, a known protein complex that was also used as an example in von Mering et al.'s recent comparative assessment of large-scale data sets of protein–protein interactions [see Box 2 in (4)].
We used the six annotated members of septin complex from von Mering et al. as input to InterDom to investigate whether the database can provide potential domain–domain interaction links between the six components to form a connected graph structure. It was reported by von Mering et al. that none of the popular high-throughput experimental methods they assessed could lead to a connected graph. By using links based on the putative protein-domain interactions from InterDom, we were able to form a fully connected graph for all six members of the complex (Fig. 1).
Figure 1.
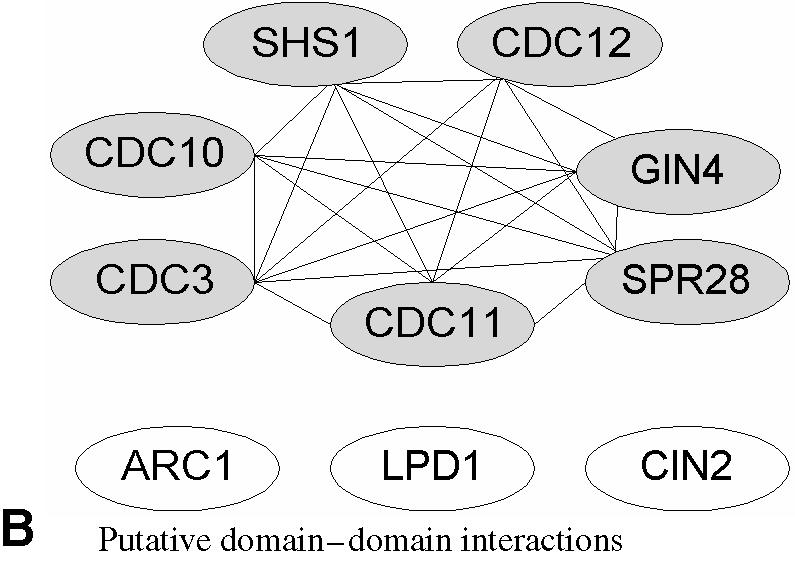
Comparison of connected graph structures for six septin-complex members (shown colored gray) based on high-throughput experimental detection methods and putative domain–domain interactions from InterDom. (A) The graph structure on the top shows the relationships between the six septin-complex members associated by high throughput detection methods as reported in Box 2 of von Mering et al., reproduced here for comparison by permission from Nature (4) copyright (2002) Macmillan Publishers Ltd. The dotted and solid boxes show the complex components detected using TAP purification and HMS-PCI purification methods respectively, and the gray links show the interacting protein components based on two-hybrid interaction. None of the three experimental methods singularly identifies the six members of the septin complex. (B) The graph structure at the bottom shows the result of using InterDoms domain–domain interactions to link up the six septin components and the three non-septin proteins. A link is established between two proteins if there is at least one putative domain–domain interaction between them. In this case, all the six septin-complex proteins were found to be fully connected with putative domain–domain interaction links.
DATABASE ACCESS
Users can access the InterDom database via the World Wide Web (http://InterDom.lit.org.sg) to (a) browse derived domain interactions with the corresponding supporting evidence in various sorted order; (b) search for potential interacting domain partners for an input molecule; and (c) validate predicted or detected protein interactions and complexes using the putative domain interaction links in the database.
FUTURE WORK
The investigation of protein interactions and complexes at the domain level provides a new granularity for the understanding, annotation and validation of predicted or detected protein interactions and complexes. Using our integrative approach, the quality of the domain interaction data in InterDom can be improved by expanding the variety and coverage of the source data. In the current version of InterDom, we have used the domains from the well-curated PfamA families (7) to decompose a protein into its domains. We can improve the domain coverage by using PfamB and other domain classifications. We will also investigate the use of larger data sets as well as additional data sources and methods to arrive at better quality data in future.
REFERENCES
- 1.Uetz P., Giot,L., Cagney,G., Mansfield,T.A., Judson,R.S., Knight,J.R., Lockshon,D., Narayan,V., Srinivasan,M., Pochart,P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 2.Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu H., Bilgin,M., Bangham,R., Hall,D., Casamayor,A., Bertone,P., Lan,N., Jansen,R., Bidlingmaier,S., Houfek,T. et al. (2001) Global analysis of protein activities using proteome chips. Science, 293, 2101–2105. [DOI] [PubMed] [Google Scholar]
- 4.von Mering C., Krause,R., Snel,B., Cornell,M., Oliver,S.G., Fields,S. and Bork,P. (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature, 417, 399–403. [DOI] [PubMed] [Google Scholar]
- 5.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Detecting protein function and protein–protein interactions from genome sequences. Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 6.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xenarios I., Salwinski,L., Duan,X.J., Higney,P., Kim,S.M. and Eisenberg,D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res., 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bader G.D., Donaldson,I., Wolting,C., Ouellette,B.F., Pawson,T. and Hogue,C.W. (2001) BIND—The Biomolecular Interaction Network Database. Nucleic Acids Res., 29, 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S. et al. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mack R. and Hehenberger,M. (2002) Text-based knowledge discovery: search and mining of life-sciences documents. Drug Discov. Today, 7, S89–S98. [DOI] [PubMed] [Google Scholar]
- 12.Ng S.K. and Wong,M. (1999) Toward Routine Automatic Pathway Discovery from On-line Scientific Text Abstracts. Genome Inform. Ser. Workshop Genome Inform., 10, 104–112. [PubMed] [Google Scholar]