Abstract
A site-specific replication terminator, RTS1, is present at the Schizosaccharomyces pombe mating-type locus mat1. RTS1 regulates the direction of replication at mat1, optimizing mating-type switching that occurs as a replication-coupled recombination event. Here we show that RTS1 contains two cis-acting sequences that cooperate for efficient replication termination. First, a sequence of ∼450 bp containing four repeated 55 bp motifs is essential for function. Secondly, a purine-rich sequence of ∼60 bp without intrinsic activity, located proximal to the repeats, acts cooperatively to increase barrier activity 4-fold. Our data suggest that the trans-acting factors rtf1p and rtf2p act through the repeated motifs and the purine-rich element, respectively. Thus, efficient site-specific replication termination at RTS1 occurs by a complex mechanism involving several cis-acting sequences and trans-acting factors. Interestingly, RTS1 displays similarities to mammalian rDNA replication barriers.
Keywords: DNA replication barrier/mating-type switching/replication termination/rtf2/Schizosaccharomyces pombe
Introduction
Generally, eukaryotic DNA replication termination occurs randomly in the intervening regions between replication origins (Zhu et al., 1992; Santamaria et al., 2000). However, at several genetic loci, replication termination is site-specific (reviewed by Hyrien, 2000). Such replication fork barriers are found in the RNA polymerase I-transcribed rDNA gene arrays of Saccharomyces cerevisiae (Brewer and Fangman, 1988; Linskens and Huberman, 1988), Schizosaccharomyces pombe (Sanchez et al., 1998), Tetrahymena (Zhang et al., 1997), peas (Hernandez et al., 1993), Xenopus (Wiesendanger et al., 1994), mouse (Gerber et al., 1997; Lopez-Estrano et al., 1998) and human (Little et al., 1993), and the RNA polymerase II-transcribed Drosophila melanogaster histone gene arrays (Shinomiya and Ina, 1993). Replication barriers are also found at non-repetitive genetic loci such as at the polymerase III-transcribed S.cerevisiae tRNA gene (Deshpande and Newlon, 1996), in centromeric regions (Greenfeder and Newlon, 1992), in the Kluyveromyces lactis plasmid pKD1 (Fabiani et al., 2001) and in the mating-type region of S.pombe (Dalgaard and Klar, 2001).
Very diverse biological functions are attributed to replication fork barriers, as follows: (i) the prokaryotic plasmid R100 replication barrier was shown to prevent a shift to rolling circle replication (Krabbe et al., 1997). (ii) The replication fork barrier in the K.lactis plasmid pKD1 plays a role in plasmid stability (Fabiani et al., 2001). (iii) Saccharomyces cerevisiae Fob1p-dependent rDNA barrier activity is important for inducing recombination between rDNA genes leading to expansion or contraction of the gene array (Kobayashi et al., 1998). (iv) In the eukaryotic RNA polymerase I transcriptional units, the barriers are thought to prevent the interference between transcription and replication forks (Brewer and Fangman, 1988). RNA polymerase I transcription occurs during S-phase of the cell cycle and the rDNA barriers arrest replication forks entering the region in opposite directions to that of transcription. Thus, the barriers prevent ‘collisions’ between replication and transcription forks, which are thought to cause topological ‘knots’ in the DNA (Olavarrieta et al., 2002). Also, in bacteria coordination between transcription and replication is of importance; in the eubacteria Escherichia coli and Bacillus subtilis, where site-specific termination of replication was first described, several polar termination barriers are located in the region diagonal to the replication origin in the circular genomes (Kuempel et al., 1977; Weiss and Wake, 1984). Here, the orientation of transcription units with respect to the site-specific terminators and the origin is such that there is the least interference between transcription and replication forks (Tillier and Collins, 2000). (v) Finally, as described below, the S.pombe RTS1 element plays an important role in optimizing mating-type switching.
The mechanism of impediment of replication fork progression is best described for the eubacterial elements. In E.coli, the trans-acting protein tus binds asymmetrically to the cis-acting sequences terA to terF, and at each site acts as a polar barrier for the replication fork (reviewed by Bussiere and Bastia, 1999). Tus is a counter helicase that interferes with the replicative helicase DnaB by acting as a physical barrier for the progression of the replication fork (Khatri et al., 1989; Lee et al., 1989). In B.subtilis, a similar mechanism accounts for replication termination. However, here the termination protein RTP interacts specifically with the replicative helicase DnaB to inhibit DNA unwinding (Sahoo et al., 1995; Mohanty et al., 2001). The eukaryotic barriers are less studied. In S.cerevisiae, fob1 is essential for barrier activity, however the mechanism of its function remains unknown (Kobayashi and Horiuchi, 1996; Ward et al., 2000). In mammals, the transcription termination factor ttf1 is implicated in replication barrier activity in rDNA gene array (Gerber et al., 1997; Lopez-Estrano et al. 1998). The ttf1 factor catalyses termination of RNA polymerase I transcription at several ‘Sal-boxes’ located downstream from the 28S rRNA gene unit. There are 10 Sal-boxes in mouse, and in vivo they all act as barriers for replication forks entering the region in the direction opposite to that of transcription (Lopez-Estrano et al., 1998). However, in vitro, only one of them, Sal-box two, acts as a barrier (Gerber et al., 1997). In the latter case, the Sal-box sequence and a GC repeat region are essential for replication barrier function, while a stretch of 64 thymidines increases barrier activity by 30% (Gerber et al., 1997). Two factors have been shown to be important for function. Depletion of the trans-acting factor ttf1 from cell extract abolishes barrier function in vitro (Gerber et al., 1997), and purified ttf1 protein has been shown to have counter-helicase activity in SV40 DNA replication assays (Putter and Grummt, 2002). In addition, the DNA binding protein complex Ku interacts with the GC-rich region, and depletion of Ku from nuclear extract abolishes replication termination in vitro (Wallisch et al., 2002).
In this study we analyse the molecular mechanism of replication fork impediment at the RTS1 element that regulates the direction of replication at the S.pombe mat1 locus (Figure 1). In S.pombe, an imprint (modification) is made at the mat1 locus, marking cells for mating-type switching (Egel and Eie, 1987; Klar, 1987). This imprint is made in connection with replication, and only when mat1 is replicated in the centromere-proximal direction (Figure 1; Dalgaard and Klar, 1999). The chromosomal imprint is maintained for one cell cycle, and during the next round of replication a recombination event is induced at mat1 (Arcangioli, 1998; reviewed in Dalgaard and Klar, 1999; Arcangioli and de Lahondes, 2000). The induction of the recombination event is thought to occur when the replication fork encounters the imprint in the template strand; as a result fork progression is inhibited, a break is formed and the recombination event initiated. The recombination event utilizes one of the two transcriptionally silent gene cassettes, located distal to mat1, mat2P (containing P cell-type specific information) or mat3M (containing M cell-type specific information), as donors during the repair (Kelly et al., 1988). The result of the recombination event is a change of mating-type specific information at the mat1 locus. The cis-acting polar terminator RTS1 ensures that mat1 is replicated in the correct orientation for this programme of cellular differentiation (Dalgaard and Klar, 2001).
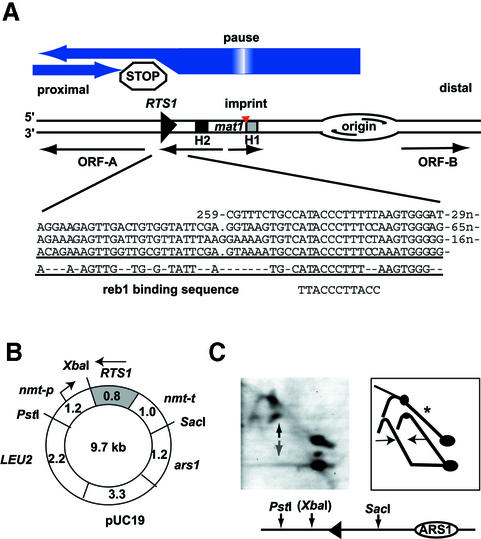
Fig. 1. (A) Graphic outline of the S.pombe mat1 region. The genetic elements and transcriptional units (thin horizontal arrows) present on the 10.6 kb HindIII fragment containing mat1 are shown. The orientation of the fragment relative to the centromere is given. Large blue arrows display the direction of replication in the region and termination at the RTS1 element. The position of the imprint that marks switchable cells is shown. swi1- and swi3-dependent pausing of the replication fork at the site of imprinting is shown as a gradient within the blue arrow, depicting replication. H1 and H2 are homology domains thought to be important during mating-type switching. ORF-A and -B are open reading frames of unknown function. An alignment of repeated motifs present in the RTS1 element is displayed below. The position of the repeats is shown relative to the EcoRI sites that flank the RTS1 element. The motif consensus sequence and the S.pombe reb1 rDNA binding sequence are shown (Melekhovets et al., 1994). (B) Graphic outline of pREP3 plasmid used in this study. Gene names, restriction sites and sizes (kbp) are given. RTS1 is shown as a grey box; the arrow indicates that the orientation is opposite of that in Figure 1A. (C) Fork-direction gel analysis of the cloned RTS1 element. Left panel: autoradiogram; right panel: interpretation of the autoradiogram. The line drawing below shows the enzymes and sites used in the experiment. XbaI, given in parentheses, was utilized for the digestion before separation in the second dimension. Intermediates from replication forks moving in both directions are detected (right panel, horizontal arrows). RTS1 barrier signal can be detected for forks moving in the non-permissive direction (left panel, vertical dark arrow), but not the permissive direction (left panel, vertical grey arrow). Only partial digestion was achieved before separation in the second dimension, allowing visualization of the original replication arc that is constituted by replication forks moving in both directions (marked with an asterisk). For a detailed explanation of this technique, please refer to Brewer and Fangman (1988).
A genetic screen for factors involved in termination of replication at RTS1 identified four genes potentially involved in the termination process: rtf1, rtf2, swi1 and swi3 (Dalgaard and Klar, 2000). Two of these genes, swi1 and swi3, were shown to be essential for RTS1 function; swi1 and swi3 are also involved in imprinting where they pause the replication fork in the vicinity of the imprint (Figure 1; Dalgaard and Klar, 2000).
Here we show that the RTS1 element consists of two types of cis-acting elements. First, a ∼450 bp region B containing the four repeated ∼55 bp motifs is essential for function, each motif contributing to barrier activity to a similar degree. Sequence homology between the trans-acting replication termination factor rtf1p and the eukaryotic transcription factors Reb1p and TTF1 suggests that rtf1p interacts directly with the repeated motifs. Secondly, an ∼60 bp purine-rich region A, with no intrinsic barrier activity, increases the activity of region B 4-fold, possibly by mediating a functional interaction between the region B motifs. Furthermore, our experiments suggest the region A activity is dependent on the trans-acting factor rtf2p.
Results
The core RTS1 element consists of ∼410 base pairs
Initially, the RTS1 element was mapped within the 859 bp EcoRI fragment located proximal to mat1. We decided to perform a deletion analysis to limit further the cis-acting sequences necessary for replication termination. Wild-type and deletion mutants of the RTS1 element were introduced adjacent to the ARS1 origin of replication in the poly-linker of the LEU2-based plasmid pREP3 (Figures 1B and 2A; Maundrell, 1993). The plasmid constructs were transformed into S.pombe strain JZ165 that carries a deletion of the chromosomal RTS1 element. The chromosomal deletion prevents interference between plasmid and chromosomal RTS1. In this study, all strains were grown in the presence of 4 µM thiamin to repress transcription from the nmt-promoter present in the plasmid (Figure 1B; Maundrell, 1993). However, initial experiments showed that transcription from the nmt-promoter does not affect the barrier activity of the wild-type RTS1 element (data not shown). Replication intermediates from each of the obtained strains were purified and analysed using native two-dimensional gel electrophoresis (Brewer and Fangman, 1987). It should be noted that the pREP3 plasmid is propagated as a multimer in S.pombe, due to replication-associated recombination at the plasmid’s origin ars1 (Segurado et al., 2002). Thus, differential initiation of replication of the multiple origins present in oligomerized pREB3 plasmids lead to bi-directional replication at the cloned RTS1 element (Figure 1C). In the study presented here we are therefore quantifying the amount of termination of replication intermediates relative to the ascending part of the replication arc, as these intermediates predominantly represent intermediates from replication forks moving in the non-permissive direction of RTS1. Using the National Institutes of Health (NIH) image software (Figures 2, 3 and 4) and Quantity One (Bio-Rad) (Figure 5), the intensities of the signals indicated in Figure 2B were quantified: T, termination signal; P, pause signal; and RI, replication intermediates of ascending arc. T and RI values were corrected for background signals, using a measurement of neighbouring areas of the same size. The P signal was corrected for the underlying replication arc by subtracting the signal from the same sized area of the ascending arc. For each gel, the ratio of paused (P) and terminated (T) signals to the signal from the ascending arc (RI) was calculated, respectively. In each set of experiments, the activities of the modified elements are shown as relative percentages of the wild-type sequence’s activity. Importantly, it should be noted that only one gel was quantified for each construct analysed (Figures 2, 3 and 4), and the results are therefore only tentative.
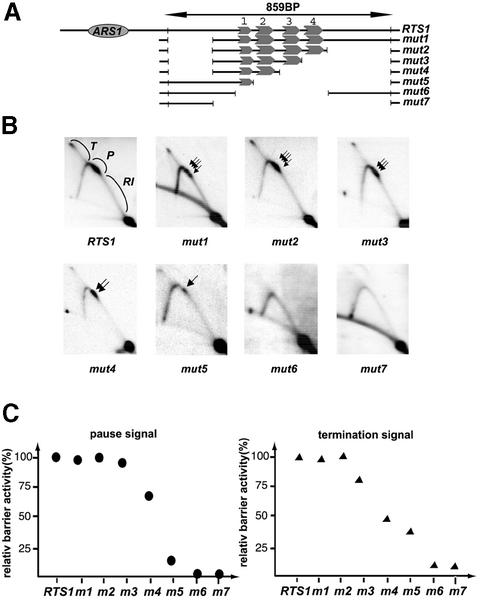
Fig. 2. The repeated motifs are essential for replication pausing and termination. (A) Graphic outline of the constructed RTS1 deletions cloned into plasmid pREB3. The length, in base pairs, of the wild-type EcoRI fragment containing RTS1 is shown. The plasmid origin of replication, ARS1, is shown as an ellipse. The conserved repeated motifs are represented as gray arrows, numbered one to four. The names of the different constructs are given to the right of the line drawing. (B) Two-dimensional gel analysis of replication intermediates of wild-type and mutant RTS1 elements. The intensity of intermediates is marked with letters indicating that termination (T), pause (P) and standard replication intermediates (RI) were used for quantifying barrier activity. Black arrows indicate that the paused signals consist of several closely spaced barriers. (C) Quantification of barrier activities. The intensities of termination and pause signals observed for each mutant construct (B) are displayed as percentages of the wild-type RTS1 element’s signals (RTS1: 100%).
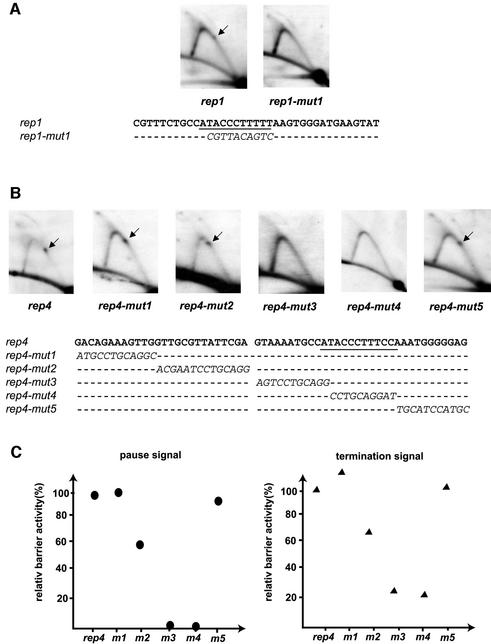
Fig. 3. Mutational analysis of motifs 1 and 4. (A) Mutational analysis of motif 1. Two-dimensional gel analysis of replication intermediates of wild-type (rep1) and mutant (rep1-mut1) motifs. The analysed sequences are given below. Only substituted bases are shown for the mutant sequence. (B) Mutational analysis of the fourth motif. Two-dimensional gel analysis of replication intermediates of wild-type (rep4) and mutant (rep4-mut1 to mut5) motifs. (C) Quantification of barrier activities. The intensities of termination and pause signals observed for each mutant construct are displayed as percentages of the wild-type rep4 motif signals (rep4: 100%).
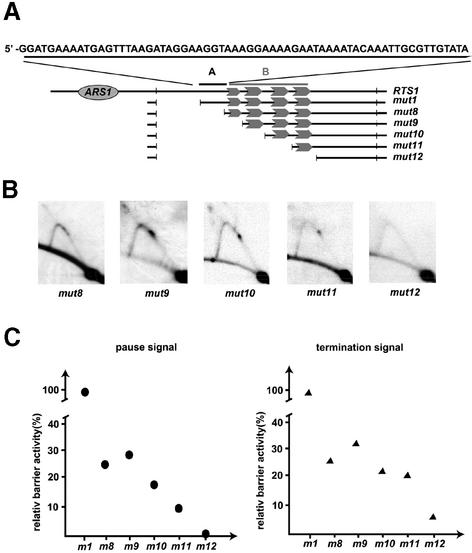
Fig. 4. Identification of a cooperatively acting region. (A) Graphic outline of the constructed deletions. The region containing the repeated motifs is indicated with the gray, upper case B and line. The identified cooperatively acting region is indicated with the black, upper case A and line. The sequence of region A is given above the line drawing. Other symbols are described in the legend to Figure 2. (B) Two-dimensional gel analysis of replication intermediates of mutant plasmids. The name of each mutant is given below the autoradiograms. (C) Quantification of barrier activities. The intensities of termination and pause signals observed for each mutant construct (B) are displayed as percentages of the wild-type RTS1 element’s signals (RTS1: 100%).
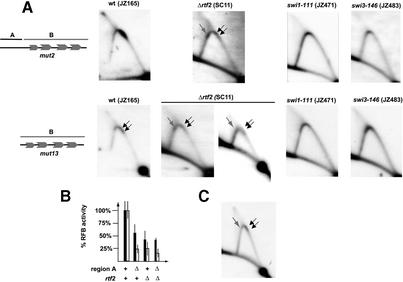
Fig. 5. The trans-acting factor rtf2 acts through the cis-acting region A. (A) Plasmids, containing region A and B (mut2) or only region B (mut13), shown to the left, were transformed into strains, carrying mutations in the indicated trans-acting factors. Strain names are given in parentheses above the autoradiograms. Two different panels are shown for the rtf2, mut13 strain. Arrows indicate replication fork barriers. The gray arrows indicate barriers only observed in the Δrtf2 genetic background. (B) Quantification of pause and termination signals observed in (A), displayed as a histogram. The open and solid bars indicate pause and termination signals, respectively. The standard deviations, based on three experiments, are shown as protruding vertical lines. Below, the strains’ genotypes are shown. (C) Δrtf2 reduces barrier activity at the 0.8-kb EcoRI fragment containing the RTS1 element. For comparison with wild type, refer to Figure 2.
Initially, two deletions were introduced: a 173 bp deletion on the proximal side and a 272 bp deletion on the distal side (Figure 2A, mut1 and mut2). Comparison between wild-type and mutant elements shows that neither of these affect RTS1 pausing and termination activities (Figure 2). These deletions define the core region of RTS1 that possesses wild-type levels of barrier activity.
The repeated motifs are essential for RTS1 function, each motif having an additive effect on barrier activity
To analyse the functional importance of the repeated motifs, they were sequentially deleted starting at the distal side. Quantification of the barrier activity of the truncated elements shows that deletion of each of the four motifs leads to a decrease in pausing and termination, suggesting that each individual motif contributes to overall RTS1 activity (Figure 2, mut3, mut4, mut5 and mut6), and deletion of all the motifs (Figure 2, mut6 and mut7) abolishes RTS1 activity, underlining that the motifs are essential for RTS1 function. Interestingly, quantification the effect the deletions have on the intensity of the pause signal shows that the sequential deletions of motifs 4, 3, 2 and 1 have dramatically different effects on the barrier; the relative drops are 8, 25, 59 and 9%, respectively (Figure 2C). There are two possible explanations; either each motif contributes unequally to the overall barrier activity or a cooperative interaction is occurring between the motifs (see below). Interestingly, in contrast to what was observed for the pause signals, quantification of the termination signals suggests that each motif has an additive effect on termination activity. We are unsure whether this difference is due to the uncertainty of the quantification of the different signals or whether it reflects a true difference in the mechanism of termination and pausing.
A single motif can act as a replication barrier
One interesting observation in the deletion analysis presented above is that the wild-type RTS1 replication barrier consists of several closely spaced barrier signals (Figure 2B, black arrows). The deletion of each motif leads to a decrease in the number of signals observed, and in mutant mut5 only one is left. This observation suggests that each of the repeated motifs can act as a barrier independently. To test this hypothesis, two synthetic DNAs of 37 and 58 bp, and containing the wild-type sequences of the first truncated motif and the fourth motif (Figure 1, alignment), were cloned and analysed as described above [Figure 3A (rep1) and B (rep4)]. Indeed, both of these short linkers can act as replication fork barriers, displaying a pause and termination activity of 5 and 9% (motif 1), and 16 and 23% (motif 4) of wild-type RTS1 levels, respectively. The differences in activity observed between the two motifs suggest that although the sequence of the truncated first motif is sufficient to act as a replication barrier, the flanking sequences present in motifs 2, 3 and 4 may play a role in optimizing barrier function.
Defining essential sequences within the repeated motifs
To define within the motifs, the sequences necessary for barrier function, a mutational analysis was performed. For this purpose, a linker of 10 base-substitutions was introduced in five different places in motif 4, rep4, scanning the motif along its full length (Figure 3B). Two of these linker substitutions totally abolish barrier activity (Figure 3B, rep4-mut3 and rep4-mut4), while one of the three other substitutions, rep4-mut2, causes a decrease in the pause and termination activity to 53 and 65%, respectively, of the wild-type rep4 motif (Figure 3B and C). Interestingly, an alignment of the four motifs (Figure 1, alignment) shows that the essential sequences display the highest degree of conservation and are similar to the S.pombe protein reb1 recognition site (Figure 1; see Discussion). The S.pombe reb1p protein is involved in transcription termination in the polymerase I-transcribed rDNA genes (Melekhovets et al., 1997; Zhao et al., 1997) and might act as a replication barrier at that locus (Sanchez et al., 1998). Finally, to verify the importance of this sequence, a 10 bp linker substitution was made in motif 1 at the reb1p-like binding sequence. Also here, such substitution leads to the abolishment of barrier activity (Figure 3A, rep1-mut1).
A purine-rich region acts cooperatively with the repeated motifs
The difference in barrier activity observed between mutant mut5 (Figure 2) and the linker containing motif 1 (Figure 3, rep1) suggests that the sequence just proximal to the repeated motifs plays a role in replication fork impediment. This 64 bp purine-rich region (Figure 4, region A) displays a 73% GA content. To analyse region A’s function, a deletion was introduced, removing the region from the RTS1 element (Figure 4A, mut8). Interestingly, this deletion leads to a dramatic drop (∼75%) in activity (Figure 4B and C) compared to that of the wild-type RTS1 and the core-mut1 elements (Figures 2B and 4C). Furthermore, when region A is analysed independently, no barrier signal is observed (Figure 2, mut6). Thus, region A cannot independently act as a barrier for replication forks. These data suggest that region A acts cooperatively with region B, containing the four repeated motifs, to increase the region’s overall activity. The subsequent sequential deletion of each of the motifs leads to a stepwise decrease in pausing and termination, highlighting the importance of the motifs for RTS1 function. However, although in this experiment there is a small increase in barrier activity when the half motif, motif 1, is deleted, the deletions of motifs 2, 3 and 4 cause relative drops in pause activities of 9, 8 and 11%, respectively (Figure 4C). This observation raises the possibility, in contrast to what was observed in the deletion analysis described above, in the absence of region A, each of the full-length region B motifs contributes in an additive manner to the overall barrier activity. The observation also suggests that in the presence of region A, cooperative interaction is occurring between the motifs and that this interaction depends on region B.
The trans-acting factor rtf2 acts through region A
To investigate whether trans-acting factors act specifically through region A, plasmids, containing both region A and region B (plasmid mut2), or only region B (plasmid mut13), were transformed into strains carrying mutations of trans-acting factors identified earlier as important for RTS1 function. The swi1-111 and swi3-146 mutations have previously been shown to abolish termination of replication at the RTS1 element (Dalgaard and Klar, 2000). In support of this, no barrier activity was observed at either of the modified RTS1 elements in these genetic backgrounds (Figure 5A). A similar lack of barrier activity was also observed in a Δrtf1 mutant (E.Sommariva, S.Mian and J.Z.Dalgaard, manuscript in preparation). However, analysis of the fourth trans-acting factor, rtf2, shows that this gene is not essential for replication termination, but increases the efficiency of the process both for plasmid and chromosomal wild-type RTS1 elements (Figure 5C; data not shown). Interestingly, similar levels of barrier activities were observed when in the Δrtf2 genetic background the barrier activity at the core element, containing both regions A and B, was compared with an element only containing region B (Figure 5B). The observation that there is no cumulative effect when combining the Δrtf2 and region-A deletions suggests that rtf2 acts through region A to mediate efficient termination of replication at RTS1.
Finally, an additional effect observed in the Δrtf2 genetic background is the appearance of new replication fork barriers (Figure 5A and C, gray arrows). These barriers are located symmetrically around the apex of the arc formed by the Y-shaped replication intermediates (see Discussion).
Overexpression of rtf1 can complement the rtf2 mutation
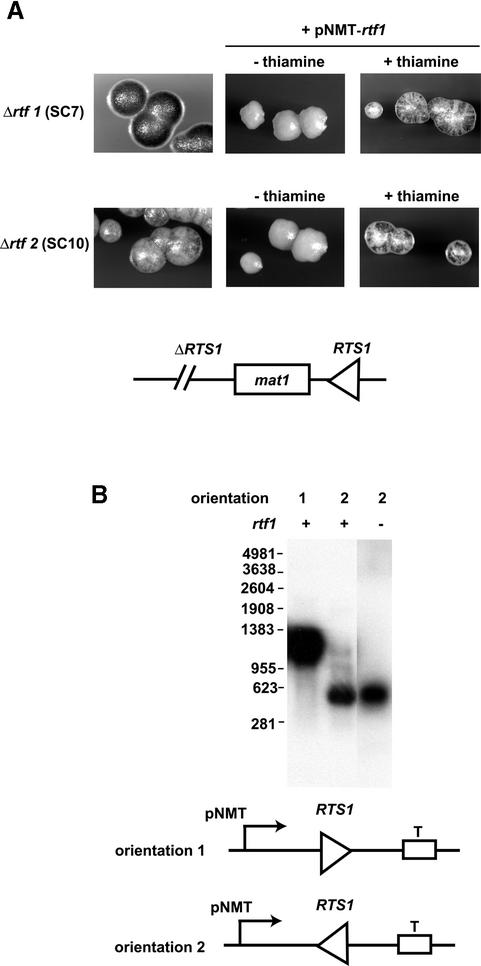
The observation that rtf2 is not essential for termination of replication made us test whether the overexpression of rtf1 could complement the rtf2 mutation. We therefore introduced a plasmid where the cDNA of rtf1 was cloned in front of the nmt-promoter. This promoter can be induced to high levels of expression by omitting thiamine from the media (Maundrell, 1990). The rtf1 overexpression plasmid was transformed into strains that carried either rtf1 or rtf2 deletion mutations. These strains also possess a rearranged mat1 locus where the RTS1 element has been transposed from the proximal side of mat1 to the SspI site at the distal side, where it had been inserted in the inverted orientation (Figure 6A, line drawing) (Dalgaard and Klar, 2001). Wild-type RTS1 activity will, at this genomic position, lead to the reversal of the direction of replication at the mat1 locus, and as a consequence inhibition of imprinting and mating-type switching. When the rtf1 or rtf2 mutations are introduced, they abolish or reduce RTS1 activity, allowing a medium level of imprinting and mating-type switching. Sporulation colonies of these strains stain brown with iodine vapour, since iodine stains starch produced in S.pombe spores. Thus, in these strains, complementation of the rtf1 and rtf2 trans-acting mutations can easily be quantified since restoration of the RTS1 function will result in non- or low-sporulating colonies that will stain yellow with iodine vapour. When the plasmid, containing rtf1 cDNA, was transformed into the Δrtf1 strain, we observed that the uninduced low level of rtf1p expression leads to complementation of the Δrtf1 mutation (Figure 6A, top right). The strain displayed a staining phenotype similar to the wild-type rtf1 strain. Overexpression of rtf1 completely inhibited sporulation (Figure 6A, top middle), suggesting that more efficient termination of replication is occurring at the RTS1 element.
Fig. 6. (A) Overexpression of rtf1p complements the rtf2 mutation. An overexpression plasmid containing the cDNA of rtf1 cloned in front of the pNMT promoter was transformed into Δrtf1 and Δrtf2 strains. These strains also carried a translocation of the RTS1 element from the proximal side of mat1 to the distal side (line drawing), allowing quantification of termination by its interference with mating-type switching and sporulation. Individual colonies were grown on sporulation media lacking leucine. Sporulation was visualized using iodine vapour staining, which stains starch present in spores in the colonies. (B) RTS1 acts as a transcription terminator in an rtf-independent manner. Northern blot analysis was performed on purified total RNA using a probe specific to RTS1. Transcription by the nmt-promoter was induced by omitting thiamine from the media of either wild-type or Δrtf1 strains, carrying either plasmid pBZ142 (lane 1) or plasmid pBZ143 (lanes 2 and 3). The molecular sizes of RNA markers used are given to the left of the panel.
When the rtf1p expression plasmid is transformed into the Δrtf2 strain, uninduced expression does not complement the speckled Δrtf2 phenotype (Figure 6A, bottom right). However, overexpression of rtf1p completely complements the Δrtf2 phenotype (Figure 6A, bottom middle). This suggests that in the presence of an excess of rtf1p, rtf2p is not required to achieve efficient replication barrier activity.
RTS1 acts as a terminator of transcription
Many features of the RTS1 element are similar to the mammalian rDNA barriers located in the RNA polymerase I transcription termination region. These regions contain repeated Sal-boxes, where the transcription factor ttf1 mediates transcription termination (Gerber et al., 1997). In mouse, however, at these Sal-boxes, TTF1 also acts as a barrier for replication forks entering the region in the direction opposite to that of transcription (Gerber et al., 1997; Lopez-Estrano et al., 1998) (see Discussion). We therefore tested whether the RTS1 element can act as a transcription terminator for polymerase II. We chose RNA polymerase II since a RNA polymerase II transcript originating from the mat1 locus has already been mapped to terminate in the RTS1 region (Figure 1) (Kelly et al., 1988). Plasmids pBZ143 and pBZ142 carry the RTS1, cloned in either orientation, between the nmt promoter and terminator (Figure 6B, line drawing). These plasmids were transformed into the strain JZ165, described above. Cells were grown in a media lacking thiamine, leading to the induction of transcription from the nmt promoter. Total RNA was purified from the strains and separated on a formaldehyde denaturing gel, next to a molecular marker, for subsequent northern blot analysis. In these plasmids, in the absence of transcription termination at the RTS1 element, transcripts will be ∼1250 bp in length due to termination at the nmt-transcription terminator. However, if transcription termination occurs at a position within the inserted 859 bp RTS1 element, it will lead to the formation of a transcript in the range of ∼200–1000 bp. Interestingly, while transcription, moving in the direction where replication forks are arrested, is allowed to pass unhindered through the RTS1 element (Figure 6B, lane 1), very efficient transcription termination occurs when the RTS1 element is transcribed in the opposite orientation (Figure 6B, lane 2). The observed transcript size of ∼600 bp suggests that transcription termination occurs within region B containing the repeated motifs. However, this transcription termination activity does not depend on the trans-acting factors rft1p (Figure 6B, lane 3) or rtf2p (not shown), as efficient transcription termination is observed in strains carrying deletion mutations of these genes.
Discussion
Here we start to address the molecular mechanism of replication termination at the polar site-specific terminator of DNA replication named RTS1. The RTS1 element was initially described as being located on a mat1-proximal, 859 bp EcoRI fragment, acting as a polar terminator of DNA replication. Although replication barriers have been observed at several other eukaryotic loci, the mechanism of site-specific replication termination remains unknown.
The data presented here reveal that the RTS1, in contrast to the bacterial elements, acts by a complex mechanism that involves at least two types of cis-acting regions, where several trans-acting factors interact to achieve efficient replication termination. Interestingly, some features of the RTS1 element are similar to the barriers observed in rDNA genes of mice and humans. At these elements the transcription termination factor TTF1 binds to repeated cis-acting Sal-boxes to mediate termination of transcription (Evers and Grummt, 1995). However, ttf1 also mediates replication termination at the Sal-boxes (Gerber et al., 1997; Lopez-Estrano et al., 1998). The S.pombe functional homologue of ttf1 is called reb1p (Melekhovets et al., 1997; Zhao et al., 1997). reb1p catalyses transcription termination of RNA polymerase I transcripts in the rDNA array. Interestingly, RTS1 contains an ∼60 bp repeated motif, with a subsequence that is similar to the rDNA binding sequence of reb1 (Figure 1, alignment). The presented RTS1 deletion analyses show that the four motifs are essential for function. Interestingly, the initial deletion analysis suggests that the motifs either contribute to a different degree to overall RTS1 barrier activity, or that the motifs cooperate to mediate their barrier function. Furthermore, a linker scanning mutagenesis of motifs 1 and 4 verified that the sequence similar to the reb1p binding sequence is essential for the motifs’ barrier function. Importantly, we know that rtf1p, a trans-acting factor identified in a genetic screen as involved in termination of replication at RTS1, is another S.pombe homologue of ttf1 (E.Sommariva, S.Mian and J.Z.Dalgaard, unpublished data). rtf1p displays 32% identity to reb1p and, as with reb1p and ttf1, contains a repeated domain similar to the myb DNA binding domain. Thus, rtf1p binding to the repeated motifs in region B is likely to be essential for RTS1 barrier function.
We also discovered that proximal to region B, another sequence is present that increases the overall barrier activity of RTS1 4-fold. This region (region A), shown in Figure 4, acts in trans over a distance to enhance the activity of region B containing the repeated motifs (Figures 2 and 5). Region A is ∼60 bp in length and consists of ∼75% guanines and adenines. This purine-rich sequence could play a similar role as the thymidine-rich sequence (please note that this element is described on the opposite strand), which enhances the activity of mammalian rDNA replication barriers by 30% (See Introduction) (Gerber et al., 1997). Interestingly, comparison of the contribution of each motif, in the presence and absence of region A, suggests that region A might act by mediating a functional cooperation between the motifs present in region B. One possible mechanism is through a trans-acting factor, or factors, that interact both with region A and the motifs in region B. Importantly, we show that the cooperative effect mediated by the RTS1 region A depends on the trans-acting factor rtf2. rtf2 is a member of an new family of proteins conserved from yeast to humans (J.Z.Dalgaard, unpublished data). Interestingly, the rtf2p protein might have an additional functional role in replication termination at RTS1: in the rtf2 mutant background, new barrier signals appear on the arc of replication intermediates (Figure 5A and C, blue arrows). The position of these new signals, in each case located symmetrically around the apex of the arc, suggests that, in the absence of rtf2p, additional sequences act as barriers for the replication forks or that rtf2p might be involved in establishing the polarity of the RTS1 element. In the latter case, the new barriers would be due to the replication fork moving in the opposite direction being paused or arrested at the repeated motifs.
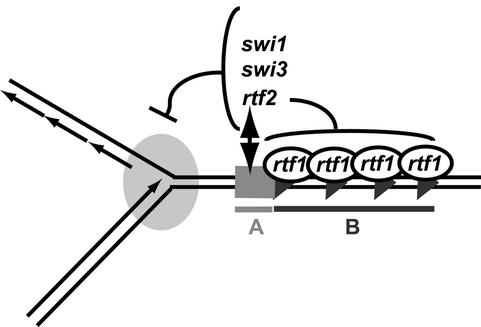
Interestingly, rtf2p is not essential for replication termination, as deletion of rtf2 only causes a decrease in RTS1 activity. Furthermore, the Δrtf2 allele can be complemented by overexpression of rtf1p. This suggests that rtf1p is a limiting factor in the termination process in the wild-type situation. Thus, rtf2p could play an important role in increasing barrier activity through its functional interaction with the region A (Figure 7).
Fig. 7. Model for the termination of replication at RTS1. A line drawing of a replication fork stalled at RTS1 is shown. Putative binding of rtf1p to the repeated motifs is illustrated. rtf2p functionally interacts with the purine-rich region A to cooperatively increase the activity of region B. Barrier activity also depends on the trans-acting factors swi1p and swi3p, which might act at the replication fork.
Several of the replication barriers described in the literature have been shown to act as barriers for the transcription apparatus. We therefore tested whether the RTS1 element can act as a transcription terminator. This is indeed the case, and similarly to the mammalian barriers, the transcription termination occurs in the opposite direction to that of replication termination. However, our analysis also showed that this transcription termination activity is independent of rtf1 and rtf2. Although our initial characterization of the plasmid used for this deletion analysis showed that nmt-promoter activity does not affect the activity of the wild-type RTS1 element, it is not known whether transcription is of importance in other situations.
Finally, the S.pombe RTS1 element acts to optimize the process of cellular differentiation by controlling the direction of replication at the mating-type locus mat1. Regulation of the direction of replication at specific genetic loci by site-specific replication termination may also play a key role in cellular differentiation in higher eukaryotes, and could involve factors and mechanisms similar to those described here.
Materials and methods
Two-dimensional gels
All two-dimensional gel procedures were performed as described by Brewer and Fangman (1987). Strains were grown in YEA supplemented with 4 µM thiamine to repress transcription from the plasmids’ nmt promoter. Cells were harvested in log phase and purified DNA (25 µg) was digested with restriction enzymes. Either PstI and SacI, or HpaI and SacI were used. HpaI and PstI cut at adjacent sites in pREB3. Replication intermediates were subsequently purified on benzoylated naphthoylated DEAE cellulose. The first and second dimension gels contained 0.5 and 1.2% agarose, respectively. Densitometry was carried out using NIH Image (Figures 2, 3 and 4) and Quantity One (Bio-Rad) (Figure 5). Refer to the main text for a description of the quantification method.
Northern blot analysis
RNA was isolated from a 10 ml overnight culture (Schmitt et al., 1990). Twenty to 30 µg were separated on a 1.2–1.5% agarose/formaldehyde gel. Five micrograms of Sigma RNA marker were used. Northerns were probed with a DNA probe specific to the RTS1 element.
Schizosaccharomyces pombe strains and plasmids
Strains were constructed using standard methods (Moreno et al., 1991) from the wild-type strain SP976 (SC8: h90, ade6-M216, leu1-32, Δrtf1::ura4+; SC11: h90, ade6-M216, leu1-32, Δrtf2::ura4+; SC99: h90, ade6-M216, leu1-32, Δrtf1::ura4+, ΔRTS1::ura4+). Other strains are described previously (Dalgaard and Klar, 1999, 2000, 2001). Plasmid RTS1 deletion mutants were constructed in pREP3 (Maundrell, 1993). The constructed plasmids contained the following nucleotides of the EcoRI fragment carrying RTS1: mut1, 173–859; mut2, 173–587; mut3, 173–527; mut4, 173–413; mut5, 1–290; mut6, 1–264/566–859; mut7, 1–168; mut8, 245–859; mut9, 291–859; mut10, 390–859; mut11, 494–859; mut12, 587–859; mut13, 245–859. For the construction of rtf1 expression plasmid, oligonucleotides Exp1 (5′-AAATGCGCAATGCAAGGGAAA AAACAATTTA-3′) and Tej5 (5′-GCATAAATCATCGGCGTTAGAA AAAG-3′) were used to amplify rtf1 cDNA using Pfu polymerase (Stratagene). The obtained PCR product was cloned into pNMT-TOPO1 (Invitrogen).
Acknowledgments
Acknowledgements
We thank our colleagues at MCRI for helpful suggestions and discussions. A special thank you to Natalie Mansfield, Trevor Eydmann, Elena Sommariva and Sonya Vengrova for technical assistance. This research was sponsored by the Association of International Cancer Research and Marie Curie Cancer Care (to J.Z.D.).
References
- Arcangioli B. (1998) A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J., 17, 4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B. and de Lahondes,R. (2000) Fission yeast switches mating type by a replication-recombination coupled process. EMBO J., 19, 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S.cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell, 55, 637–643. [DOI] [PubMed] [Google Scholar]
- Bussiere D.E. and Bastia,D. (1999) Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol., 31, 1611–1618. [DOI] [PubMed] [Google Scholar]
- Dalgaard J.Z. and Klar,A.J. (1999) Orientation of DNA replication establishes mating-type switching pattern in S.pombe. Nature, 400, 181–184. [DOI] [PubMed] [Google Scholar]
- Dalgaard J.Z. and Klar,A.J. (2000) swi1 and swi3 perform imprinting, pausing and termination of DNA replication in S.pombe. Cell, 102, 745–751. [DOI] [PubMed] [Google Scholar]
- Dalgaard J.Z. and Klar,A.J. (2001) A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S.pombe. Genes Dev., 15, 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A.M. and Newlon,C.S. (1996) DNA replication fork pause sites dependent on transcription. Science, 272, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Egel R. and Eie,B. (1987) Cell lineage asymmetry for Schizosaccharomyces pombe: Unilateral transmission of a high-frequency state of mating-type switching of a homothallic fission yeast. Curr. Genet., 12, 429–433. [Google Scholar]
- Evers R. and Grummt,I. (1995) Molecular coevolution of mammalian ribosomal gene terminator sequences and the transcription termination factor TTF-I. Proc. Natl Acad. Sci. USA, 92, 5827–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani L., Irene,C., Aragona,M. and Newlon,C.S. (2001) A DNA replication origin and a replication fork barrier used in vivo in the circular plasmid pKD1. Mol. Genet. Genomics, 266, 326–335. [DOI] [PubMed] [Google Scholar]
- Gerber J.K., Gogel,E., Berger,C., Wallisch,M., Muller,F., Grummt,I. and Grummt,F. (1997) Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell, 90, 559–567. [DOI] [PubMed] [Google Scholar]
- Greenfeder S.A. and Newlon,C.S. (1992) Replication forks pause at yeast centromeres. Mol. Cell Biol., 12, 4056–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P., Martin-Parras,L., Martinez-Robles,M.L. and Schvartzman,J.B. (1993) Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J., 12, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. (2000) Mechanisms and consequences of replication fork arrest. Biochimie, 82, 5–17. [DOI] [PubMed] [Google Scholar]
- Kelly M., Burke,J., Smith,M., Klar,A. and Beach,D. (1988) Four mating-type genes control sexual differentiation in the fission yeast. EMBO J., 7, 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri G.S., MacAllister,T., Sista,P.R. and Bastia,D. (1989) The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell, 59, 667–674. [DOI] [PubMed] [Google Scholar]
- Klar A.J. (1987) Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature, 326, 466–470. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Horiuchi,T. (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells, 1, 465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck,D.J., Nomura,M. and Horiuchi,T. (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev., 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbe M., Zabielski,J., Bernander,R. and Nordstrom,K. (1997) Inactivation of the replication-termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol. Microbiol., 24, 723–735. [DOI] [PubMed] [Google Scholar]
- Kuempel P.L., Duerr,S.A. and Seeley,N.R. (1977) Terminus region of the chromosome in Escherichia coli inhibits replication forks. Proc. Natl Acad. Sci. USA, 74, 3927–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.H., Kornberg,A., Hidaka,M., Kobayashi,T. and Horiuchi,T. (1989) Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl Acad. Sci. USA, 86, 9104–9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M.H. and Huberman,J.A. (1988) Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell Biol., 8, 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R.D., Platt,T.H. and Schildkraut,C.L. (1993) Initiation and termination of DNA replication in human rRNA genes. Mol. Cell Biol., 13, 6600–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Estrano C., Schvartzman,J.B., Krimer,D.B. and Hernandez P. (1998) Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J. Mol. Biol., 277, 249–256. [DOI] [PubMed] [Google Scholar]
- Maundrell K. (1990) nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem., 265, 10857–10864. [PubMed] [Google Scholar]
- Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- Melekhovets Y.F., Good,L., Elela,S.A. and Nazar,R.N. (1994) Intragenic processing in yeast rRNA is dependent on the 3′ external transcribed spacer. J. Mol. Biol., 239, 170–180. [DOI] [PubMed] [Google Scholar]
- Melekhovets Y.F., Shwed,P.S. and Nazar,R.N. (1997) In vivo analyses of RNA polymerase I termination in Schizosaccharomyces pombe. Nucleic Acids Res., 25, 5103–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K., Bussiere,D.E., Sahoo,T., Pai,K.S., Meijer,W.J., Bron,S. and Bastia,D. (2001) Structural and functional analysis of a bipolar replication terminus. Implications for the origin of polarity of fork arrest. J. Biol. Chem., 276, 13160–13168. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Olavarrieta L., Hernandez,P., Krimer,D.B. and Schvartzman,J.B. (2002) DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol., 322, 1–6. [DOI] [PubMed] [Google Scholar]
- Putter V. and Grummt,F. (2002) Transcription termination factor TTF-I exhibits contrahelicase activity during DNA replication. EMBO Rep., 3, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T., Mohanty,B.K., Lobert,M., Manna,A.C. and Bastia,D. (1995) The contrahelicase activities of the replication terminator proteins of Escherichia coli and Bacillus subtilis are helicase-specific and impede both helicase translocation and authentic DNA unwinding. J. Biol. Chem., 270, 29138–29144. [DOI] [PubMed] [Google Scholar]
- Sanchez J.A., Kim,S.M. and Huberman,J.A. (1998) Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res., 238, 220–230. [DOI] [PubMed] [Google Scholar]
- Santamaria D., Viguera,E., Martinez-Robles M.L., Hyrien,O., Hernandez,P., Krimer,D.B. and Swartzman,J.B. (2000) Bi-directional replication and random termination. Nucleic Acids Res., 28, 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M., Gomez,M. and Anteguera,F. (2002) Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol. Cell, 10, 907–916. [DOI] [PubMed] [Google Scholar]
- Shinomiya T. and Ina,S. (1993) DNA replication of histone gene repeats in Drosophila melanogaster tissue culture cells: multiple initiation sites and replication pause sites. Mol. Cell. Biol., 13, 4098–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier E.R. and Collins,R.A. (2000) The contributions of replication orientation, gene direction and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol., 50, 249–257. [DOI] [PubMed] [Google Scholar]
- Wallisch M., Kunkel,E., Hoehn,K. and Grummt,F. (2002) Ku antigen supports termination of mammalian rDNA replication by transcription termination factor TTF-I. Biol. Chem., 383, 765–771. [DOI] [PubMed] [Google Scholar]
- Ward T.R., Hoang,M.L., Prusty,R., Lau,C.K., Keil,R.L., Fangman,W.L. and Brewer,B.J. (2000) Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol. Cell. Biol., 20, 4948–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A.S. and Wake,R.G. (1984) Impediment to replication fork movement in the terminus region of the Bacillus subtilis chromosome. J. Mol. Biol., 179, 745–750. [DOI] [PubMed] [Google Scholar]
- Wiesendanger B., Lucchini,R., Koller,T. and Sogo,J.M. (1994) Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res., 22, 5038–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Macalpine,D.M. and Kapler,G.M. (1997) Developmental regulation of DNA replication: replication fork barriers and programmed gene amplification in Tetrahymena thermophila. Mol. Cell Biol., 17, 6147–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A., Guo,A., Liu,Z. and Pape,L. (1997) Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res., 25, 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Newlon,C.S. and Huberman,J.A. (1992) Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae.Mol. Cell Biol., 12, 4733–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]