Abstract
Lac+ revertants of Escherichia coli that occur after prolonged nonlethal selection display a high frequency of transposon loss when the transposon Tn10 and the reverting lacI33 allele are linked on an F′128 episome. As many as 20% of the Lac+ revertants are sensitive to tetracycline, about half because of transposon loss, nearly all by precise excision, and the remainder because of amplification of both the transposon and the linked lac allele. Lethality of the amplified products in the presence of tetracycline is a peculiarity of the tetA gene at high gene dosage. The selective conditions on lactose medium result in 10% transposon-free revertants, whether or not a requirement for conjugal DNA transfer is imposed. In addition, a similar fraction, about 5% of Lac− unreverted colonies that are products of transfer between cells experiencing nonlethal selection are also tetracycline-sensitive, and all are attributable to loss of the Tn10 transposon. These results suggest the possibility that the high frequency of transposon loss is a consequence of conjugal transfer, making this loss a marker for that transfer. We suggest that conjugal DNA transfer may be a prominent feature in the mutability process that occurs during nonlethal selection and that the subset of bacteria displaying hypermutability are those that experience such transfer.
Escherichia coli FC40, carrying the episomal lacI33 allele (1), which is Lac− by virtue of a +1 frameshift, accumulates Lac+ revertant mutants, sometimes called adaptive mutants, during nonlethal selection (2). The mechanism responsible for the accumulation of new Lac+ revertants with time remains unsolved, although some features of the process have been elaborated (3–9). Among those features are the presence and capability of transfer of the F′128 episome (7, 10–12). During an inquiry concerning the reversion, we noticed that late-appearing Lac+ revertants of this allele, in this case linked to a transposon Tn10 conferring tetracycline resistance (TetR), were frequently Tet-sensitive (TetS). This observation prompted us to examine the effect of nonlethal selection on the loss of the TetR phenotype. We describe some features of the TetS bacteria found among the Lac+ revertants and of those found among the unreverted Lac− cells that had experienced conjugal transfer during nonlethal selection.
In our investigation, we used the strains PR324, an FC40 derivative carrying the Tn10 linked to the Lac− lacI33 allele on the episome, and VG201, an episome-free FC40 derivative in which the two linked markers are on the chromosome. The scavenger bacteria, also present on the selective plates, bear an episome isogenic with that of FC40 but with a lac deletion and no transposon. They are used to metabolize any usable carbon source, other than lactose, that may be present on the lactose-selective plates (2). We asked whether the Tet resistance (TetR) phenotype encoded on the transposon or the transposon itself is lost under nonlethal selection, either in Lac+ or in lacI33 colonies, after prolonged incubation on selective plates. Bacteria may become Tet-sensitive (TetS) either by losing the Tet coding sequences (i.e., by losing the Tn10) or by amplification of the copy number of these sequences (13), i.e., by DNA amplification, a suggested intermediate in the Lac+ reversion under investigation (9, 14, 15). The TetS phenotype of the bacteria with DNA amplification that includes the transposon is presumably attributable to jamming of the cells secretory apparatus caused by induction of the tetA gene product at high copy number in the presence of the inducer Tet (13). For the episomal allele, the analysis was carried in the presence of streptomycin (Sm), conditions in which conjugal transfer to scavenger cells is required for detection of reversion (10), because the lacI33-carrying cells are sensitive to Sm and the scavenger cells are Sm-resistant (SmR). These results were compared with those obtained in the absence of Sm, when no such requirement was imposed. Conjugal transfer is understood here as episomal DNA transfer from the lacI33-containing bacteria to the scavenger cells as well as self-transfer between lacI33 bacteria.
We found that TetR was frequently lost in episomal Lac+ revertant colonies that accumulate during selection with and without an imposed requirement for conjugal transfer. This loss was found many orders of magnitude more frequently than had been observed for spontaneous transposon loss in the past (16–21). Moreover, even lacI33 Lac− cells, products of conjugal transfer to scavenger, frequently had become TetS. About 10% of the late-appearing Lac+ revertants that are TetS have lost the transposon DNA, most often precisely, and an equal number show evidence of amplification of the Lac/Tn genetic regions. This is true both for Lac+ revertants that had been products of conjugal transfer and are in scavenger hosts as well as for the Lac+ revertants that have accumulated without a requirement of conjugal transfer imposed on them. For Lac− bacteria that have experienced prolonged selection on lactose minimal medium and had experienced conjugal transfer, a similar fraction, approximately 5%, are transposon-free and the loss seems precise. Few amplification products would be expected here, because they would be likely to be Lac+ because of the leakiness of the lacI33 allele.
In the course of this work, no TetS colonies were found among the products of conjugal transfer between Lac− cells that had not experienced nonlethal selection. We also have found that DNA amplification, spanning both the transposon and the lac genes, is responsible for all of the few late-appearing revertants derived from a chromosomal lacI33 allele. Neither the amplification nor the Tn10 loss appears to be associated with heritable mutators in the population under selection.
These results suggest that transfer of DNA by conjugation followed by either recombination or slow replication of the transferred single-stranded DNA may be a feature whereby Lac+ revertants are formed and transposon DNA lost. In addition, we suggest that those bacteria that have experienced conjugal transfer, whether transfer to the scavenger or probably, more frequently, self-transfer, may constitute the subset of hypermutable bacteria that are evident under the nonlethal conditions of selection (8, 22).
Materials and Methods
Strains and Reagents.
The strains used in this study were E. coli DH5α (lab collection); P90C (1) and its SmR derivative (lab collection); and FC40, a rifampicin-resistant derivative of P90C with an F′128 episome carrying the lacI33 mutation (2). This lac allele is a constitutive lacI-lacZ fusion that contains all but the last four codons of the repressor gene fused to the 25th codon of the lacZ gene. The frameshift mutation (an extra “C” in the 320th “CCC” codon of lacI) lies at the end the lacI gene, creating a +1 frameshift polar to lacZ. The phenotype of a strain bearing the lacI33 allele is therefore Lac−, but because of translational readthrough, the allele is leaky and the colonies have a pale blue phenotype in plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The strain PR324, an isogenic derivative of FC40, contains the zah-281∷Tn10 (23) on the episome (10) linked to the lacI33 allele. The strain FC29 is an isogenic derivative of P90C bearing the same F′128 episome but with a deletion in the lacI-lacZ gene, F′ΔlacIZ (2). Therefore, homologous recombination per se between the F′ΔlacIZ and the lacI33 episome does not give rise to Lac+ revertants. FC29 SmR (10) is an isogenic derivative of FC29 used as scavenger in experiments described below. The VG10 strain is an isogenic derivative of PR324 with a replacement of the Tn10 linked to the lacI33 allele by a Tn10Kan on the F′128 episome, and VG201 is a derivative of MG4792, an isogenic strain with P90C except for the Lac− allele (the lacI33 allele on the chromosome in MG4792 was constructed by homogenotization and was provided by M. Marinus, University of Massachusetts Medical School, Worcester) with the zah-281∷Tn10 linked to lacI33 on the chromosome. This strain was isolated after P1 transduction (1) of MG4792 from strain 12080 (zah-281∷Tn10 lac+pro+, provided by R. Kolter, Harvard Medical School, Boston).
Bacterial cultures were routinely grown in M9 minimal liquid medium with 0.1% limiting glucose or in complete LB medium (1). Plating was done on M9 minimal plates (1) as indicated below. The antibiotic concentrations used were 100 μg/ml rifampicin, 100 μg/ml Sm, 12 μg/ml Tet, 30 μg/ml kanamycin (Kan), and 150 μg/ml ampicillin. The chromogenic substrate X-Gal was used at a final concentration of 40 μg/ml. Primers were custom-synthesized by the Tufts Core Facility, Physiology Department, Tufts School of Medicine (Boston, MA).
Isolation of Lac+ Revertant Colonies and Lac− Cells That Have Experienced Selection.
Lac+ reversion experiments were carried out as described in Radicella et al. (10). Selective conditions for which conjugal transfer was required involved the presence of Sm in the minimal lactose selection plates and FC29 SmR scavenger cells as in Radicella et al. (10). In these experiments, the Lac+ revertants are products of conjugation between the starving Sm-killed PR324/F′lacI33 zah-281∷Tn10 Sm-sensitive bacteria and the scavenger FC29/F′ΔlacIZ SmR cells. As previously shown (10), the yield of Lac+ revertants is reduced under these conditions as compared with the yield in the absence of the antibiotic. Early Lac+ revertant colonies appeared after 2–3 days of incubation for experiments in the absence of a requirement for transfer, and after 3–4 days of incubation for experiments where conjugal transfer is required for detection of revertants. Late Lac+ revertants were those that appeared on the 7th day. The revertant colonies were picked from the plates with a platinum wire and resuspended in 150 μl of M9 salt solution in 96-well dishes. The same procedure was followed for the chromosomal Lac+ revertants. Unreverted Lac− cells were isolated after 7 days of incubation on the selective plates by resuspending 2-mm plugs and plating on minimal glucose medium with Sm and X-Gal for single colonies. Pale blue colonies reflecting the presence of the leaky lacI33 allele were isolated.
Tet sensitivity was determined for Lac− and Lac+ isolates that had experienced nonlethal selection and for bacteria that had not undergone the lactose selection. Among the former were Lac+ revertants isolated as indicated above, with and without a requirement for transfer during selection, and the Lac− bacteria present in the lawn of selected cells. The episomal transfer products that had not gone through selection were P90C SmR exconjugants bearing the unreverted Lac− leaky lacI33 Tn10-zah-281∷Tn10 episome from PR324. They are products of transfer between exponentially grown cultures of P90C SmR and PR324 containing the Tn10-lacI33 linked alleles, following the mating procedure of Peters and Benson (24).
Substituting the KanR Marker for the Tn10 TetR Marker and Cloning of the Tn10 Insertion Site on the Episome.
The Tn10 used in this study, zah-281∷Tn10 (23), carries the tetA gene. This gene is positively regulated by Tet, resulting in cell death (i.e., TetS phenotype) when the gene dosage number is high and the inducer Tet is present. This lethality may be the consequence of jamming the secretory pathway of the cell (13). To avoid this complication, the TetR marker was replaced by the Kan resistance marker (KanR) through recombination as described in Singer et al. (23) by using the λ phage 1104 as the Tn10Kan source (provided by N. Kleckner, Harvard University, Boston; ref. 25). The replacement of the Tn10 by Tn10Kan was confirmed genetically by PCR amplification and Southern blotting (see Analysis of the Products of Tn10 Loss).
The Tn10Kan transposon was cloned to locate the site of Tn10 insertion on the episome. This procedure was accomplished by digesting total DNA isolated from VG10 with EcoRI/ClaI and ligating this mixture to pBluescript digested with the same enzymes. The ligation reaction was used to transform DH5α selecting for KanR/ampicillin-resistant colonies. The resulting plasmid, pVRC1123, contains Tn10Kan and the respective flanking sequences. The IS10-containing arms with the flanking sequences were subcloned in the same vector by digesting with EcoRI and HindIII, creating pVRC108, and with HindIII and ClaI, creating pVRC103. These two plasmids then were sequenced in the Massachusetts Institute of Technology Biopolymers Laboratory with an IS10 outward primer, 5′-GACAAGATGTGTATCCACCT-3′.
Screening for Tn10 DNA Sequences in Lac+ Colonies.
Lac+ TetS colonies were tested for the presence of Tn10 sequences by Southern blot hybridization (26). Total DNA isolated from the TetS Lac+ revertants was digested with BglI, transferred to nylon membranes, and hybridized to an IS10R probe isolated from pNK214 (provided by the N. Kleckner lab) as a 2,163-bp BglII fragment containing the complete IS10R and a piece from the pBR333 backbone.
To quantify the Tn10 loss among the Lac+ revertants, late-appearing colonies were picked from selection plates and resuspended in 150 μl of M9 salt solution in 96-well dishes as described previously. The bacterial suspensions were transferred by using a 96-multipoint device (27), carrying between 2 μl and 5 μl of each suspension onto lactose minimal plates with and without Tet. Aliquots of 50 μl of these cell suspensions were transferred as well into a 96-well dot-blot apparatus. The colony suspensions were filtered through the wells and treated as for filter hybridization as described by Ausubel et al. (26). The membranes with the fixed total DNA were hybridized to a MutY probe used to standardize the amount of total DNA present in each dot, followed by stripping and rehybridizing with the IS10R or to Tet or Kan probes to screen for Tn10 sequences. The Tet probe was obtained from the pNK214 plasmid as a 2,885-bp BglII fragment. The Kan probe used was a HindIII fragment of 3,424 bp isolated from the plasmid pNK1022 (provided by the N. Kleckner lab), and the MutY probe was isolated as a 719-bp BstEII-BglI fragment from the D601 plasmid (28). These probes were radioactively labeled with 32P-dCTP following the protocol suggested by the manufacturer.
A Lac+ colony is scored as transposon-free when there is no growth on the antibiotic-containing plate and no signal on the respective autoradiograph hybridized with the Tet/Kan/IS10 probe as compared with the signal present on the autoradiograph hybridized with the MutY probe. This assay, quantified as shown, detects only clonal transposon-free colonies. A Lac+ colony containing amplified sequences spanning the transposon is detected as having a signal on the autoradiograph hybridized to the MutY probe and a stronger signal on the autoradiograph hybridized with the Tet/Kan or IS10 probe. It should be pointed out that there are no steps of colony purification between sampling and testing of colonies by hybridization.
The dot blots were exposed to Kodak film NAR for 2–20 h, exposed to a phosphorimager screen, and scanned. The quantification was done with the imagequant application from Molecular Dynamics.
Analysis of the Products of Tn10 Loss.
A collection of 29 purified Lac+ transposon-free TetS or KanS colonies (based on Southern hybridization or dot-blot assay), isolated at early or late times after plating, were analyzed by PCR. The primers used for the PCR reactions were VG1031, 5′-TGAATCAGCTTTTATCGTCAGCC-3′, and VG1032, 5′-ATACCAATCAGATCGCGCAG-3′. These two primers flank the transposon insertion site, giving an amplicon of 498 bp when PR324 DNA is used as template. The conditions for PCR were as follows: one cycle of 5 min at 94°C followed by 30 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 56°C, and 1 min of polymerization at 72°C. The PCR products were separated by electrophoresis, and 10 of them, apparently precisely excised, also were sequenced with VG1032 in the Biopolymers Laboratory at Massachusetts Institute of Technology; 6 of them were the products of Tn10 loss and the other 4 were from Tn10Kan loss.
Results
TetS Colonies Among Lac+ Revertants.
Late-appearing Lac+ revertants of the F′128 episome-borne lacI33 allele were tested for the TetR phenotype endowed by the linked Tn10 allele in strain PR324. When the Lac+ revertants appearing on days 7 and 8 were isolated under conditions that required conjugal transfer to the scavenger population, about 20% were TetS (Table 1). A similar fraction of late-appearing Lac+ revertants are TetS when no requirement for conjugal transfer is imposed during selection in lactose minimal medium (Table 1). For Lac+ revertants appearing early, fewer, about 4–6%, are TetS, whether or not a requirement for conjugal transfer is imposed.
Table 1.
Loss of Tet resistance in bacteria experiencing nonlethal selection
| Strain | Early Lac+ revertants, Lac+ TetS/total
|
Late Lac+ revertants, Lac+ TetS/total*
|
||
|---|---|---|---|---|
| Transfer† | No transfer‡ | Transfer | No transfer | |
| FC40 | 4% | 6% | 22% | 20% |
| (18/413) | (16/269) | (168/754) | (34/170) | |
| VG201 | — | <0.04% | — | 1.8% |
| — | (0/255) | — | (10/565) | |
Lac+ revertants isolated late on days 7–8 after plating.
Lac+ revertants isolated early on days 3–4 after plating.
Lac+ revertants isolated early on days 2–3 after plating.
In contrast, when the lacI33 and Tn10 alleles are chromosomal, many fewer Lac+ revertants appear, and among the 255 early Lac+ clones that were examined, none were TetS. About 2% of the late-appearing revertants displayed a TetS phenotype (Table 1).
These observations show that the loss of the drug phenotype is characteristic of revertants when the transposon and lac alleles are located on an episome. The marker loss may be the consequence of transfer under these conditions or may be associated with mutation to Lac+ or both may be associated with the process of conjugal transfer.
TetS Among Selected Cells That Had Not Reverted.
Examination of the Lac− PR324 bacteria still rifampicin-resistant and isolated from the unreverted lawn of selection plates, yielded only TetR colonies. Thus, 1,200 Lac− colonies analyzed after 2 days of selection, without a requirement for transfer, were all TetR as were 1,300 colonies that were examined from the unreverted population after 7 days of selection. Furthermore, with the chromosomal lacI33 allele, no TetS colonies were found among 2,078 unreverted chromosomal Lac− colonies tested after 7 days of selection.
TetS Among lacI33 Lac− Cells That Had Transferred During Lactose Selection.
To determine whether conjugal transfer played a role in the occurrence of sensitivity to Tet, we examined the products of transfer to the FC29 SmR scavenger cells for their Tet phenotype among selected but unreverted Lac− cells. Melting point capillary tubes were used to take 2-mm plugs from the areas of the plates with unreverted Lac− cells after 7 days of lactose selection.
To assess the frequency of transfer among the unreverted cells, plugs derived from lactose selection plates without Sm of PR324 and the scavenger FC29 were resuspended, diluted, and plated on minimal glucose plates with Sm and X-Gal. The exconjugants make pale blue colonies characteristic of the leaky Lac− lacI33 allele on these plates as compared to the white color of the colonies containing the FC29 episome (lacIZ deletion). None of the pale blue colonies grew in the presence of rifampicin (100 μg/ml), supporting the view that they were products of transfer occurring during the nonlethal selection. We found that the frequency of transfer of the PR324 episome by this criterion to FC29 is between 1% and 7% after 7 days of selection.
The products of transfer between the PR324 lacI33 bacteria and the scavenger cells, after 7 days of selection, were isolated from plugs taken from selection plates containing Sm. The plugs were resuspended, and the bacterial suspension was plated on glucose minimal plates with X-Gal. Among 851 pale blue SmR colonies isolated this way, 41 were TetS (≈5%), all caused by absence of the transposon. The similarity of this frequency with the 10% of the Lac+ bacteria that had lost the transposon suggests that there might be a relationship between transposon loss and DNA transfer during nonlethal selection.
Origin of the TetS Phenotype.
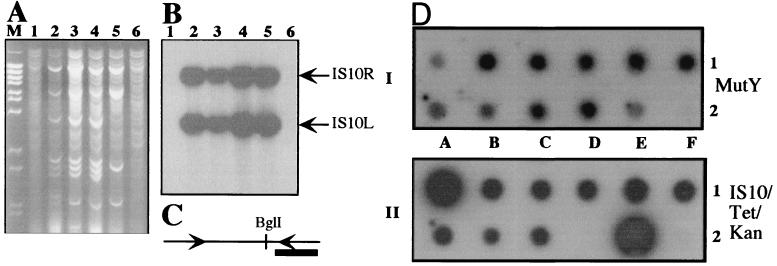
The high frequency of Lac+ colonies that were TetS provoked us to determine whether the TetS phenotype was caused by amplification or by transposon loss. We examined, by Southern blotting, a set of early- and late-appearing Lac+ TetS revertant colonies that had been isolated under conditions requiring conjugal transfer. Total DNA isolated from these colonies was digested with BglI and probed with a DNA fragment containing IS10R (see diagram of Tn10 map and location of probe in Fig. 1C). We found that some were TetS because of transposon loss and some were TetS because of amplification of the transposon sequences (Fig. 1 A and B). For Lac+ revertants that appeared after prolonged selection (7 days), the transposon-free category constitutes approximately 43% (25/58) of the Lac+ TetS revertants when DNA transfer to the scavenger was required. DNA amplification was responsible for the remaining late Lac+ TetS clones examined.
Figure 1.
The TetS phenotype is the result of Tn10 loss or DNA amplification of Tn10 sequences. (A) Agarose gel electrophoresis of total DNA isolated from late Lac+ TetS cultures digested with BglI and stained with ethidium bromide. M, λ BstEII size marker; lane 1, FC40; lanes 2–6, late Lac+ and TetS revertants. (B) Southern blotting of the gel presented in A, hybridized with the IS10R probe shown in C. (C) Schematic of Tn10. The arrows represent the IS10 inverted sequences, left and right, respectively. The thick line represents the IS10R probe used in these experiments. (D) Assessment of Tn10 sequences among the Lac+ revertants. Panel I is an autoradiograph of total DNA obtained by in situ lysis of late Lac+ revertants picked directly from selective plates and hybridized to a MutY probe. Panel II depicts the same samples shown in Panel I, but hybridized to an IS10/Tet/Kan probe.
Investigation of late Lac+ TetS clones that occurred in the absence of Sm (transfer to scavenger was not required) yielded similar results. Tn10 loss was found in 26 of 59 TetS revertants, and the remaining 33 colonies displayed transposon DNA amplifications.
Among late-appearing chromosomal Lac+ TetS revertant colonies, all of the 10 examined (Table 1) were products of DNA amplification.
The DNA amplification observed in the DNA isolated from PR324 is so striking that it can even be seen in the ethidium bromide-stained agarose gel (see Fig. 1 A, lanes 2–5, and corresponding autoradiograph in B). From this ethidium bromide-stained gel photograph, we note that the region of DNA spanning the amplified sequences varies from revertant to revertant. For example, in Fig. 1A, the amplified region of DNA present in the total DNA prepared from the Lac+ isolate in lane 2 clearly is different from the one in lane 5.
To assess both transposon loss and DNA amplification in a larger fraction of the Lac+ population rather than being limited to the TetS revertant colonies, further testing was carried out by dot-blot analysis. Lac+ revertants, isolated from lactose selection plates in which conjugal transfer to the scavenger had been required, were picked directly from the reversion plates. To normalize the amount of total DNA loaded in each dot, the blotted membranes were probed with a sequence derived from the mutY gene located on base pairs 3,101,031–3,102,083 of the E. coli chromosome. To test for the transposon sequences, the IS10R or a Tet probe was used. An example of the results obtained in these experiments is shown in Fig. 1D. Panel I is the result of hybridization of the total DNA present in late-appearing Lac+ revertants blotted on the membrane and probed with a MutY probe. Panel II shows the results of the same blot stripped and rehybridized with the IS10 or Tet sequences. Both probes gave similar results. These blots show that the amount of DNA loaded on the membrane, the product of lysis in situ, is in general comparable from dot to dot. A Lac+ revertant easily can be scored in pure colonies as transposon-free (see panel I, dot 2D, as compared with panel II) or as DNA amplifications (see isolates 1A and 2E and compare between panels I and II). The results are summarized in Table 2.
Table 2.
Lac+ revertants often are the result of transposon loss and DNA amplification
| Strain | Transposon loss
|
DNA amplification
|
|
|---|---|---|---|
| Days 3–4 | Days 7–8 | Days 7–8 | |
| Wild-type Tn10 | 7% | 10% | 11% |
| (9/125) | (33/344) | (37/344) | |
| Tn Kan | — | 9% | 8% |
| — | (14/163) | (13/163) | |
Data are presented as percentage of transfer at indicated days after plating. Numbers in parentheses indicate number of Lac+ revertants that had lost or amplified the transposon/total number of revertants examined. Tn Kan, Tn10 containing the Kan resistance marker.
There is a high frequency of Tn10 loss among late-appearing Lac+ revertants, about 10%, during the nonlethal selection. We also found an about equal fraction of Lac+ revertants with amplification of the transposon sequences. These amplified products were readily evident in the late-appearing Lac+ revertant colonies as shown in Table 2 and Fig. 1. Summarizing, we have shown that nonlethal selection results in revertants with transposon loss and in those with DNA amplification when the transposon is on an episome. With the chromosome-borne lacI33 linked to Tn10, the TetS revertants are all products of DNA amplification.
Replacement of TetR by KanR.
The drug resistance determinant of Tn10 was replaced by a sequence encoding the KanR marker to eliminate the possibility that the observed Tn10 loss and DNA amplifications were the result of some feature of the tetA gene and to eliminate the uncertainty regarding the copy number required for Tet sensitivity. The replacement was carried out by homologous recombination with a copy of Tn10 containing a Kan marker delivered by the replication-deficient bacteriophage λ 1104 vector. We obtained 50 VG10 KanR colonies, after λ infection, that also were TetS, suggesting that the replacement had been successful. P1 phage grown in four independent isolates of PR324/F′ zah-281∷Tn10 lacI33 (i.e., carrying the Tet marker) were used to infect VG10/zah-281∷Tn10Kan lacI33 isolates. The P1 transductions in all cases resulted in colonies resistant to Tet and sensitive to Kan, providing additional support for the conclusion that the Tn10Kan had replaced the original Tn10. In addition, PCR amplification with primers IS10 out and VG1031 or VG1032 flanking the transposon insertion site, gave the same expected band sizes of approximately 250 bp (see Materials and Methods for primer sequences) using PR234/F′ zah-281∷Tn10 lacI33 or VG10/zah-281∷Tn10Kan lacI33 as templates (data not shown).
We expected to find that the fraction of Lac+ revertants with a KanS phenotype would be similar to that with transposon loss (tested by hybridization assay) for both Tn10 and Tn10Kan. Similarly, we expected a comparable fraction of Lac+ revertants to bear DNA-amplified regions, which in this case would still be phenotypically KanR. The analysis of late-appearing Lac+ revertants that were products of transfer fulfilled these expectations (see Table 2). We found 18 late-appearing Lac+ revertants (of 163 analyzed) with a KanS phenotype, but 4 of them were mixtures of KanS and KanR bacteria. The clonal fraction of KanS colonies (14/163) agrees with the observed fractions of transposon-free Lac+ revertants, about 10%, for both Tn10Kan and Tn10 (Table 2).
Moreover, the frequency of revertants under these conditions bearing an amplified Tn10Kan is similar to the frequency found when using the prototype Tn10. We observed that 8% (13/163) of late-appearing Lac+ revertants with Tn10Kan have evidence of DNA amplification, which is comparable with 11% (37/344) found previously in Lac+ revertants carrying Tn10 (see Table 2). In this case, however, the increase in transposon copy number does not result in a phenotype as it does with Tn10.
Genetic Mutators.
It has been reported that absence of methyl-directed mismatch repair in mutator strains, caused by mutations in mutH or mutL genes, results in moderate increases in the frequency of Tn loss (20, 29). We found 1 in 62 of the transposon-free late Lac+ revertants with a mutator phenotype, suggesting that a role for genetic mutators is not a prominent feature of this process.
Tn10 Loss Is Precise.
Spontaneous Tn10 loss has been reported to occur by precise excision and, more prominently, by excision that is nearly precise. The latter usually leaves behind a residue about 50 bp at the insertion site (13, 21, 30). The former leaves no traces of transposon insertion in the host DNA and is approximately 100-fold less frequent than is nearly precise excision (21).
The sequence obtained through cloning of the episomal Tn10Kan locates the transposon at the base pair 371,180 of the corresponding E. coli chromosome. The site of insertion contained a 9-bp repeat, one of the signatures of Tn10 insertion (13). Based on this sequence, two PCR primers were generated (see Materials and Methods). These primers were used to amplify the insertion region on the episome of transposon-free Lac+ revertants. A product of excision that is nearly precise shows, by agarose electrophoresis, a larger size PCR band as compared with the transposon-free control. PCR analysis of 28 Lac+ revertants that were transposon-free (including Lac+ revertants derived from either Tn10 or Tn10Kan hosts) showed that 3 of them seemed to be, based on size, nearly precise (data not shown). Detection of these seemingly imprecise events suggests that the transposon loss observed may be through a replication mechanism of the transferred single-stranded DNA in either the donor or the recipient hosts. We have shown in our laboratory that the Klenow fragment of DNA Polymerase I is able in vitro to replicate template molecules containing a secondary structure formed by Tn10, omitting all but 50 bp sequences comprised by the IS10 ends (S. Chen, N. An, and M.S.F., unpublished results). Nevertheless, the remaining 25 samples appeared to be precise; 10 of these 25 PCR fragments were sequenced. They were all, in fact, precise, showing no alteration from the wild-type sequence, which is what one would expect if the transposon were lost by recombination.
Discussion
Examination of the late-appearing Lac+ revertants of the lacI33 allele resident on the F′128 episome and linked to the TetR-carrying transposon of zah-281∷Tn10 (PR324) revealed a large fraction (≈20%) of revertants that was no longer resistant to Tet. This high frequency of TetS revertants was equally high in either the absence or the presence of an imposed requirement for conjugal transfer of the F′128 to scavenger cells on the selection plate. We do not know whether the products that we observe are the result of complete episome replacement or of partial episome transfer followed by recombination with the resident F′.
About half of these revertants are sensitive to Tet because the transposon is absent. The other half are drug-sensitive because the DNA sequences, including the lac gene, have experienced amplification. In addition to finding revertants that were transposon-free, we found that a similar fraction, about 5%, of 851 Lac− clones that were isolated on day 7 as products of conjugal transfer between the PR324 and the FC29 scavenger were TetS, and all had lost the transposon. Our rough estimate of the frequency of such episome transfer is between 1% and 7% after 7 days of lactose selection, when 1–2 × 109 scavenger cells are present.
Such a high level of transposon loss has not been reported so far. In general, transposon loss occurs at frequencies of 10−6 or less (13) with no more than about 10-fold increase when the bacteria are defective in some function involved in DNA metabolism (19, 20, 29) or have been exposed to UV light or other treatments that enhance transposon loss (31). In this paper, we report transposon loss at levels at least 104-fold greater than in earlier reports. These data could be evidence that favors the notion of transposon loss by a recombinational mechanism.
This loss seems to be characteristic of conjugal episome transfer under nutritionally sparing conditions, in this case during lactose selection for Lac+ revertants of the leaky lacI33 allele. These starvation conditions relax the exclusion of transfer between episome-carrying strains. We therefore suggest that the transposon loss can be used as a marker for conjugal transfer in these experiments. It should be noted that we could only detect conjugal transfer when it was to the FC29 scavenger. It could be that transfer is much more common between genetically like bacteria, either between sisters or by direct transfer to the same bacterium under the nutritionally sparing conditions for selection of Lac+ revertants (32, 33).
If this suggestion has merit, the similarity in the levels of transposon loss among episomes that have been transferred and among those that have transferred and reverted as well as those that have reverted without Sm selection for transfer to scavenger, suggests the proposal that conjugational transfer is a requirement for much of the accumulation of adaptive Lac+ revertants that is evident. This proposal could account for the evidence of a subset of the selected bacteria that displays hypermutability (4, 12, 34). For those occasional episomes that transfer, the slowly replicating single-stranded DNA, either in the donor or in the recipient bacterium, or the possibility of recombination with the recipient episome could be the signal for elevated mutability, perhaps by derepression of the SOS system (35), for both the lacI33 allele and the bacterium as a whole (12).
The high frequency of transposon loss that we see among Lac− and Lac+ bacteria that have experienced conjugational transfer is consistent with the observation that loss of the exclusion phenotype, traS, moderately enhances Tn10 loss (36). Under these nongrowing conditions, nonlethal selection, F′-containing cells are no longer responding to the exclusion phenotype and could be able to prepare for DNA transfer (7, 10) and to conjugate. With the limited metabolism that is characteristic of selection (8), the slow replication of the transferred single-stranded DNA may provide the conditions and the time to allow the formation of a stable hairpin DNA structure by hybridization of the inverted terminal IS10 sequences within the recipient cell. We suggest the slow replication may pass over this hairpin structure and result in the formation of a heteroduplex product with the Tn10 present only in one strand. The resolution of that heteroduplex may be similar to that of the bacteriophage λ DNA that is heteroduplex for a Tn10 sequence that has been used to transfect E. coli cells. The artificially created λ DNA heteroduplex contains a copy of Tn10 only in one strand, inserted either between the genes Q and P or within the cI gene. The products of single transfection of mut+ and mutL, mutS, mutH, or uvrD backgrounds reveal the presence of sites from both strands, demonstrating the biological activity of both strands of such heteroduplexes. However, none of the λ progeny from 25 transfection events showed evidence of the Tet marker. For the case of Tn10 in the cI gene, all of the cI products are cI+, suggesting that Tn10 had been lost before λ DNA replication (37). The latter observation is consistent with the view that the transposon has been precisely excised. We also have shown in our laboratory that, in vitro, the large fragment of DNA polymerase I, the Klenow product, is able to replicate and bypass a hairpin structure when single-stranded DNA containing Tn10 is used as template (S. Chen, N. An, and M.S.F., unpublished results).
The conjugal transfer that is evident in these experiments is discrepant with the reports of Foster and Trimarchi (7). However, there can be little doubt that revertants accumulate in the presence of Sm, a drug to which the scavenger is resistant, but which kills the FC40 or PR324 rapidly. Sm-killed cells still may engage in conjugal transfer (10).
Postconjugational recombination could be prominent in these experiments. If so, the RecA dependence that is evident for adaptive mutability could be accommodated (2).
Anderson et al. (9) have provided evidence for gene amplification as an intermediate in the formation of Lac+ revertants that accumulate under conditions of nonlethal selection. Although they used a strain defective in conjugal transfer, our observations provide support for their proposal. We have observed highly amplified Tn10/lac sequences in approximately 10% of the late-appearing Lac+ revertants and in all of the chromosomal Lac+ revertants. In the latter case, the DNA amplification occurred in the absence of conjugal transfer. These observations support the notion that gene amplification is at least one of the pathways that contributes to the mutability that is observed during nonlethal selection (9). The sensitivity to Tet in the amplified products is presumably caused by the lethal effect of the overexpression of the tetA gene in the presence of its inducer, Tet (13). The fraction of Lac+ revertants bearing DNA amplification does not depend on the tetA gene (Table 2), because replacing it by the KanR marker confirms the results obtained with the transposon bearing the Tet marker for both Tn10 loss and DNA amplification.
As with bacteria incubated under starvation conditions (38), new reactions are evident in bacteria that are incubated under the sparing metabolic conditions of nonlethal selection. We have shown that both Lac+ and Lac− cells, which have experienced demonstrable conjugal transfer under nonlethal selection, display a remarkably high frequency of transposon loss. Because transposon-free colonies are found at a similar frequency in both Lac+ and Lac− bacteria, transposon loss appears to be a marker for conjugation. Therefore, conjugal transfer would seem to be an important element in creating the subset of hypermutable bacteria that is evident under nonlethal conditions of selection (8, 12, 22). The possibility that recombination may play a role in the formation of the products of conjugal transfer suggests a way in which the mutability could display RecA dependence. Sorting out these reactions will be interesting projects for the future.
Acknowledgments
We thank N. Kleckner, R. Kolter, and M. Marinus for bacterial strains, plasmids, and phages. V.G.G. thanks F. Stahl for a helpful discussion, P. Radicella for encouragement, advice, and ideas throughout this work, and R. Gallegos for his constant support. This work was supported by National Institutes of Health Grant R37AI 05388 and a Merck fellowship to V.G.G.
Abbreviations
- Tet
tetracycline
- TetR
Tet-resistant
- TetS
Tet-sensitive
- Sm
streptomycin
- SmR
Sm-resistant
- Kan
kanamycin
- KanR
Kan-resistant
- KanS
Kan-sensitive
- X-Gal
5bromo-4-chloro-3-indolyl-β-d-galactopyranoside
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130186597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130186597
References
- 1.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, New York: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 2.Cairns J, Foster P L. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg S M, Longerich S, Gee P, Harris R S. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg S M, Thulin C, Harris R S. Genetics. 1998;148:1559–1566. doi: 10.1093/genetics/148.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg S M, Harris R S, Longerich S, Galloway A M. Mutat Res. 1996;350:69–76. doi: 10.1016/0027-5107(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 6.Foster P L. Annu Rev Microbiol. 1993;47:467–504. doi: 10.1146/annurev.mi.47.100193.002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster P L, Trimarchi J M. J Bacteriol. 1995;177:6670–6671. doi: 10.1128/jb.177.22.6670-6671.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster P L. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson D I, Slechta E S, Roth J. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- 10.Radicella J P, Park P U, Fox M S. Science. 1995;268:418–420. doi: 10.1126/science.7716545. [DOI] [PubMed] [Google Scholar]
- 11.Galitski T, Roth J R. Science. 1995;268:421–423. doi: 10.1126/science.7716546. [DOI] [PubMed] [Google Scholar]
- 12.Godoy V G, Gizatullin F S, Fox M S. Genetics. 2000;154:49–59. doi: 10.1093/genetics/154.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 14.Tlsty T D, Albertini A M, Miller J H. Cell. 1984;37:217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi T, Horiuchi S, Novick A. Genetics. 1963;48:157–169. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopkins J D, Clements M B, Liang T Y, Isberg R R, Syvanen M. Proc Natl Acad Sci USA. 1980;77:2814–2818. doi: 10.1073/pnas.77.5.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haniford D B, Chelouche A R, Kleckner N. Cell. 1989;59:385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- 18.Roberts D, Kleckner N. Proc Natl Acad Sci USA. 1988;85:6037–6041. doi: 10.1073/pnas.85.16.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundblad V, Taylor A F, Smith G R, Kleckner N. Proc Natl Acad Sci USA. 1984;81:824–828. doi: 10.1073/pnas.81.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lundblad V, Kleckner N. Basic Life Sci. 1982;20:245–258. doi: 10.1007/978-1-4613-3476-7_16. [DOI] [PubMed] [Google Scholar]
- 21.Foster T J, Lundblad V, Hanley-Way S, Halling S M, Kleckner N. Cell. 1981;23:215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- 22.Torkelson J, Harris R S, Lombardo M J, Nagendran J, Thulin C, Rosenberg S M. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters J E, Benson S A. J Bacteriol. 1995;177:847–850. doi: 10.1128/jb.177.3.847-850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 26.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. Vol. 1. New York: Wiley; 1999. pp. 2.9.16–2.9.20. [Google Scholar]
- 27.Lovelace T E, Colwell R R. Appl Microbiol. 1968;16:944–945. doi: 10.1128/am.16.6.944-945.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai-Wu J-J, Radicella J P, Lu A-L. J Bacteriol. 1991;173:1902–1910. doi: 10.1128/jb.173.6.1902-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundblad V, Kleckner N. Genetics. 1985;109:3–19. doi: 10.1093/genetics/109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross D G, Swan J, Kleckner N. Cell. 1979;16:733–738. doi: 10.1016/0092-8674(79)90089-8. [DOI] [PubMed] [Google Scholar]
- 31.Eichenbaum Z, Livneh Z. Genetics. 1998;149:1173–1181. doi: 10.1093/genetics/149.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayes W. The Genetics of Bacteria and Their Viruses. New York: Wiley; 1968. [Google Scholar]
- 33.Achtman M. J Bacteriol. 1975;123:505–515. doi: 10.1128/jb.123.2.505-515.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosche W A, Foster P L with Cairns, J. (Appendix) Proc Natl Acad Sci USA. 1999;96:6862–6867. doi: 10.1073/pnas.96.12.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith B T, Walker G C. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syvanen M, Hopkins J D, Griffin T J, IV, Liang T-Y, Ippen-Ihler K. Mol Gen Genet. 1986;203:1–7. doi: 10.1007/BF00330376. [DOI] [PubMed] [Google Scholar]
- 37.Raposa S, Fox M S. Genetics. 1987;117:381–390. doi: 10.1093/genetics/117.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkel S E, Kolter R. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]