Abstract
Background
DNA microarrays are now routinely used to monitor the transcript levels of thousands of genes simultaneously. However, the array fabrication method, hybridization conditions, and oligodeoxyribonucleotide probe length can impact the performance of a DNA microarray platform.
Results
We demonstrate solution-phase hybridization behavior of probe:target interactions by showing a strong correlation between the effect of mismatches in probes attached to a three dimensional matrix of a microarray and solution-based, thermodynamic duplex melting studies. The effects of mismatches in the probes attached to the microarray also demonstrate that most, if not all, of the oligodeoxyribonucleotide is available for hybridization. Kinetic parameters were also investigated. As anticipated, hybridization signals increased in a transcript concentration-dependent manner, and mismatch specificity increased with hybridization time. Unexpectedly, hybridization time increased the accuracy of fold changes by relieving the compression observed in expression ratios, and this effect may be more dramatic for larger fold changes.
Conclusions
Taken together, these studies demonstrate that a three-dimensional surface may enable use of shorter oligodeoxyribonucleotide probes and that hybridization time may be critical in improving the accuracy of microarray data.
Background
DNA microarrays have emerged as a powerful tool to monitor the transcript levels of thousands of genes simultaneously [1,2]. This parallel analysis permits tumor prognosis and classification [3,4], drug target validation [5], toxicology evaluations [6,7], and functional discovery [8,9].
The microarray fabrication method can play a key role in the performance of a DNA microarray platform. For example, oligodeoxyribonucleotide probes can be covalently attached to a surface [10], synthesized in situ [11-13], or retained via electrostatic interactions with a positively charged surface [14]. A recent study examining the effect of mismatches along the length of in situ synthesized 60 mer oligodeoxyribonucleotides demonstrated a lack of an effect of mismatches for the first ten to fifteen bases at the 3' (surface) end of the oligodeoxyribonucleotide, suggesting that these bases may not be accessible during the hybridization reaction [12]. Other problems may exist when oligodeoxyribonucleotides are adsorbed to a positively-charged surface. Oligodeoxyribonucleotide probes have been found to form duplexes with non-helical properties on positively charged surfaces [14]. These duplexes are highly asymmetrical and unwound, possibly incurring a significant loss in base stacking which may subsequently affect the energetics of duplex formation. Such noncanonical structures may not be restricted to oligodeoxyribonucleotide arrays. Spotting of amplified cDNAs onto glass is another method commonly used to fabricate arrays [15]. Interestingly, multi-stranded DNA structures have been found to form on the surface of such arrays [16]. Furthermore, a low concentration of amplified cDNA in the dispense plate can generate a compression in the expression ratios, underscoring the importance of this parameter [15]. Although PCR preparation is not a factor in the fabrication of oligodeoxyribonucleotide arrays, a similar problem exists, namely, surface probe density. Probe density may result in steric effects [17] and could affect the efficiency of duplex formation and kinetics of target capture [18].
The microarray hybridization conditions can also affect the performance of a DNA microarray platform. An early study on DNA microarray hybridization found that hybridization was strongly dependent on the rate constants for DNA adsorption/desorption in the non-probe covered regions of the surface, the two-dimensional diffusion coefficient, and the size of probes and targets and also suggested that sparse probe coverage may provide results equal to or better than those obtained with a surface totally covered with DNA probes [19]. A theoretical analysis of the kinetics of DNA hybridization demonstrated that diffusion was important in determining the time of required to reach equilibrium and was proportional to the equilibrium binding constant and to the concentration of binding sites [20]. A recent theoretical and experimental analysis of competitive hybridization in a two-color system demonstrated the need for the hybridization kinetics of the two probes to be the same [21]. An elegant study on the use of hybridization kinetics to differentiate specific from non-specific binding was recently published [22]. This study found that the hybridization kinetics for specific and non-specific binding of labeled cRNA to surface-bound oligodeoxyribonucleotides were significantly different, with specific binding requiring longer to reach hybridization equilibrium than non-specific binding. This property was exploited to estimate and correct for the level of hybridization contributed by non-specific binding, enabling the selection of optimal oligodeoxyribonucleotides and the reduction of false positives in exon identification. Lastly, a longer hybridization time was previously shown to marginally increase relative fluorescence, potentially increasing detection of rare transcripts [23].
Lastly, oligodeoxyribonucleotide length can impact the performance of a DNA microarray platform. Early studies with high density arrays suggested the use of 25 mer in situ synthesized oligodeoxyribonucleotides [1]. A more recent study using covalently attached oligodeoxyribonucleotides found that 30- and 35 mer oligodeoxyribonucleotides generated signals two- to five-fold higher than 25 mers, and signals obtained form 60 mers were only two-fold higher than those obtained from 30 mers but ten-fold higher than those obtained from 25 mers [24]. In fact, in situ synthesized 60 mer oligodeoxyribonucleotides, hybridized in 30–32% formamide, have been shown to represent a compromise between maximal sensitivity and specificity compared to other lengths and hybridization conditions [12]. Lastly, a study employing in situ synthesis via maskless photolithography demonstrated an increase in signal intensity with increasing oligodeoxyribonucleotide length, up to 50 mers, followed by a plateau above that length [13].
Besides the effects on performance parameters such as sensitivity and specificity, fabrication methods can also affect the accuracy of a microarray platform. A sophisticated analysis of the Affymetrix platform was recently presented [25]. The authors used a relative error calculation to describe the dependence of the accuracy of the platform on the number of probe pairs per transcript. Furthermore, data was gathered, but not presented, regarding the increase in the relative error as a function of the fold-change (expression ratio). Such issues are important in all microarray platforms and impact oligodeoxyribonucleotide design as well as microarray data interpretation.
Thus, it is apparent that many factors can impact the performance of a microarray system and that these factors should be assessed prior to gathering large amounts of microarray data. The fact that the hybridization kinetics of specific and non-specific binding differ [22] raises the questions: what is the optimal hybridization time and could the hybridization time affect the accuracy of microarray data? Moreover, in order to take advantage of the large body of solution-phase hybridization data when designing oligodeoxyribonucleotide probes for microarrays and when optimizing hybridization conditions, the probes attached to the microarray surface or matrix must show solution-phase hybridization properties. In this report, we show solution-phase hybridization behavior on a three-dimensional microarray surface [10] and demonstrate that hybridization conditions, specifically the time allowed for hybridization, can impact the accuracy of fold-change calculations.
Results
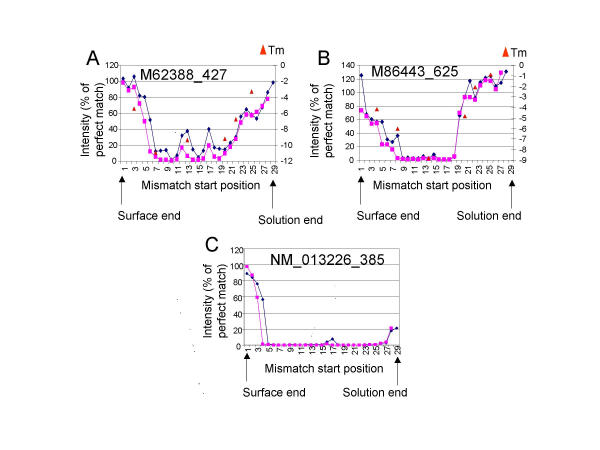
The specificity on the CodeLink platform was previously demonstrated to result in less than 5% of the initial signal being retained when a 3-base mismatch is present in the middle of the 30-mer oligodeoxyribonucleotide probe [10]. We used this information to test whether the DNA:RNA duplex formation is affected by the probe's attachment to the DNA microarray surface and to determine how much of the probe is accessible for hybridization. A series of 2-base and 3-base mismatches were designed across the length of 30-mer probes to assess its availability for hybridization. Labeled cRNA, prepared from total RNA isolated from kidney, was hybridized to DNA microarrays consisting of a series of 2-base and 3-base mismatches across the length of 30-mer probes containing the mismatches (Fig. 1). The profile of these mismatches showed that mismatches near the 3' (solution) or 5' (surface) end of the probe affected the hybridization signal nearly equivalently, demonstrating that the surface did not generate steric effects in the hybridization. For example, M62388 showed a symmetrical profile, with the greatest effect of the mismatches in the middle of the oligodeoxyribonucleotide (Figure 1A). M86443 showed approximately eight bases at the 3' (solution) end which exhibited only a small effect of mismatches (Figure 1B). This lack of an effect could be due to the fact that the mismatches at those positions did not significantly destabilize the duplex or due to the effect of the surface. The latter possibility is unlikely but will be addressed later. Lastly, NM013226 showed approximately 3 bases at the 5' (surface) end which did not affect the signal intensity significantly (Figure 1C). This lack of an effect could be due to the same reasons outlined for Figure 1B. A total of ten genes were designed and tested in this manner. Three of those are shown in Figure 1, and the other seven genes showed patterns similar to those shown in Figure 1 and, therefore, were not shown for the sake of brevity.
Figure 1.
Comparison of microarray hybridization versus solution-phase melting temperatures. DNA microarrays containing 2-base (diamonds) or 3-base (squares) mismatches across the entire length of the 30-mer oligonucleotide probe for multiple transcripts were hybridized to complex target prepared from total RNA isolated from kidney. The intensity for each mismatch, represented as a percent of the perfect match signal, for three probes, (A) M62388, (B) M86443, and (C) NM013226, is shown. The change in melting temperature of a 3-base mismatch of a DNA oligonucleotide with a complementary RNA oligonucleotide (triangles) is plotted for measurements made in stringency wash buffer in (A) and (B). Bases are numbered starting from the 5' end. The surface (5') and solution (3') ends of the oligonucleotide probe are indicated by the arrows.
To show that these hybridization results were similar to a solution phase hybridization, melting curve measurements were taken in solution using a standard temperature-controlled spectrophotometer. The Tm of the perfect match and 6 mismatches scanning across the 30-mer probe were measured in both the hybridization buffer and in the stringency wash buffer. Significantly, the difference in the Tm measurements (ΔTm) between the perfect match and the mismatches in solution followed the same pattern as the decrease in the hybridization signal on the arrays (Fig. 1). The strong correlation in the array and solution data in Figure 1B demonstrate that the mismatches at the 3' end of the oligodeoxyribonucleotide did not destabilize the duplex significantly, explaining the lack of a decrease in the signal intensity of these mismatches compared to the perfect match. Additionally, the patterns for the ΔTm calculations from measurements in the hybridization buffer (data not shown) and in the stringency wash buffer were nearly identical, suggesting that both the hybridization and the stringency wash conditions produced equivalent stringency. Due to the expense of oligoribonucleotide synthesis, a total of two genes (both of which are shown in Figure 1) were examined for concordance of microarray and melting temperature data. However, all six data points for the first gene and all six data points for the second gene (12 out of 12 or 100%) showed good concordance between the two measures. We conclude that the three-dimensional surface enables solution phase-type hybridization behavior and that most, if not all, of the oligodeoxyribonucleotide probe is available for hybridization with the target in solution.
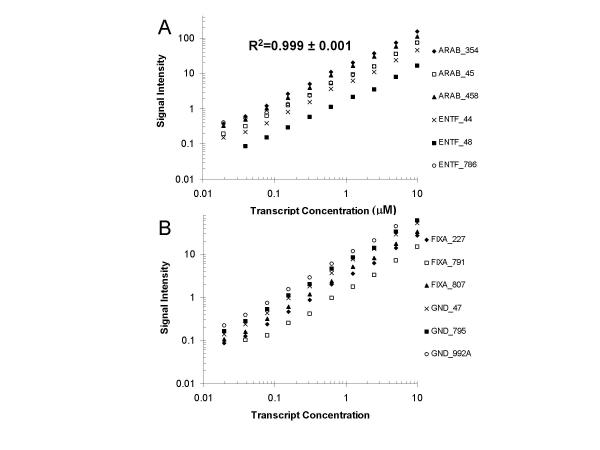
The solution phase-type hybridization behavior on the microarray surface suggested that probes on this surface should exhibit typical hybridization kinetics, where the reaction rate is dependent on the second order rate constant, the target concentration, and the probe concentration [19,20]. In other words, increases in the target concentration should generate proportional increases in the signal intensity. We therefore prepared serial dilutions where four control transcripts were spiked into a total RNA sample. In subsequent samples, two-fold serial dilutions of this spiked total RNA into fresh total RNA generated a dilution series consisting of ten concentrations of each transcript for a final range of 1:100,000 to 1:51,200,000 (10 microM to 20 nM). We prepared labeled cRNA targets from each of these samples and hybridized them to Codelink microarrays. These microarrays contained three bacterial probes which were designed to hybridize to each of the four spiked transcripts, for a total of twelve signals (six shown in Figure 2A and six shown in Figure 2B) which were measured at each serial dilution. Any errors in the measurements will include all aspects of the microarray platform. We plotted the signal intensity as a function of the spiked transcript concentration for oligodeoxyribonucleotide probes on the array that were complementary to these control transcripts. We found a linear response in the signal intensity from a transcript concentration of 20 nM to 10 microM, approximately three logs (Figures 2A and 2B). This signal linearity is consistent with a previously published report on the Codelink platform [10]. The average R2 value was found to be 0.998 ± 0.001. The fact that all twelve of these probes (100%) demonstrated this behavior suggests that this observation is general. However, we could not ascertain what portion of the probes which were designed to measure the endogenous human transcripts showed this behavior because it is not possible to vary the target concentration for each of those transcripts in a controlled, quantitative manner. In summary, these data verified the array signal was indeed dependent on the transcript concentration, as anticipated. They also demonstrated the precision and low variability of the entire microarray process (target prep, labeling, hybridization, and detection).
Figure 2.
Standard curves for the CodeLink DNA microarray. The signal obtained for a 2-fold serial dilution series (1:100,000 through 1:51,200,000 = 10 μM through 19.53 nM) is plotted for multiple probes for (A) araB and entF, and (B) fixA and gnd.
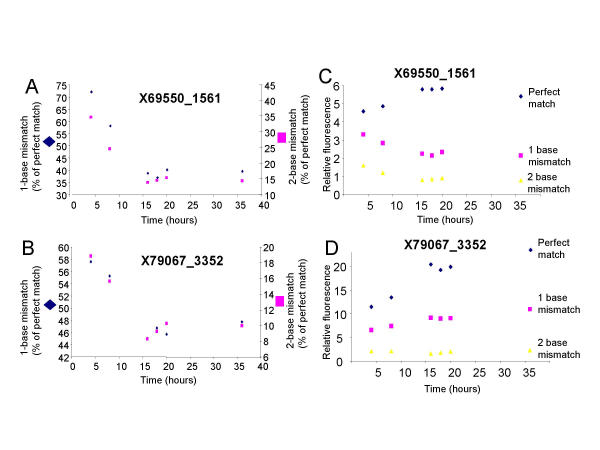
Mismatched duplexes generally exhibit larger dissociation rates than their perfectly matched counterparts [27]. We therefore examined the effect of one-, two-, and three-base mismatches as a function of hybridization time in two different probe sequences. A hybridization time course was performed by hybridizing the same cRNA sample to separate arrays, each array hybridized for different times, and examining the hybridization intensities for endogenous transcripts. The data in Figure 3 demonstrate that the effect of a one- and two-base mismatch is maximal using a hybridization time equal to or greater than sixteen hours. For example, in probe sequence X69550 (Figure 3A), the hybridization signal of a one-base mismatch was 72% of that of the perfect match after four hours but was only 39% of that of the perfect match after sixteen hours. Similarly, the hybridization signal of a two-base mismatch was 35% of that of the perfect match after four hours but was only 14% of that of the perfect match after sixteen hours. Identical results were seen for the effects of a three-base mismatch (data not shown). Consistent with previous findings [10], the effect of a one-base mismatch was found to be variable, reducing the hybridization signal to 39% (Figure 3A) or 47% (Figure 3B) of that of the corresponding perfect match signal. Also consistent with previous findings [10], a two-base mismatch had a greater effect, reducing the hybridization signal to 14% (Figure 3A) or 9% (Figure 3B) of that of the corresponding perfect match signal. We note that other array platforms may generate different effects of mismatches due to probe length or to the fact that the cRNA used in our experiments was fragmented to about 100 bases. Therefore, the actual probe/target duplex length was primarily dictated by the length of the 30 base probe (the shorter of the two single strands).
Figure 3.
Specificity against mismatches increases with hybridization time. (A and B) The intensity of a one-base (diamonds, left y-axis) or two-base (squares, right y-axis) mismatch, as a percent of the perfect match signal, is plotted as a function of time for two different probe sequences. (C and D) The intensity of the perfect match (diamonds), one-base mismatch (squares), or two-base mismatch (triangles) are plotted as a function of time for the same probe sequences shown in A and B.
Two scenarios could generate the increased specificity with time. First, at longer hybridization times, a greater proportion of mismatched duplexes (versus perfectly matched duplexes) may have dissociated, resulting in lower hybridization intensities for the mismatched duplexes. Secondly, the intensity of the perfect match may increase more significantly than that of the mismatch. To determine whether one or both scenarios exist, we plotted the intensity versus time for the perfect match, one base mismatch and two base mismatch probes (Figures 3C and 3D). While the intensity of the perfect match probe for X69550_1561 increases with time, reaching a plateau around 16 hours, the intensity of both the mismatch probes decreases with time, reaching a plateau around 16 hours (Figure 3C). In contrast, while the intensity of the perfect match probe for X79067_3352 increases by 80% between 4 and 16 hours, the intensity of the one base mismatch probe increases only 40% between 4 and 16 hours and the intensity of the two base mismatch probe decreases by 20% between 4 and 16 hours (Figure 3D). Thus, specificity can be generated in different ways. These data provide further evidence of solution phase hybridization behavior in the Codelink microarray system and demonstrate that short hybridization times could result in decreased specificity.
It is important to note that the data in Figures 2 and 3 do not address the accuracy of the microarray platform. For example, the results in Figure 2 showed a relative error ([fold change expected – fold change observed]/fold change expected) of 17% which is low compared with higher errors reported elsewhere [25]. However, the data exhibited a low but consistent compression of the ratios (obtained from comparing the signal obtained from one transcript concentration to that obtained from the next dilution). Therefore, a hybridization time course was performed to find the time which would generate the lowest compression in the ratios. A simple approach to calculate ratios for multiple genes at once (>1000 genes) is to vary the amount of cRNA used in the hybridization and compare the calculated and expected ratios of every probe. This approach eliminates bias which might be introduced when solely using control transcripts and it prevents systematic errors introduced by the sample preparation method. Importantly, the total nucleic acid concentration was kept constant by varying the amount of labeled cRNA and supplementing the lower amount of labeled cRNA with unlabeled cRNA. Therefore, the only variable in these experiments was the hybridization time.
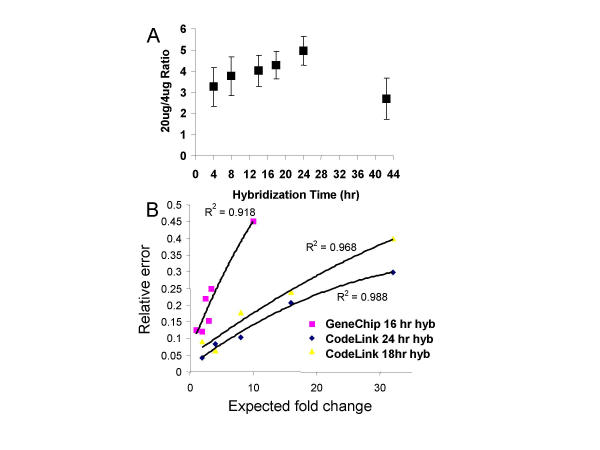
Using this method, a ratio of five would be expected from each probe if the signal intensity generated from 20 micrograms of labeled cRNA was compared to that obtained from 4 micrograms of labeled cRNA. We acknowledge that this is an oversimplification given that some probes will reach equilibrium faster than others and that saturation of pixels may arise for high signals. However, as a global approach, the compression of the ratio, as a function of the hybridization time, was investigated. A hybridization time of 24 hours generated a ratio of 5 (Fig. 4A). Significant compression was found if the hybridization time was too short or was too long. These results suggested that hybridization time could affect the accuracy of ratios by introducing a compression effect at shorter times.
Figure 4.
Accuracy of expression ratios: (A) Accuracy of expression ratios as a function of hybridization time. The incubation time was varied from 4 to 42.5 hours for hybridizations containing a total of 20 μg of cRNA target (4 μg of labeled cRNA + 16 μg of unlabeled cRNA) versus 20 μg of labeled cRNA. (B) Examination of the relative error for two platforms as a function of the fold change. The relative error for the Affymetrix GeneChip data from Zhou and Abagyan is shown versus the relative error for the CodeLink data from the 2-fold dilution series in Figure 2. The GeneChip data be obtained at the following web site: http://carrier.gnf.org/publications/MOID/spike.html.
How does the hybridization time affect ratios smaller or larger than five? Does this compression effect (or the relative error) increase with larger ratios? Lastly, how do these data compare with other platforms? To address these questions, we repeated the dilution series experiments presented in Figure 2 but performed the hybridization for 24 hours instead of the 18 hours that were used to generate the data in Figure 2. We analyzed both sets of dilution series data according to the method of Zhou and Abagyan [25], using the highest concentration of the dilution series and determining how the relative error changed over a large range of differential expression ratios. We also analyzed the Affymetrix data from the Zhou and Abagyan publication in this format. We plotted the relative error as a function of the fold change for the Codelink platform, using either an 18- or 24-hour hybridization time (Figure 4B). The data points were fit with a second order polynomial, and the R2 values (0.968 and 0.988 for the 18- and 24-hour hybridization times, respectively) showed a good fit of the data to the curve. Lastly, when data from an analogous Affymetrix experiment were plotted in this format, the relative error also increased as a function of the fold change. The Affymetrix data plotted were based on twelve to 154 data points for each expected fold change, thus representing a total of 366 data points. We note that, due to the very different conditions used to generate the Affymetrix data and to the fact that it is derived from only one publication, these data should not be compared with the Codelink data. However, these data do demonstrate that the increase in relative error with increasing fold changes may be a common feature of multiple microarray platforms.
The data in Figure 4B highlighted several points. First, the data using the spiked control transcripts (Figure 4B) agreed with the data presented in Figure 4A using the endogenous transcripts when the 18 and 24 hour timepoints were compared and demonstrated that a 24 hour hybridization time produced more accurate data (less compression or lower relative error) than an 18 hour hybridization time. We note that the 42 hour time point in Figure 4A can not be compared to a similar time point in Figure 4B. Secondly, a 24 hour hybridization time consistently outperformed an 18 hour hybridization time, with respect to the relative error, for all fold changes examined on the Codelink platform. Thirdly, the relative errors in all three conditions (Codelink 18- and 24-hour and Affymetrix platform) increased with the larger ratios. There are two important points that should accompany these conclusions. First, different platforms use different oligonucleotide lengths (from 25 to 60 bases in length) and even amplified cDNA products [1,2,10,12,13,22,24], with different accessibilities of the array-bound nucleic acid. Therefore, it is impossible to generalize that a time which is optimal on one platform will be optimal on the second platform. Secondly, more studies on the Codelink arrays will be required to verify that 24 hours is indeed the optimal. We only know that it outperforms the 18 hour time points. We conclude that a longer hybridization time (e.g., 33% longer) may generate microarray data with lower relative errors (while hybridization times of 42 hours could generate more compression), and this kinetic parameter merits further investigation as a simple method to increase both performance (signal intensity and mismatch specificity) and accuracy.
Discussion
Much debate exists regarding the optimal oligodeoxyribonucleotide probe length. However, a more functional way of thinking about the probe length may be how many bases of the probe are actually available for hybridization and if these bases are exhibiting solution phase biophysical behavior. In this manner, linker length and surface effects must also be considered. For example, bases at the surface end of some in situ synthesized 60 base oligodeoxyribonucleotide probes may not be accessible for hybridization [12] while other in situ synthesized probes may require linkers for optimal performance [13]. We present evidence, using 30 base oligodeoxyribonucleotide probes, that most, if not all, of the probe is available for hybridization and that the surface does not introduce significant steric effects. We note that the three-dimensional Codelink arrays used in this study are different from standard surface-bound arrays, and, therefore, it is not unexpected that the observations presented in this report are different from those generated on other array platforms. Furthermore, we show, for the first time on a microarray platform, a strong concordance of microarray hybridization mismatch data with solution phase duplex melting experiments (Figure 1).
Another important consideration in microarray experiments is the time allowed for the hybridization reaction. Recent data has shown that specific binding takes longer to reach equilibrium than non-specific binding [22], suggesting that a longer hybridization time may be beneficial. Implicit in such findings is the fact that the accuracy of a fold change may also increase with a longer hybridization time. We present evidence demonstrating that hybridization time can in fact increase the accuracy of expression ratios (fold changes), relieving the observed compression in ratios, and that this effect may be more dramatic for larger fold changes (Figure 4). In retrospect, these data make sense from a biophysical perspective because, at the longer hybridization times, the mismatched duplexes will have dissociated due to their faster dissociation rates (Figure 3), leaving primarily the perfectly matched duplexes. The optimal hybridization time on different platforms could vary, depending on probe length and accessibility, diffusion coefficients, and detection methods, but the basic premise of increased accuracy with increased hybridization time should hold. Thus, we believe that this parameter merits further investigation.
Various computational and statistical measures have been used to improve and filter microarray data. For example, locally weighted linear regression (lowess) normalizations have been used to correct the systematic dependence of the log2 of the red/green expression ratios on hybridization intensity [28]. The finding that competitive hybridization in a two-color system requires the hybridization kinetics of the two targets to be the same [21] may help explain the need for such normalizations. Thus, understanding the hybridization behavior of probes and targets in a microarray platform may obviate the need for large amounts of data manipulation.
Moreover, a recent study found that both cDNA and oligodeoxyribonucleotide arrays underestimated the relative changes in mRNA expression between experimental and control samples, as determined by quantitative reverse transcriptase polymerase chain reaction [29]. This underestimation (or ratio compression) increased as the relative change increased, consistent with our observations (Figure 4B). Such comparative studies underscore the need to understand the root cause(s) for ratio compression in microarray platforms in order to design effective solutions. The fact that specific binding takes longer to reach equilibrium [22] and that a longer hybridization time may alleviate compression in expression ratios (this report) is one example of how fundamental studies may eventually improve microarray data.
Conclusions
The data in this report demonstrate, for the first time, a strong concordance between the effect of mismatches in probes attached to a three dimensional matrix of a microarray and solution-based, thermodynamic duplex melting studies. Moreover, an increased hybridization time was shown to increase the accuracy of fold changes by relieving the compression observed in expression ratios, and this effect may be more dramatic for larger fold changes. Studies such as these may ultimately help improve microarray data quality.
Methods
Array experiments
Target preparation, CodeLink™ DNA microarray hybridization, and processing were performed as described previously [26] except as described. A single, labeled nucleotide, biotin-11-UTP, was used in the cRNA labeling reactions at a concentration of 1.25 mM. Unlabeled UTP was present at 3.75 mM, while GTP, ATP, and CTP were at 5 mM. cRNA was fragmented prior to the hybridization reaction as previously described [10,26]. Hybridization time studies followed the above procedures with the following exceptions. The first sample consisted of 20 ug of labeled, cRNA target. The second sample consisted of 4 ug of labeled cRNA target and 16 ug of unlabeled cRNA target for a total of 20 ug of cRNA. Each target was hybridized to an array for 4, 8, 14, 18, 24, or 42.5 hours.
Serial dilutions were prepared by adding the ≅1000 base transcripts from the E. coli genes araB, entF, fixA, and gnd at a final concentration of 10 μM for each transcript in kidney total RNA. This 10 μM dilution was then diluted into kidney total RNA in a 2-fold series to a lowest concentration of 19.53 nM for each transcript. The 78 nM dilution is approximately equal to 1 copy per cell, assuming an mRNA population which is 2.5% of the total RNA, 300,000 mRNAs/cells, and an average mRNA length of 1,000 bases [10].
Array design
The data shown in Figure 1 were generated using the mismatch scanning array. This array consisted of ten probe sets where each probe set was designed to hybridize to either a spiked, bacterial transcript or an endogenous transcript present in a complex human polyA+ RNA sample (e.g., from human liver or human brain). Each probe set consisted of the perfect match to the targeted transcript and two subsets. The first subset consisted of thirty probes, each having a two-base mismatch. The position of the mismatch was shifted one base for each probe in this subset, generating a subset of probes with two-base mismatches scanning the length of the 30 base sequence. The second subset also consisted of thirty probes, each having a three-base mismatch. The position of the mismatch was shifted one base for each probe in this subset, generating a subset of probes with three-base mismatches scanning the length of the 30 base sequence.
The data shown in Figures 2 through 4 were generated using the Codelink™ Human Uniset I arrays. These arrays contain 9,589 probes (representing approximately 9200 unique accession numbers) designed to hybridize to human transcripts present in polyA+ RNA and approximately 386 control probes (designed to hybridize primarily to bacterial transcripts). The noncontrol probes (those designed to measure relative expression levels of the endogenous human transcripts) were each designed based on the paradigm of one probe per gene. However, three to ten bacterial control probes were designed to hybridize to each bacterial transcript. These bacterial probes can be used as negative controls or a subset of these can be used as positive controls when the corresponding bacterial transcripts are spiked into the human polyA+ RNA. The latter scenario, in which four bacterial transcripts were spiked into the polyA+ RNA, was used to generate the data shown in Figure 2. The hybridization signal from each of the three bacterial probes on the array was measured for each of the four spiked transcripts, for a total of twelve signals which were measured at each serial dilution. In addition, the set of control probes contains five probe sets, each of which contain the perfect match, one-, two-, three-, and four-base mismatches to either a bacterial transcript or an endogenous human transcript. The intensities of these probe sets was measured after different array hybridization times to generate the data shown in Figure 3. Lastly, the probes on the Human Uniset I arrays which were designed to measure the endogenous human transcripts in polyA+ RNA were measured after different array hybridization times and with different amounts of labeled cRNA to generate the data shown in Figure 4A. The twelve probes on these arrays which were designed to hybridize to the four spiked bacterial transcripts were measured after different hybridization times and with serial dilutions of the bacterial transcripts to generate the data shown in Figure 4B.
Tm determination
The solution-phase melting temperatures were measured with an Agilent 8453 UV-VIS spectroscopy system with added Peltier thermostated single cell holder using a 1.5 ml quartz cuvette. Each probe-target set contained a perfect DNA:RNA 30-mer match and 6 DNA:RNA pairs with 3-base mismatches incorporated into the oligodeoxyribonucleotide. The oligoribonucleotides (IDT Technologies) were incubated at room temperature with equimolar amounts of the oligodeoxyribonucleotide in either in stringency wash solution (75 mM Tris-Cl, 112.5 mM NaCl) or in hybridization buffer (50% formamide/6 × SSPE), then the melting profile was performed in 1°C increments with constant monitoring at 260 nm. The Tm was determined for each DNA:RNA pair by calculating the first derivative of the A260 profile.
Authors' Contributions
DD and AN conducted data analysis and helped with experimental design. LG, AL, MP, ET, and RE performed all of the experiments. RL and TJS conducted all of the bioinformatics, designed probes, and helped with the fabrication of arrays used. AM wrote the paper, generated most of the figures, suggested most of the experiments, and provided overall technical guidance. All authors read and approved the final manuscript.
Contributor Information
David R Dorris, Email: daviddorris@yahoo.com.
Allen Nguyen, Email: allenn1999@yahoo.com.
Linn Gieser, Email: linngieser@yahoo.com.
Randall Lockner, Email: rlockner@hotmail.com.
Anna Lublinsky, Email: a_lublinsky@yahoo.com.
Marcus Patterson, Email: marcuskpatterson@yahoo.com.
Edward Touma, Email: edwardtouma@yahoo.com.
Timothy J Sendera, Email: tsendera@usa.net.
Robert Elghanian, Email: randelghanian@prodigy.net.
Abhijit Mazumder, Email: rmazumder@msn.com.
References
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–42. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts C, Stoughton R, Burchard J, Slade D, Dai H, Bassett DE, Jr, Hartwell LH, Brown PO, Friend SH. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- Waring JF, Ciurlionis R, Jolly RA, Heindel M, Ulrich RG. Microarray analysis of hepatotoxins in vitro reveals a correlation between gene expression profiles and mechanisms of toxicity. Toxicol Lett. 2001;120:359–68. doi: 10.1016/S0378-4274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, Bennett L, Tennant R, Stoll R, Barrett JC, Blanchard K, Paules RS, Afshari CA. Gene expression analysis reveals chemical-specific profiles. Toxicol Sciences. 2002;67:219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R, Dorris DR, Lublinsky A, Nguyen A, Domanus M, Prokhorova A, Gieser L, Touma E, Lockner R, Tata M, Shippy R, Sendera T, Mazumder A. An assesment of Motorola CodeLink™ microarray performance for gene expression profiling applications. Nucleic Acids Res. 2002;30:e30. doi: 10.1093/nar/30.7.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, Kobayashi S, Davis C, Dai H, He YD, Stephaniants SB, Cavet G, Walker WL, West A, Coffey E, Shoemaker DD, Stoughton R, Blanchard AP, Friend SH, Linsley PS. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Nuwaysir EF, Huang W, Albert TJ, Singh J, Nuwaysir K, Pitas A, Richmond T, Gorski T, Berg JP, Ballin J, McCormick M, Norton J, Pollack T, Sumwalt T, Butcher L, Porter D, Molla M, Hall C, Blattner F, Sussman MR, Wallace RL, Cerrina F, Green RD. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeshko SV, Powdrill T, Belosludtsev YY, Hogan M. Oligonucleotides form a duplex with non-helical properties on a positively charged surface. Nucleic Acids Res. 2001;29:3051–8. doi: 10.1093/nar/29.14.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Eastman PS, Wang BB, Minor J, Doctolero MH, Nuttall RL, Stack R, Becker JW, Montgomery JR, Vainer M, Johnston R. An evaluation of the performance of cDNA microarrays for detecting changes in global gene expression. Nucleic Acids Res. 2001;29:e41. doi: 10.1093/nar/29.8.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SJ, Scheffer A, Bjeldanes E, Reynolds MA, Arnold LJ. DNA exhibits multi-stranded binding recognition on glass microarrays. Nucleic Acids Res. 2001;29:4251–6. doi: 10.1093/nar/29.20.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepinov MS, Case-Green SC, Southern EM. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AW, Heaton RJ, Georgiadis RM. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan V, Graves DJ, McKenzie SE. The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys J. 1995;69:2243–2255. doi: 10.1016/S0006-3495(95)80095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits MA, Mirzabekov AD. Theoretical analysis of the kinetics of DNA hybridization with gel-immobilized oligonucleotides. Biophys J. 1996;71:2795–2801. doi: 10.1016/S0006-3495(96)79473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, Guo S-W, Ghosh S. Conditions to ensure competitive hybridization in two-color microarray: a theoretical and experimental analysis. Biotechniques. 2002;32:1342–1346. doi: 10.2144/02326mt05. [DOI] [PubMed] [Google Scholar]
- Dai H, Meyer M, Stepaniants S, Ziman M, Stoughton R. Use of hybridization kinetics for differentiating specific from non-specific binding to oligonucleotide microarrays. Nucleic Acids Res. 2002;30:e86. doi: 10.1093/nar/gnf085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudin E, Walker R, Kosaka A, Wu SX, Rabert D, Chang TK, Kreder DE. Assessment of the relationship between signal intensities and transcript concentration for Affymetrix GeneChip arrays. Genome Biology. 2001;3:0005.1. doi: 10.1186/gb-2001-3-1-research0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relogio A, Schwager C, Richter A, Ansorge W, Valcarcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Abagyan R. Match-only integral distribution (MOID) algorithm for high-density oligonucleotide array analysis. BMC Bioinformatics. 2002;3:3. doi: 10.1186/1471-2105-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris DR, Ramakrishnan R, Trakas D, Dudzik F, Belval F, Zhao C, Nguyen F, Domanus M, Mazumder A. A highly reporducible, linear, and automated sample preparation method for DNA microarrays. Genome Res. 2002;12:976–984. doi: 10.1101/gr.227402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S, Wagner RW. Hybridization and dissociation rates of phosphodiester or modified oligodeoxynucleotides with RNA at near-physiological conditions. Nucleic Acids Res. 1991;19:2463–2470. doi: 10.1093/nar/19.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nature Genetics Suppl. 2002;32:496–502. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48. doi: 10.1093/nar/30.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]