Abstract
Recent availability of extensive genome sequence information offers new opportunities to analyze genome organization, including transposon diversity and accumulation, at a level of resolution that was previously unattainable. In this report, we used sequence similarity search and analysis protocols to perform a fine-scale analysis of a large sample (≈17.2 Mb) of the Arabidopsis thaliana (Columbia) genome for transposons. Consistent with previous studies, we report that the A. thaliana genome harbors diverse representatives of most known superfamilies of transposons. However, our survey reveals a higher density of transposons of which over one-fourth could be classified into a single novel transposon family designated as Basho, which appears unrelated to any previously known superfamily. We have also identified putative transposase-coding ORFs for miniature inverted-repeat transposable elements (MITEs), providing clues into the mechanism of mobility and origins of the most abundant transposons associated with plant genes. In addition, we provide evidence that most mined transposons have a clear distribution preference for A + T-rich sequences and show that structural variation for many mined transposons is partly due to interelement recombination. Taken together, these findings further underscore the complexity of transposons within the compact genome of A. thaliana.
Transposons are fundamental components of most eukaryotic genomes, with important contributions to their size, structure, and variation. Based on mode of mobility, transposons are divided into two classes. Class I transposons move through an RNA intermediate and are reverse transcribed before their integration at another location in the genome. Retroelements can be further subdivided into retrotransposons (e.g., copia-like and gypsy-like) which are flanked by long terminal repeats (LTRs) and non-LTR retroelements (e.g., long and short interspersed nuclear elements). Class II elements are characterized by terminal inverted repeats (TIRs) and move directly through a DNA form by a “cut and paste” mechanism (1). Structural features, shared sequence similarity, and the size and sequence of the target site duplication (TSD) generated upon insertion, serve to further distinguish transposon superfamilies. As mutational agents, much of transposon impact may be deleterious to their hosts, although some insertions appear to play a significant role in adaptive evolution (2–4).
Historically, transposon discovery and analysis have been primarily conducted through the molecular genetic characterization of transposon-induced morphological mutations (1). Whereas these studies have allowed for the characterization of many mobile element groups, they do not allow for fine-scale investigation into the extent of transposon diversity and into the forces driving this variability. As genome sequencing projects increase in scale and number, detailed analysis of the patterns of transposon diversity and abundance, and their contribution to genome organization, becomes possible (5). The evidence thus far indicates that these patterns may be extremely variable among eukaryotic genomes (6, 7), suggesting the importance of such studies across diverse organisms.
The genome of the model plant Arabidopsis thaliana is small, with a correspondingly low repetitive DNA content relative to other higher plants (8), and is currently targeted for complete sequencing. Previous studies using computer-based sequence similarity searches have revealed numerous repetitive elements within the Arabidopsis genome (9, 10). Similarly, the complete sequences of Arabidopsis chromosomes 2 and 4 have also allowed for the identification of transposon-related sequences (11, 12). In this report, we performed a systematic and fine-scale search of Arabidopsis genome sequences to identify and characterize transposons. Our study differs from previous mining attempts in that we not only have compiled repetitive sequences but also provide evidence that many are in fact transposons by structural analysis and demonstration of past mobility. In this way, we provide insight into Arabidopsis transposon structure, mobility, distribution, and diversity.
Materials and Methods
Transposon Mining.
Arabidopsis genomic sequences from 243 clones (approximately 17.2 Mb) with representation from all five linkage groups, were accessed from GenBank (National Center for Biotechnology Information, NCBI; http://www.ncbi.nlm.nih.gov/), between June and December 1998. Intergenic and intron sequences longer than 500 bp in length were used as blast search queries (version 2.0, http://www.ncbi.nlm.nih.gov/blast/) (13). Repetitive sequences with similarity to the query (score >80) were compiled into groups and used as queries in additional database searches. A total of 770 repetitive sequences belonging to 197 groups were identified. Of these, 444 mined sequences belonging to 135 groups were determined to be members of previously described transposon superfamilies by virtue of shared sequence composition and/or structural features such as TIRs, LTRs, a terminal poly(A)-rich sequence, coding capacity for mobility-related proteins, and flanking direct repeats (i.e., TSDs). Another 7 repetitive sequence groups representing 179 elements were determined to be mobile elements by documentation of a mobile history (see below). In many cases, some members of each mobile element group lacked one or both terminal sequences and were defined as truncated. Lastly, 147 repetitive sequences belonging to 55 groups could not be classified as transposons based on structural and sequence analysis. A detailed description of the mined mobile elements in our survey can be accessed from our relational database (http://soave.biol.mcgill.ca/clonebase/). A subset of the mined transposons was previously identified and/or annotated to be repetitive DNA, putative transposons, or transposon-related sequences by other researchers (6, 9–12, 14–22).
Data Analysis.
When necessary, further sequence analysis and alignments were performed by using clustal w (23), diverge, translate, pileup, bestfit, and gap from the University of Wisconsin Genetics Computing Group suite of programs (version 10.0) or additional blast search tools provided at NCBI (13, 24). Visualization of amino acid alignments was done by using genedoc (25). A perl program was written to compile sequences immediately flanking transposon insertion and to calculate the average G + C content using a 20-bp sliding window (written by C. Olive, McGill University). Only flanking sequences of intact, and not truncated, elements were included in the calculations. As a control, 50 positions within intergenic regions were randomly selected and submitted to the program. A copy of this program is available upon request.
Documentation of Mobile History.
Sequences immediately flanking the element were used as queries in database searches. Sequences sharing similarity to the queries typically represented either orthologous or, more frequently, paralogous regions (e.g., multigene families, transposons, duplicated genomic regions, or other repetitive sequences). In many cases, a pairwise comparison could be used to identify a gap corresponding to the absence of the insertion. This mined sequence with high nucleotide sequence similarity to the original query but lacking the insertion is referred to as a related to empty site (RESite). Examination of RESites also served to delimit element termini and to identify corresponding TSDs when this information could not be obtained from sequence and structural analysis. Alternatively, when a computer-assisted approach failed, RESites were amplified from other ecotypes of Arabidopsis thaliana, Arabidopsis lyrata, or Brassica spp. by using a previously described PCR approach (ref. 26; data not shown).
Results and Discussion
To obtain a complete transposon profile of the Arabidopsis genome, we systematically surveyed a large sample (≈17.2 Mb) of genomic sequences for transposon insertions by using sequence similarity search algorithms (13, 24). Demonstration of past mobility of the mined elements was provided through the identification of RESites, which are sequences similar to the empty site of an insertion (Fig. 1). This approach allows a rapid and efficient means to delimit element termini and highlight putative target site duplication events. We suggest that RESites provide convincing evidence that many mined interspersed repetitive sequences in Arabidopsis are in fact transposons. Together, mined repetitive sequences and their corresponding RESites strongly support a transposition-based insertion mechanism.
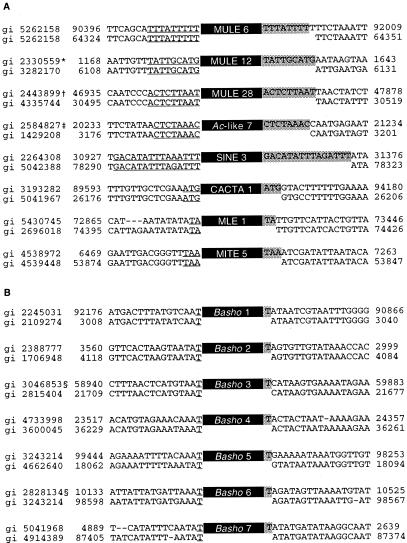
Figure 1.
RESites corresponding to mined Arabidopsis elements. (A) Examples of RESites for different groups of mined elements. All of the RESites illustrated were identified by computer-assisted database searches. In total, 34 RESites were found by computer-assisted database searches, and 13 were identified by cross-ecotype PCR analysis. The target sequences are underlined, and the TSDs are shaded. GenBank geninfo identifier (gi) numbers and nucleotide position on clones are indicated. *, Inserted into a Basho III element; †, inserted into a Basho III element; ‡, inserted into a MITE IX element. (B) RESites found for Basho insertions confirm mononucleotide TSD (shaded). §, Inserted into a Basho V element.
Based on shared sequence and structural similarities and the analysis of RESites, the majority of repetitive sequences mined in our survey (82%) could be compiled and categorized into 142 groups of putative transposons. The majority of the mined transposons corresponded to known superfamilies according to shared structural and sequence similarities (Table 1, Fig. 1A). Among the groups of mined transposons, 28 have members that are structurally reminiscent to the maize Mutator element and/or harbored ORFs with significant similarity to the maize Mutator transposase (i.e., MURA). Mutator-like elements (MULEs) were in part defined as transposons with long TIRs (TIR-MULEs) as in the case of the maize Mu family of elements (30). However, some MULEs lack long TIRs (non-TIR-MULES) but have other features characterizing them as MULEs such as a 8- to 10-bp TSD and, for 5 elements, an ORF encoding a MURA-like transposase. In fact, sequence comparison and analysis of Arabidopsis TIR- and non-TIR-MULEs indicate that they share a common evolutionary history (Z.Y., S.W., and T.B., unpublished data).
Table 1.
Transposons in 17.2 Mb of the Arabidopsis thaliana (Columbia) genome
| Type | Superfamily | Number of groups | Number of transposons |
|---|---|---|---|
| Class I | SINEs* | 3 | 16 |
| LINEs† | 28 | 31 | |
| copia-like Retrotransposons | 27 | 40 | |
| gypsy-like Retrotransposons | 23 | 45 | |
| Undetermined‡ | 2 | 2 | |
| Class II | Ac-like | 7 | 38 |
| CACTA-like | 1 | 3 | |
| MULEs | 28 | 108 | |
| MITEs | 15 | 105 | |
| MLEs | 1 | 56 | |
| Class? | Basho | 7 | 179 |
| Total | 142 | 623 |
Short interspersed nuclear elements (SINEs) are defined as elements that lack coding capacity or have no similarity to coding regions and have either a putative pol III promoter, a long TSD, and/or a poly A+T-rich tail at one terminus (27, 28).
Long interspersed nuclear elements (LINEs) are defined as having members with sequence similarity to the coding domains of previously reported LINEs.
These elements structurally resemble LTR-retrotransposons (i.e., an element with LTRs and 4-bp TSDs but no coding capacity and a putative solo-LTR) but lack signature sequences typical of copia-like or gypsy-like retrotransposons (29).
Interestingly, over one-fourth of the elements identified from 7 distinct groups did not belong to any of the previously described superfamilies but appeared related based on common structural features. The identification of 9 RESites indicate that these elements, which we have named Basho (after the nomadic Japanese haiku poet), have defined termini (5′-CHH … CTAG-3′, where H = A, T, or C) and a target site preference for the mononucleotide “T.” Basho-like elements have been previously described as repetitive sequences and putative insertion sequences (9, 16), but no evidence (e.g., RESites, defined element termini, and target site sequence) was presented to indicate whether they are in fact mobile elements. Curiously, Basho elements appear to insert in a preferred orientation relative to the sequence context of the target site, namely 5′-AT-3′ (Fig. 1B). In addition, we have identified a Basho-like group in maize (data not shown), suggesting that Basho defines a new superfamily of transposons. Although high sequence similarity and RESites attest to past mobility, the mechanism by which Basho elements have transposed is presently unknown.
Our survey permitted an examination of the genomic patterns of transposons within Arabidopsis. For many eukaryotes, there appears to be a relationship between the number of transposons, primarily class I elements (i.e., LTR and non-LTR retrotransposons), and genome size (31). In very large genomes, for instance, the transposon content can account for the majority of the genome (32). In agreement with the small size of the Arabidopsis genome, approximately 5% of the genomic sequences surveyed were composed of transposons of which only 2% were class I elements. Consistent with previous estimates of the retrotransposon content in Arabidopsis (10, 11, 18), we found significantly fewer class I elements than reported in the genomes of other higher plants (29). This contrast in abundance of transposon type between various genomes may suggest differential success depending on genomic environment.
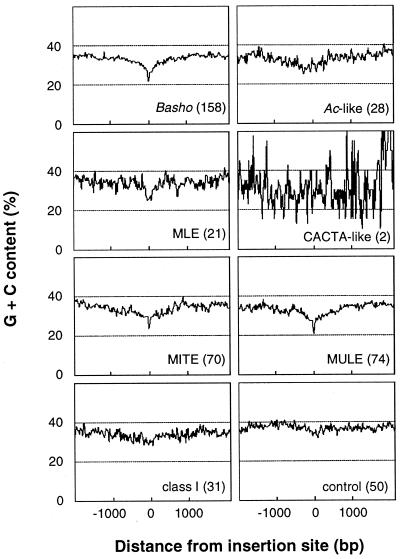
The mined transposons were predominantly found in intergenic regions, with 5% located within introns of predicted genes, and approximately 8% were nested insertions. The prevalence of mined transposons in noncoding regions is similar to the patterns which have been observed in other organisms; for example, heterochromatic regions in Drosophila and intergenic regions in maize typically contain numerous nested transposons (32, 33). A previous report also suggests that transposons are highly enriched within centromeric heterochromatin in Arabidopsis (34). In this report, only four of the clones surveyed in our study correspond to centromeric heterochromatin. Therefore, a meaningful comparison between these regions and euchromatic regions could not be achieved. Whereas many insertions appeared to have a random target site sequence context, preferences for specific A + T-rich target sites were observed for three of the most abundant types of elements: miniature inverted-repeat transposable elements (MITEs) (TA, TAA, ATA), Basho (T), and Mariner-like elements (MLE; TA) (Fig. 1). In addition to sequence-specific target sites, these elements appear to be distributed preferentially in A + T-rich regions. Up to 300 bp of sequences immediately flanking the site of insertion show a G + C content of ≈25% (Fig. 2), which is lower than previous estimations for the Arabidopsis genome [36.7% (35) and 35.8% (11)] and lower than observed in our control (Fig. 2). Arabidopsis MULEs, which do not appear to have target sequence preference are also preferentially distributed in A + T-rich regions.
Figure 2.
A majority of mined transposons show an insertion preference for A + T-rich regions. The number of transposons used in the calculations is indicated in parentheses. The average G + C content was determined by using a 20-bp sliding window over 2000 bp of sequences flanking the transposon insertion sites. Only two CACTA-like elements were used, resulting in a low signal-to-noise ratio. Individual groups (see Table 1) of class I elements showed a similar profile (data not shown) as indicated for the entire class I average.
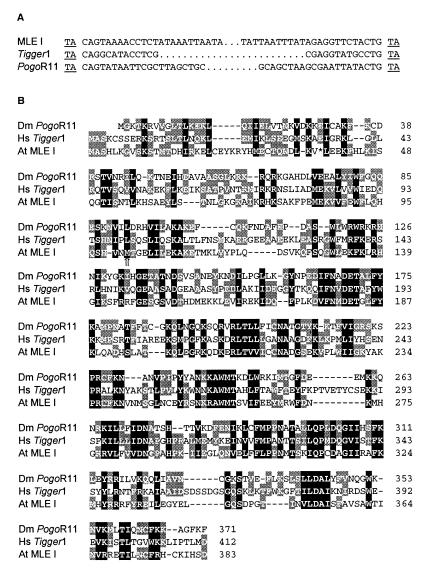
For a subset of mined transposons, the high level of shared nucleotide sequence similarity observed (>90%) suggests that elements have been recently active. Shared sequence identity between flanking regions of elements and matching RESites also attest to recent mobility (Fig. 1). Although the majority of mined elements lacked any coding capacity, some members of mined groups were found harboring corresponding ORFs, such as reverse transcriptase, Ac-like transposase, CACTA-like transposase, or maize Mutator transposase (29, 30, 36). In addition, one group of mined elements (MLE I) not only shares structural features with the Tc1/Mariner transposon superfamily (Fig. 3A), but also has at least one member located on chromosome 2 that harbors an ORF with up to 46% amino acid sequence similarity with the transposase of Tc1/Mariner-like elements, PogoR11, and Tigger1 (Fig. 3B). Furthermore, MLE I elements have the conserved terminal bases (5′-CAGT-3′) necessary for the efficient transposition of other typical Tc1/Mariner-like elements (37). Some members of the MLE I have previously been reported to belong to a novel family of MITEs, referred to as Emigrant (14), based on their small size and target site preference for the dinucleotide TA. However, the MLE I elements clearly have more in common with transposons of the Tc1/Mariner superfamily (Fig. 3) than to elements belonging to the MITE superfamily (38). The mined MLE I transposase shares no significant sequence similarity with two Tc1/Mariner-like transposases reported by Lin et al. (11) also on chromosome 2.
Figure 3.
Structure of an Arabidopsis Tc1/Mariner-like transposon. (A) Similarities between TIRs and TSDs (underlined) of an Arabidopsis MLE I member and Tc1/Mariner-like elements Pogo (Drosophila, gi 8354) and Tigger (human, gi 2226003). (B) Putative transposase for the Arabidopsis MLE I (gi 4262216) aligned with transposases from Drosophila melanogaster PogoR11 (gi 2133672) and from human Tigger1 (gi 2226004). Amino acid residues are shaded based on the level of structural and functional similarities. Residues conserved between three or two sequences are shaded black and gray, respectively. The arrow (⇑) indicates the predicted start of the Arabidopsis MLE I ORF as annotated in GenBank. The first methionine of the Arabidopsis MLE I transposase was inferred from the reading frame and sequence similarity with the human Tigger1 element. The stop (*) was introduced by a single nucleotide substitution (at position 85709 in gi 4262209) from GAG (glutamine) to TAG (stop).
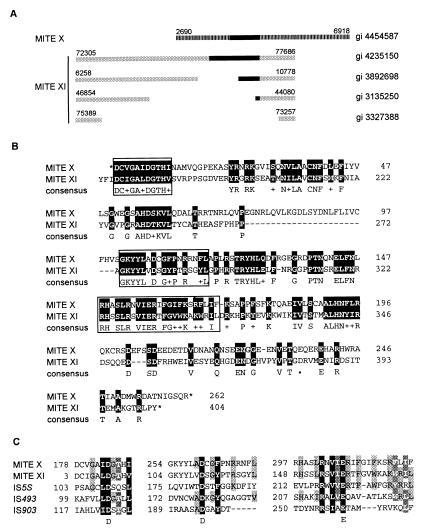
Whereas the mechanism of MITE mobility was previously unclear, we found evidence that some groups of MITEs have unusually large members that potentially encode for a transposase (Fig. 4A). Specifically, putative ORFs found in members of two distinct MITE groups (X and XI) share 51% amino acid sequence similarity (Fig. 4B). In addition, the ratio of synonymous to nonsynonymous nucleotide substitutions of 2.82 suggests the possibility of functional constraint on this coding sequence. These putative ORFs also share sequence similarity with the transposases of cyanobacterial and other prokaryotic insertion sequences (IS) (Fig. 4C). Because MITE X and XI members are structurally similar to other MITE groups in Arabidopsis, as well as the Tourist-like elements in grasses (43), we suggest that the MITE X and XI ORFs represent the transposases characteristic of the Tourist-like family of elements.
Figure 4.
MITE transposases (A). Diagram depicting structural similarities between members of MITE XI elements (gray boxes) and a member of MITE X (striped box). Black boxes represent ORFs corresponding to a putative transposase: MITE XI ORFs share >98% amino acid sequence similarity. MITE X and XI are distinct groups because no significant internal nucleotide sequence similarity is observed between MITE X and XI nor for the consensus sequence of their TIRs (5′-GG(G/T)GGTGTTATTGGTT-3′ for MITE X and 5′-GGCCCTGTTTTGTTTG-3′ for MITE XI). However, putative transposase ORFs, length of TIRs (16 bp), and TSD (5′-TTA-3′) of MITE X and XI are similar, indicating a related mechanism of transposition and possibly a common origin for both groups. GenBank gi numbers of the clones from which elements were mined are indicated to the right. Nucleotide positions of transposon termini are indicated above each element. (B) Amino acid sequence similarity between the conceptual translation for MITE X (gi 4454587, nucleotide position 4650–5865) and the corresponding region of MITE XI ORF (gi 4585884). Identical amino acids and functionally or structurally related residues are shaded in black. *, Translational stops. Boxed sequences indicate the region containing the DDE motif. (C) Alignment of conserved regions corresponding to the functionally important DDE motif found in transposases and integrases of many transposable elements (39–42). Transposases are from MITE X (gi 4454587, conceptual translation of nucleotides 4650–5865), MITE XI (gi 4585884), IS5S (gi 1256580) from Synechocystis sp., IS493 (gi 1196467), and IS903 (gi 136129) from Escherichia coli. Amino acid residues are shaded based on the level of structural and functional similarities. Residues conserved between all, four, or three sequences are shaded black, dark gray, and light gray, respectively.
We observe extensive diversity among mined elements, both within and among groups. The large number of mined transposon groups (Table 1) illustrates a high level of evolutionary divergence and is indicative of long-term persistence or the result of horizontal transfer (44). Differences in size between related elements suggests that insertions and deletions may account for a significant proportion of variation within groups and is reflected by the high occurrence of apparently truncated elements (38%). In addition, interelement recombination appears to generate sequence diversity among some of the mined elements. For example, the mosaic structure of members of some MULE groups is due to the exchange of terminal sequences (Fig. 5). Such mosaic elements are apparently capable of transposition, as suggested by both their copy number, sequence similarity, and presence of perfect TSDs. Another possible factor driving sequence diversity in MULEs is the acquisition of truncated cellular genes. For example, a member of MULE I harbors sequences that share ≈85% nucleotide sequence similarity to a region spanning exon 1, intron 1, and exon 2 of the Arabidopsis homeobox gene Athb-1 (Fig. 6; ref. 45; Z.Y., S. W., and T.B., unpublished data). Despite the sequence diversity observed, some conserved motifs among elements exist, which suggests a common interaction with host factors (e.g., general transcription factors and gene regulatory proteins). We have identified a subterminal sequence motif (5′-TTTTCCCGCCAAAA-3′) shared between MITE XI and Ac-like VII. This sequence may be analogous to the motifs shared between the maize Ac and Mutator, which are thought to be important in the regulation of mobility (46). Furthermore, a motif determined to be a matrix attachment region (5′-CAAATTTATTTTTATA-3′) (47) within a member of MULE XXIX is conserved among all group members.
Figure 5.
Interelement recombination in non-TIR-MULEs generates mosaic transposons. (A) Diagrammatic alignment of four members of MULE VI. Black and gray boxes delimit regions of nucleotide sequence similarity (>90%) between members of MULE groups VI. GenBank gi numbers are marked to the right. Nucleotide positions on clones are noted above each element. (B) Expanded view (as boxed in A) indicating the sequence of the putative recombination region.
Figure 6.
Acquisition of a truncated cellular gene by a member of MULE-I. (A) MULE-I (gi 2182289) shares 85% nucleotide similarity with the first two exons and first intron of the homeobox gene Athb-1 (gi 6016704). Conserved nucleotides are shaded in black, and positions on clones are indicated on the right of the alignment. Sequences for the first two exons are boxed. The first ATG is underlined, and an in-frame stop codon of the MULE-I ORF is double underlined. (B) Diagram illustrating the regions of nucleotide similarity (85%) between the MULE-I and the genomic sequence of Athb-1 (shaded in gray). Positions for the Athb-1 gene and MULE-I element on their respective clones are indicated. MULE-I TIRs are represented by black triangles, and the TSD sequences are indicated flanking both termini.
Our report documents the features and characteristics of the surprisingly diverse forms of transposons residing in, for the most part, noncoding regions of the relatively small and compact genome of Arabidopsis. Lack of insertions into coding regions and prevalence in A + T-rich sequences/regions may reflect purifying selection against deleterious mutations and/or insertion preference of many mined elements. In either case, richness of elements in nongenic regions may represent merely genomic “junk,” or, in some cases, elements may have a host function, as has been observed in other organisms (2). Clearly, the actual impact of transposons in genome evolution will require further molecular dissection of the factors important in gene expression, as well as analysis of population dynamics of element insertions. Our survey also provides additional clues as to the origin and evolution of transposons. Significant similarity between transposases of bacterial insertion sequences and the mobility-related proteins found in this (MITE X and XI) and other studies (Mutator or MULEs, Tc1/Mariner, and retroelements) (39–41, 48) suggests that the majority of element superfamilies are ancient components of genomes. In addition, recombination appears to be important in giving rise to novel forms of MULEs and possibly other types of elements in Arabidopsis.
By providing a fine-scale approach in the mining of transposons from sequence databases, we have demonstrated that the abundance and variation of transposons in Arabidopsis is higher than previously reported. This observation obviously highlights the need for continued analysis of genomic sequence information to provide a high-resolution image of the Arabidopsis genome (49). Finally, information on transposon structures, mobile histories, and genomic coordinates presented in our report will expedite the development of novel biotechnologies for mapping purposes, systematic gene disruption, and functional analysis.
Acknowledgments
We thank Dr. Daniel J. Schoen for comments on the manuscript. We are grateful to Chris Olive, Chengbin Feng, Hala Razi Khan, and Sylva Petrova for setting up and maintaining our complete transposon profile database, which can be accessed at http://soave.biol.mcgill.ca/clonebase/. This work was funded by a National Science and Engineering Research Council of Canada grant to T.E.B.
Abbreviations
- gi
geninfo identifier
- IS
insertion sequence
- MITE
miniature inverted-repeat transposable element
- MLE
Mariner-like element
- MULE
Mutator-like element
- RESite
related to empty site
- TIR
terminal inverted repeat
- TSD
target site duplication
- LTR long terminal repeat.
References
- 1.Berg D E, Howe M M, editors. Mobile DNA. Washington, DC: American Society for Microbiology; 1989. [Google Scholar]
- 2.Britten R J. Proc Natl Acad Sci USA. 1996;93:9374–9377. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidwell M G, Lisch D. Proc Natl Acad Sci USA. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald J F. Trends Ecol Evol. 1995;10:123–126. doi: 10.1016/s0169-5347(00)89012-6. [DOI] [PubMed] [Google Scholar]
- 5.Jordan I K, McDonald J F. Genetics. 1999;151:1341–1351. doi: 10.1093/genetics/151.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurka J. Curr Opin Struct Biol. 1998;8:333–337. doi: 10.1016/s0959-440x(98)80067-5. [DOI] [PubMed] [Google Scholar]
- 7.Smit A F A. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 8.Leutwiler L S, Hough-Evans B R, Meyerowitz E M. Mol Gen Genet. 1984;194:15–23. [Google Scholar]
- 9.Surzycki S A, Belknap W R. J Mol Evol. 1999;48:684–691. doi: 10.1007/pl00006512. [DOI] [PubMed] [Google Scholar]
- 10.Wright D A, Voytas D F. Genetics. 1998;149:703–715. doi: 10.1093/genetics/149.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X, Kaul S, Rounsley S, Shea T P, Benito M-I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 12.Mayer K, Schüller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Düsterhöft A, Stiekema W, Entian K-D, Terryn N, et al. Nature (London) 1999;402:769–777. [Google Scholar]
- 13.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Casacuberta E, Casacuberta J M, Puigdomènech P, Monfort A. Plant J. 1998;16:79–85. doi: 10.1046/j.1365-313x.1998.00267.x. [DOI] [PubMed] [Google Scholar]
- 15.Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, et al. Nature (London) 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 16.Doutriaux M P, Couteau F, Bergounioux C, White C. Mol Gen Genet. 1998;257:283–291. doi: 10.1007/s004380050649. [DOI] [PubMed] [Google Scholar]
- 17.Chye M-L, Cheung K-Y, Xu J. Plant Mol Biol. 1997;35:893–904. doi: 10.1023/a:1005947804227. [DOI] [PubMed] [Google Scholar]
- 18.Konieczny A, Voytas D F, Cummings M P, Ausubel F M. Genetics. 1991;127:801–809. doi: 10.1093/genetics/127.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pélissier T, Tutois S, Deragon J M, Tourmente S, Genestier S, Picard G. Plant Mol Biol. 1995;29:441–452. doi: 10.1007/BF00020976. [DOI] [PubMed] [Google Scholar]
- 20.Tsay Y-F, Frank M J, Page T, Dean C, Crawford N M. Science. 1993;260:342–344. doi: 10.1126/science.8385803. [DOI] [PubMed] [Google Scholar]
- 21.Klimyuk V I, Jones J D. Plant J. 1997;11:1–14. doi: 10.1046/j.1365-313x.1997.11010001.x. [DOI] [PubMed] [Google Scholar]
- 22.Wright D A, Ke N, Smalle J, Hauge B M, Goodman H M, Voytas D F. Genetics. 1996;142:569–578. doi: 10.1093/genetics/142.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas K B, Nicholas H B, Jr, Deerfield D W II. EMBNEW NEWS. 1997;4:14. [Google Scholar]
- 26.Bureau T E, Wessler S R. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert N, Labuda D. Proc Natl Acad Sci USA. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshioka Y, Matsumoto S, Kojima S, Oshima K, Okada N, Machida Y. Proc Natl Acad Sci USA. 1993;90:6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walbot V. Genetic Engineering. 1991. , Setlow, J. K., ed. (Plenum, New York), pp. 1–37. [DOI] [PubMed] [Google Scholar]
- 31.Orgel L E, Crick F H C. Nature (London) 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 32.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, et al. Science. 1996;274:765–767. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 33.Vaury C, Bucheton A, Pelisson A. Chromosoma. 1989;98:215–224. doi: 10.1007/BF00329686. [DOI] [PubMed] [Google Scholar]
- 34.Copenhaver G P, Nickel K, Kuromori T, Benito M-I, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell L D, et al. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 35.Barakat A, Matassi G, Bernardi G. Proc Natl Acad Sci USA. 1998;95:10044–10049. doi: 10.1073/pnas.95.17.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fedoroff N V. Cell. 1989;56:181–191. doi: 10.1016/0092-8674(89)90891-x. [DOI] [PubMed] [Google Scholar]
- 37.Fischer S E J, van Luenen H G A M, Plasterk R H A. Mol Gen Genet. 1999;262:268–274. doi: 10.1007/pl00008641. [DOI] [PubMed] [Google Scholar]
- 38.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 39.Grindley N D F, Leschziner A E. Cell. 1995;83:1063–1066. doi: 10.1016/0092-8674(95)90132-9. [DOI] [PubMed] [Google Scholar]
- 40.Lohe A R, De Aguiar D, Hartl D L. Proc Natl Acad Sci USA. 1997;94:1293–1297. doi: 10.1073/pnas.94.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capy P, Vitalis R, Langin T, Higuet D, Bazin C. J Mol Evol. 1996;42:359–368. doi: 10.1007/BF02337546. [DOI] [PubMed] [Google Scholar]
- 42.Tavakoli N P, DeVost J, Derbyshire K M. J Mol Biol. 1997;274:491–504. doi: 10.1006/jmbi.1997.1410. [DOI] [PubMed] [Google Scholar]
- 43.Bureau T E, Wessler S R. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kidwell M. Curr Opin Genet Dev. 1992;2:868–873. doi: 10.1016/s0959-437x(05)80109-1. [DOI] [PubMed] [Google Scholar]
- 45.Ruberti I, Sessa G, Lucchetti S, Morelli G. EMBO J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker H-A, Kunze R. Mol Gen Genet. 1996;251:428–435. doi: 10.1007/BF02172371. [DOI] [PubMed] [Google Scholar]
- 47.van Drunen C M, Oosterling R W, Keultjes G M, Weisbeek P J, van Driel R, Smeekens S C M. Nucleic Acids Res. 1997;25:3904–3911. doi: 10.1093/nar/25.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisen J A, Benito M-I, Walbot V. Nucleic Acids Res. 1994;22:2634–2636. doi: 10.1093/nar/22.13.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karp P D. Bioinformatics. 1998;14:753–754. doi: 10.1093/bioinformatics/14.9.753. [DOI] [PubMed] [Google Scholar]