Abstract
SNARE proteins participate in recognition and fusion of membranes. A SNARE complex consisting of vti1b, syntaxin 8, syntaxin 7, and endobrevin/VAMP-8 which is required for fusion of late endosomes in vitro has been identified recently. Here, we generated mice deficient in vti1b to study the function of this protein in vivo. vti1b-deficient mice had reduced amounts of syntaxin 8 due to degradation of the syntaxin 8 protein, while the amounts of syntaxin 7 and endobrevin did not change. These data indicate that vti1b is specifically required for the stability of a single SNARE partner. vti1b-deficient mice were viable and fertile. Most vti1b-deficient mice were indistinguishable from wild-type mice and did not display defects in transport to the lysosome. However, 20% of the vti1b-deficient mice were smaller. Lysosomal degradation of an endocytosed protein was slightly delayed in hepatocytes derived from these mice. Multivesicular bodies and autophagic vacuoles accumulated in hepatocytes of some smaller vti1b-deficient mice. This suggests that other SNAREs can compensate for the reduction in syntaxin 8 and for the loss of vti1b in most mice even though vti1b shows only 30% amino acid identity with its closest relative.
Lysosomes are acidic organelles containing hydrolytic enzymes which function in the degradation of proteins, lipids, polysaccharides, and DNA. Soluble lysosomal enzymes are imported into the endoplasmic reticulum during biosynthesis, bind to mannose-6-phosphate receptors in the trans-Golgi network (TGN) via mannose-6-phosphate residues as sorting signals, and traffic to endosomes. The mannose-6-phosphate receptors are recycled back to the TGN, while their cargo reaches the lysosome (23, 30). Newly synthesized lysosomal membrane proteins can traffic by different routes (20). LAMP-1 and LIMP-1 travel via the TGN and endosomes directly to lysosomes. By contrast, LAP (lysosomal acid phosphatase) moves from the TGN to the plasma membrane, is endocytosed into early endosomes, and recycles for multiple rounds before delivery to late endosomes and lysosomes. Endocytosis of proteins can occur as fluid phase uptake or by using specific receptors in receptor-mediated endocytosis via clathrin-coated vesicles (37). After decoating, these vesicles fuse with early endosomes, which serve as a sorting organelle. Traffic can be directed either back to the plasma membrane, in some cases via a recycling endosome, or forward to the late endosome. Membrane proteins targeted for lysosomal degradation are sorted into internal vesicles in early endosomes and late endosomes, forming multivesicular bodies (32). These internal vesicles are degraded after fusion of the outer membrane with lysosomes. Lysosomes also fuse with autophagic vacuoles. Autophagocytosis is a basic mechanism to turn over cytosol and organelles which is stimulated under starvation conditions (22). Cytosol and organelles are engulfed by double membranes to form autophagosomes or early autophagic vacuoles (Avi). They mature to late autophagic vacuoles (Avd) through fusion with late endosomes or lysosomes, rendering their interiors acidic and degradative (12).
These trafficking events require budding of transport vesicles and fusion between membranes. SNARE proteins on both membranes are integral parts of the machinery required for recognition and fusion between membranes (11, 21). These SNAREs form complexes bridging the gap between the membranes. SNAREs are conserved in evolution and posses a common domain structure. Most SNAREs contain a C-terminal transmembrane domain. The conserved SNARE motif consists of 60 amino acid residues and is sufficient for SNARE complex formation. Four different SNARE motifs form an extended four-helix bundle (3, 40). Most amino acid side chains pointing into the interior of the bundle are hydrophobic. In the middle of the SNARE motif, however, the side chains of one arginine and three glutamine residues interact in a plane perpendicular to the axis of the helical bundle which is called the 0 layer. SNAREs can be subdivided into R-, Qa-, Qb-, and Qc-SNAREs according to 0-layer residues and further sequence homologies (7).
We are interested in SNAREs involved in endosomal trafficking. In yeast, a single Qb-SNARE, Vti1p, is utilized throughout the endosomal system as part of four different SNARE complexes. Vti1p is required for traffic from the Golgi to the endosome, for traffic to the vacuole (lysosome), for retrograde traffic to the cis-Golgi, and for homotypic TGN fusion (9, 15, 17). While Caenorhabditis elegans has one ortholog, Arabidopsis thaliana, Drosophila, and mammals express two proteins related to yeast Vti1p. Mammalian vti1a and vti1b share only 30% of their amino acid residues with each other, as well as with yeast Vti1p (1, 16, 27). vti1a and vti1b have different but overlapping subcellular localizations (25). vti1a is found mostly in the Golgi and TGN. vti1b is localized predominantly to endosomes, vesicles, and tubules in the TGN area and the TGN. Both proteins are part of distinct SNARE complexes. vti1a is in a complex with the R-SNARE VAMP-4, syntaxin 16 (Qa), and syntaxin 6 (Qc) (25, 28). vti1b complexes with endobrevin/VAMP-8, syntaxin 7 (Qa), and syntaxin 8 (Qc). vti1a and vti1b function in distinct trafficking steps, as indicated by antibody inhibition experiments in vitro. Antibodies directed against vti1a inhibit transport of vesicular stomatitis virus G protein through the Golgi (49), fusion of early endosomes (4), and transport from early and recycling endosomes to the TGN (28). Antibodies directed against vti1b inhibit fusion of late endosomes (4).
In this study, we inactivated the mouse gene coding for vti1b by targeted disruption to investigate the role of vti1b in the whole organism. Absence of vti1b resulted in reduced protein levels of one SNARE partner, syntaxin 8, but levels of endobrevin and syntaxin 7 remained unchanged. vti1b-deficient mice did not suffer from serious defects. However, some vti1b-deficient mice were smaller than their littermates. Lysosomal degradation of an endocytosed protein was slightly delayed, and multivesicular bodies and autophagic vacuoles accumulated in hepatocytes of some of these smaller mice.
MATERIALS AND METHODS
Isolation of genomic DNA for vti1b and construction of targeting vector.
A vti1b cDNA expressed sequence tag clone (GenBank accession number AA105524; obtained from the American Type Culture Collection) containing the coding sequence for vti1b plus a 70-bp 5′ untranslated region and a 200-bp 3′ untranslated region was used to screen a lambda FIX II library with genomic DNA derived from mouse strain 129/SvJ (Stratagene, La Jolla, Calif.). A phage clone with a 15-kb insert containing exons 3 to 6 of vti1b was isolated. To generate the targeting construct, a 7-kb SpeI fragment with exon 4 in the center was subcloned into pBluescript, a SalI site was introduced by site-directed mutagenesis, and a neo expression cassette (derived from pMC1neopA [Stratagene]) was inserted into the SalI site, generating opposing open reading frames (Fig. 1B).
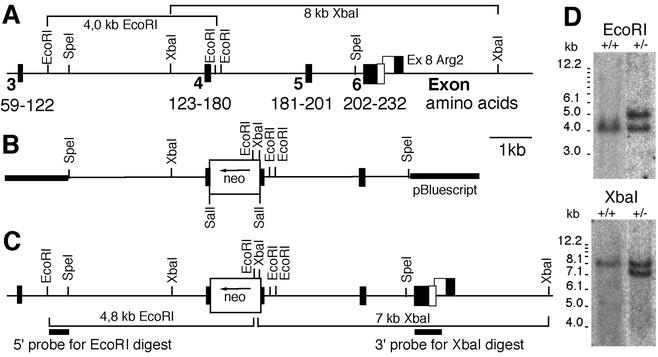
FIG. 1.
Targeted disruption of vti1b. (A) Genomic DNA for vti1b isolated from a phage library. According to information from the Mouse Genome Sequencing Consortium, exon 1 encodes 100 bp of 5′ untranslated region and amino acid residues 1 to 38 and exon 2 encodes amino acid residues 39 to 58. (B) Seven-kilobase targeting vector with exon 4 disrupted by insertion of the neomycin resistance cassette (neo). (C) vti1b locus after homologous recombination. Sections of DNA used as probes for Southern blot hybridizations are marked, and fragments detected after EcoRI and XbaI digestions are indicated for the targeted and wild-type (A) alleles. (D) Southern blots of ES cell DNA after EcoRI and XbaI digestions. An EcoRI fragment of 4.0 kb is detected in the wild-type (+/+) allele, and a 4.8-kb band is detected in the targeted allele. Insertion of the neo gene reduces an 8-kb wild-type XbaI fragment to 7 kb.
Targeted disruption of vti1b in mice.
The targeting construct was linearized with NotI and electroporated into the mouse 129/SvJ embryonic stem cell line ES 14-1. ES cell clones resistant to G418 were screened for homologous recombination by Southern blot analysis. A 5′ external probe and a 3′ external probe were used after EcoRI or XbaI digestion, respectively (Fig. 1A and C). Cells from two targeted clones (114 and 177) were microinjected into C57BL/6J blastocysts and implanted into pseudopregnant recipients. Chimeric animals were bred to C57BL/6J mice. Genotypes were determined by PCR using DNA isolated from tail biopsy specimens. A 375-bp fragment derived from the neo gene was amplified using the primers CGGATCAAGCGTATGCAGCCG and CAAGATGGATTGCACGCAGG. The primers CTCTTCTATGATTTCTGTACC and GAGGGATCCAATACCTTCTC were used to amplify a 520-bp fragment of the wild-type vti1b genomic region including exon 4 and a 1,620-bp fragment after neor insertion. All mice were housed in the animal care unit of the Department of Biochemistry II, Georg-August University of Goettingen, according to animal care guidelines.
Western blot analysis.
Tissue homogenates of liver, brain, kidney, and spleen were prepared by homogenization in lysis buffer (1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 5 mM iodoacetic acid in Tris-buffered saline [TBS] containing 150 mM NaCl and 50 mM Tris-HCl, pH 7.4) in an Ultra-Turrax homogenizer. Cultivated hepatocytes and mouse embryonic fibroblasts (MEFs) were scraped off in TBS with 0.1% Triton X-100 and sonicated for 3 s. One milligram of protein from liver homogenate was extracted with Triton X-114 (8), and the detergent fraction was concentrated by acetone precipitation to enrich membrane proteins and to facilitate detection of endobrevin. Twenty micrograms of total protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to immunoblotting using enhanced chemiluminiscence (Pierce, Rockford, Ill.). Polyclonal antisera directed against vti1a, vti1b, SNAP-29, syntaxin 7, syntaxin 8, and endobrevin were described previously (4, 6, 14).
Northern blotting.
Total RNA was isolated from brain and kidney using the RNeasy kit (Qiagen, Hilden, Germany). Ten micrograms of total RNA was separated on an agarose formaldehyde gel and transferred onto a Hybond-N membrane (Amersham, Braunschweig, Germany). cDNA probes were labeled with [α-32P]dCTP using the Megaprime DNA-labeling kit (Amersham). The membranes were hybridized with labeled vti1b cDNA in RapidHyb hybridization solution (Amersham) according to the manufacturer's instructions. Hybridization with a syntaxin 8 cDNA probe encoding the cytoplasmic domain (4) was performed in the following buffer: 48% (vol/vol) formamide, 125 mM NaCl, 12.5 mM trisodium citrate, 1% SDS, 10 mM Tris-HCl (pH 7.5), and 10% dextran sulfate in Denhardt's solution at 42°C.
Syntaxin 8 immunoprecipitation and stabilization in MEFs.
MEF lines were established from day 13.5 embryos by continuously passaging the cells (42). MEFs were plated onto 3-cm-diameter plastic culture dishes and kept in culture till they reached ∼50% confluency. To immunoprecipitate newly synthesized syntaxin 8, the cells were washed twice with phosphate-buffered saline (PBS) and incubated for 12 or 24 h with 200 μCi of [35S]methionine/ml in Dulbecco's modified Eagle medium without methionine and containing 5% dialyzed fetal calf serum. After being labeled, the cells were washed twice with PBS at 4°C, scraped off in 400 μl of TBS plus 0.1% Triton X-100 supplied with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 mM iodoacetic acid, 1 mM EDTA), sonicated, and centrifuged. The supernatants were preabsorbed with protein A-Sepharose and incubated with 4 μl of antiserum directed against syntaxin 8 at 4°C overnight and for an additional 2 h after the addition of 50 μl of protein A-Sepharose. The pellet was washed five times with lysis buffer, and the proteins were eluted with 60 μl of 2× stop buffer and separated by SDS-PAGE.
To inhibit lysosomal proteases, MEFs on 3-cm-diameter plastic culture dishes were incubated for 8 or 16 h in medium with or without 100 μM leupeptin and scraped off in 500 μl of PBS with 0.1% Triton X-100. Syntaxin 8 and vti1a protein levels were checked by immunoblotting.
Activities of lysosomal enzymes in tissue homogenates.
Homogenates of tissues (adjusted to 0.05% Triton X-100) were assayed for β-hexosaminidase, β-mannosidase, and β-glucuronidase using 4-methylumbelliferyl substrates (24). Arylsufatase A was measured using p-nitrocatechol sulfate as a substrate (34). The measurements were done in a fluorescence spectrophotometer at 365-nm excitation and 410-nm emission wavelength. The measured values were corrected with the help of 4-methylumbelliferone solution as a reference. The activities of enzymes in homogenates were calculated as milliunits per gram of tissue.
Preparation of primary mouse hepatocytes.
Mouse hepatocytes from 10- and 15-month-old vti1b-deficient and wild-type mice were prepared using a collagenase-independent method described previously (29). The cells were enriched using a Percoll gradient (58% [wt/vol]) and plated on gelatin-treated plastic culture dishes at 6.5 × 105 per 3-cm-diameter dish. The hepatocytes were cultured overnight in RPMI 1640 containing 10% fetal calf serum and penicillin-streptomycin (Gibco) before the experiments.
Uptake and degradation of 125I-asialofetuin in hepatocytes.
125I-asialofetuin degradation was measured as described previously (44). Asialofetuin was iodinated using IODO-Gen tubes (Pierce) according to the manufacturer's instructions. Hepatocytes were serum starved in RPMI with 0.1% bovine serum albumin for 4 h and incubated with 10 nM 125I-asialofetuin in RPMI with 0.1% bovine serum albumin (600 μl; 120,000 cpm) for 20 min. The plates were washed once and supplemented with new medium for the appropriate chase periods. At the end of each incubation, the medium was collected and the cells were scraped off in PBS plus 0.2% Triton X-100. The medium and the cell lysates were precipitated with 10% trichloroacetic acid (TCA) for 10 min on ice. TCA pellets were obtained by centrifugation and dissolved in 0.5 M NaOH. The soluble (degraded 125I-asialofetuin) and insoluble (intact proteins) radioactivities in the medium and cells were measured in a gamma counter. The rates of lysosomal degradation were calculated as percentages of soluble radioactivity in relation to the sum of precipitated cellular 125I-asialofetuin and soluble radioactivity for each time point.
Electron microscopy.
Ultrathin cryosections were prepared as described previously (43). Hepatocytes were fixed with 2% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M sodium phosphate, pH 7.4, for 30 min at room temperature directly on cell culture dishes. The cells were postfixed with 4% paraformaldehyde and 0.1% glutaraldehyde for 2 h on ice and removed from the dishes with a cell scraper. After being washed twice with PBS- 0.02% glycine, the cells were embedded in 10% gelatin, cooled on ice, and cut into small blocks. The blocks were infused with 2.3 M sucrose overnight and stored in liquid nitrogen. For immunolabeling, ultrathin sections were incubated with antibodies against LAMP-1 (1:30), followed by protein A-gold (10-nm diameter). Sections were contrasted with uranyl acetate-methyl cellulose for 10 min on ice, embedded in the same solution, and examined with a Phillips CM120 electron microscope.
The isolated hepatocytes were fixed in 2% glutaraldehyde in 0.2 M HEPES, pH 7.4, for 2 h. The cells were scraped off the culture dish, stored in 0.2 M HEPES, pH 7.4, postfixed in 1% OsO4 for 1 h, dehydrated in ethanol, and embedded in Epon. Autophagosome profiles were counted under the microscope. The cell area was estimated by point counting, using negatives taken at ×400 magnification. Three separate grid openings were counted from each sample.
RESULTS
Targeted disruption of vti1b.
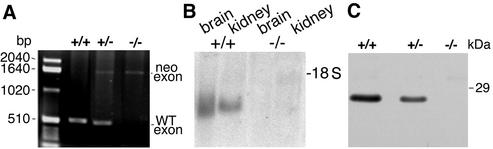
Genomic DNA for vti1b was isolated from a phage library using a full-length vti1b expressed sequence tag clone as a probe. A phage was isolated with an insert of 15 kb containing exons 3 to 6 of vti1b encoding amino acid residues 59 to 122, 123 to 180, and 202 to 232, plus the 3′ untranslated region, respectively (Fig. 1A). The 3′ untranslated region of vti1b overlapped with the 3′ untranslated region of arginase 2 on the opposite strand. Exon 4 encodes most of the SNARE motif, which is crucial for SNARE function. Therefore, exon 4 was interrupted by insertion of a neomycin resistance gene (Fig. 1B). This targeting construct was electroporated into the mouse embryonic stem cell line ES 14-1. G418-resistant clones were tested by Southern blotting (the strategy is shown in Fig. 1C). Clones with homologous recombination were identified by a 4.8-kb band after EcoRI digestion with a 5′ probe and by a 7-kb band after XbaI digestion with a 3′ probe, in addition to the wild-type bands of 4.0 and 8 kb, respectively (Fig. 1D). Seven out of 96 clones tested carried the targeted allele in the vti1b locus. These ES cells were injected into blastocysts, and chimeric males were crossed with C57BL/6 females; heterozygous (vti1b+/−) animals were crossed, and their offspring were genotyped using a PCR strategy (Fig. 2A). vti1b-deficient (vti1b−/−) mice were found with almost Mendelian inheritance (20.8% of 245 mice). These data indicate that vti1b-deficient mice do not suffer from reduced viability. vti1b mRNA was undetectable in the brains and kidneys of vti1b-deficient mice (Fig. 2B). The vti1b protein was not found in the tissues of vti1b-deficient mice, and reduced levels were detected in vti1b+/− animals (Fig. 2C).
FIG. 2.
vti1b mRNA and protein were absent in animals homozygous for the targeted vti1b allele. (A) Genotyping of genomic DNA using PCR. A band of 500 bp including exon 4 of vti1b was amplified from the wild-type (WT) allele. After insertion of the neo gene, the fragment shifts to 1,600 bp. In addition, the neo gene was amplified using specific primers (not shown). (B) Northern blot for vti1b mRNA. RNA was isolated from the brains and kidneys of wild-type (+/+) and vti1b-deficient (−/−) animals, separated by agarose gel electrophoresis, blotted, and probed for vti1b. (C) Western blot for vti1b protein. Protein extracts from wild-type, heterozygous (+/−), and vti1b-deficient embryonic fibroblasts were separated by SDS-PAGE and stained with an antiserum directed against vti1b. The vti1b protein was not detected in vti1b−/− cells. Less vti1b was present in heterozygous than in wild-type cells.
Loss of syntaxin 8 protein.
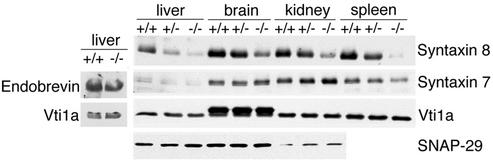
The absence of vti1b may affect other SNAREs in endosomal membrane trafficking. Different tissues were homogenized, and proteins were separated by SDS-PAGE and analyzed for levels of different SNAREs by immunoblotting. Homogenates of liver, brain, kidney, and spleen derived from vti1b-deficient mice contained much less syntaxin 8 than homogenates derived from wild-type mice (Fig. 3). Syntaxin 8 protein levels were already reduced in vti1b+/− mice, most dramatically in the liver. Syntaxin 8 forms a SNARE complex with vti1b, syntaxin 7, and endobrevin (5). However, levels of syntaxin 7 and endobrevin remained unaffected in vti1b-deficient tissues. The amounts of vti1a and SNAP-29 were also not significantly changed. These two SNAREs are not found in a complex with vti1b.
FIG. 3.
Syntaxin 8 protein levels are reduced in vti1b-deficient mice. Homogenates were prepared from livers, brains, kidneys, and spleens of wild-type (+/+), heterozygous (+/−), and vti1b-deficient (−/−) animals; separated by SDS-PAGE; and stained with antisera directed against the indicated SNAREs. Liver homogenates from wild-type and vti1b-deficient animals were extracted with Triton X-114, and the detergent phase was separated by SDS-PAGE to allow the detection of endobrevin. No reduction in protein levels was observed for endobrevin, syntaxin 7, vti1a, and SNAP-29.
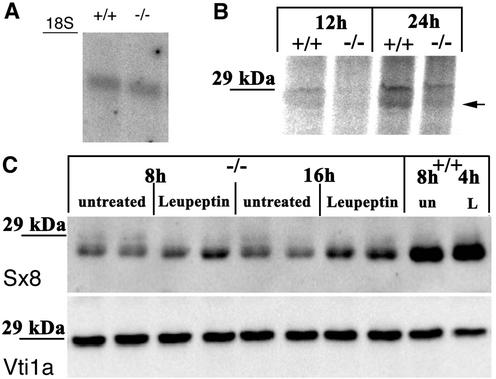
A reduction in syntaxin 8 protein levels could be due to different processes influencing either the mRNA or the protein levels. Northern blots were prepared to investigate whether transcription or mRNA stability was affected (Fig. 4A). Similar amounts of syntaxin 8 mRNA were detected in the kidneys of wild-type and vti1b-deficient animals.
FIG. 4.
Syntaxin 8 mRNA was stable in vti1b-deficient mice, but the syntaxin 8 protein was degraded in lysosomes. (A) Similar levels of syntaxin 8 mRNA were detected in wild-type and vti1b-deficient mice. RNA was isolated from the kidneys of wild-type (+/+) and vti1b-deficient (−/−) animals, separated by agarose gel electrophoresis, blotted, and probed for syntaxin 8. (B) Syntaxin 8 protein was slowly synthesized in wild-type embryonic fibroblasts. Wild-type (+/+) and vti1b-deficient (−/−) embryonic fibroblasts were incubated with [35S]methionine for 12 or 24 h. Syntaxin 8 was immunoprecipitated, and the fractions were analyzed by SDS-PAGE and autoradiography. Even though syntaxin 8 protein could be precipitated, incorporation of radioactivity was detected only after 24 h (arrow), indicating that the protein is very stable. The upper band at 29 kDa was nonspecific. (C) Treatment with the lysosomal protease inhibitor leupeptin stabilized syntaxin 8 protein in vti1b-deficient fibroblasts. vti1b-deficient (−/−) and wild-type (+/+) embryonic fibroblasts were treated with 100 μM leupeptin as indicated. Extracts containing equal amounts of protein were analyzed by immunoblotting, using antiserum directed against syntaxin 8 or vti1a as a loading control. un, untreated; L, leupeptin.
Thus, the synthesis or stability of syntaxin 8 protein was reduced in vti1b-deficient cells. To monitor the synthesis of syntaxin 8, wild-type (vti1b+/+) and vti1b-deficient (vti1b−/−) embryonic fibroblasts were incubated with [35S]methionine for different periods of time. Syntaxin 8 was immunoprecipitated from cell extracts. Sufficient incorporation of [35S]methionine into newly synthesized syntaxin 8 required 24 h in wild-type cells (Fig. 4B). This was not due to problems with the immunoprecipitation or labeling, as syntaxin 8 contains four methionine residues. These data indicate that syntaxin 8 is a stable protein with a low synthesis rate. [35S]methionine-labeled syntaxin 8 was not detected in vti1b-deficient cells after 24 h. However, due to the long labeling periods required, it cannot be determined whether the lack of syntaxin 8 was due to reduced synthesis or increased degradation. To circumvent this problem, protein degradation was inhibited using protease inhibitors blocking protein degradation either in the lysosome or by the proteasome. Lysosomal proteases were inhibited in embryonic fibroblasts by the addition of leupeptin. More syntaxin 8 was detected by immunoblotting in vti1b-deficient cells after incubation with leupeptin for 16 h than in untreated vti1b−/− cells (Fig. 4C). However syntaxin 8 protein levels remained much lower than in wild-type cells. The amount of vti1a was not affected by leupeptin treatment, excluding a more general effect. The proteasomal inhibitor lactacystin did not stabilize syntaxin 8 in vti1b-deficient cells (data not shown). This small stabilization of syntaxin 8 by leupeptin indicates that syntaxin 8 was degraded in lysosomes in vti1b-deficient cells.
Heterogeneity of phenotypes among vti1b-deficient mice.
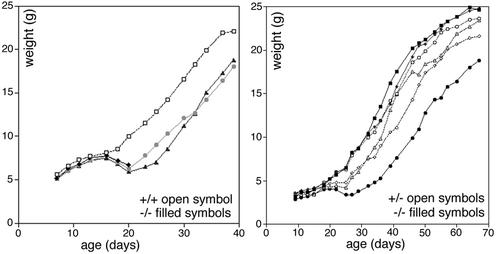
We noticed that some vti1b-deficient mice were smaller than their littermates. The weights of mice from the same litter and of the same sex were compared (Fig. 5). All mice gained weight at similar rates until days 16 to 18. Around this time, pups start to feed on a solid diet. Afterward, some vti1b-deficient mice lost weight and remained lighter than their littermates. A few small mice even died at an age between 3 and 5 weeks (Fig. 5, left). Twenty-two percent (33 out of 149) of the vti1b-deficient mice were considerably smaller than their littermates, two-thirds of them males (21). These mice had considerably less fat tissue than other mice of the same age.
FIG. 5.
vti1b-deficient mice showed a heterogeneous growth phenotype. Male mice from the same litter were compared. All of the mice gained weight at similar rates until day 16. Afterward, a few died (left, solid diamonds), and ∼20% of the vti1b-deficient mice lost weight and remained smaller than their littermates (left, shaded circles and solid triangles; right, solid circles). Most vti1b-deficient mice (right, solid squares and diamonds) grew at rates similar to those of their heterozygous (right, open symbols) or wild-type (left, open symbols) littermates.
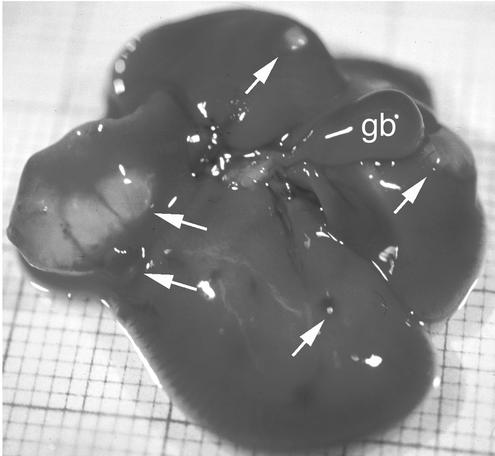
The morphologies of different tissues were investigated to find a basis for these differences. Paraffin sections were obtained and stained with hematoxylin and eosin or with periodic acid-Schiff's reagent (PAS) to detect accumulation of mucopolysaccharides or glycogen. Little PAS staining was observed in the livers of wild-type and normal-size vti1b-deficient mice. Large amounts of PAS-positive globules were found in hepatocytes of two small vti1b-deficient mice (data not shown). This material was identified as glycogen, as it was removed by pretreatment with diastase. Morphological changes were not observed in the other tissues investigated (data not shown). In 3 out of 16 smaller vti1b-deficient mice, the gall bladder was enlarged. Gall stones were found in one vti1b−/− mouse. Multiple cysts were present in the livers of 8 out of 23 normal-size vti1b-deficient mice older than 15 months (Fig. 6). Liver cysts are not among the spontaneous diseases reported for mouse strains C57BL or 129 (http://www.informatics.jax.org/external/festing/search_form.cgi).
FIG. 6.
Liver cysts in aged normal-size vti1b-deficient mice. Eight out of 23 vti1b-deficient mice between 15 and 21 months old had multiple liver cysts (arrows). These cysts were filled with a clear fluid, except for two small cysts containing a yellow liquid. gb, gall bladder.
Lysosomal transport in vti1b-deficient hepatocytes.
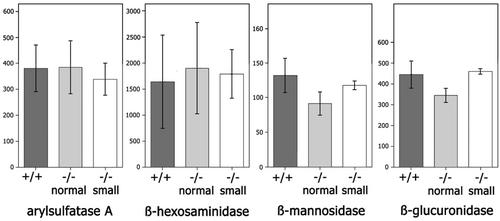
We wanted to determine whether the morphological changes observed were connected with changes in lysosomal function. Therefore, several experiments were performed using liver homogenates and isolated hepatocytes. At first, steady-state concentrations of lysosomal proteins were analyzed. Activities of soluble lysosomal hydrolases were determined in liver extracts derived from wild-type mice, as well as low- and normal-weight vti1b-deficient mice. No significant differences between the activities of arylsulfatase A, β-hexosaminidase, β-mannosidase, and β-glucuronidase of wild-type and vti1b-deficient mice of normal and small size were found (Fig. 7). These data demonstrate that loss of vti1b did not affect steady-state levels of lysosomal enzymes.
FIG. 7.
Levels of lysosomal hydrolases were not affected by deletion of vti1b. The activities (in milliunits per gram of tissue) of the lysosomal hydrolases arylsulfatase A, β-hexosaminidase, β-mannosidase, and β-glucuronidase were determined in liver homogenates derived from wild-type (+/+; n = 5), normal-size (n = 3), and small (n = 2) vti1b-deficient (−/−) mice. The error bars indicate standard deviations.
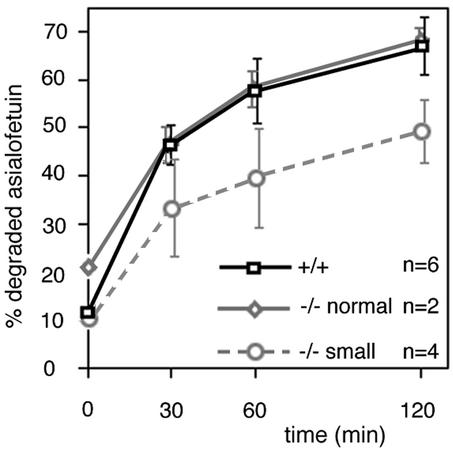
However, a subtle change in transport kinetics cannot be detected in this way. Transport kinetics were investigated using cultured primary hepatocytes derived from these mice. The processing of pulse-chase-labeled cathepsin D from the Golgi proform to the intermediate form found in endosomes and lysosomes (33) was measured to monitor biosynthetic transport from the Golgi to endosomes and lysosomes. Cathepsin D processing occurred with similar kinetics in wild-type and vti1b-deficient hepatocytes (data not shown). Lysosomes receive material via endocytosis, in addition to traffic from the Golgi. Asialofetuin was used to follow receptor-mediated endocytosis and lysosomal degradation. Asialofetuin binds to the asialoglycoprotein receptor, which is found exclusively in hepatic parenchymal cells. It recognizes carbohydrate moieties with a terminal galactose. After internalization of the complex, the empty receptor is recycled back to the cell surface. The ligand is transported to lysosomes for degradation. 125I-asialofetuin was used to quantify endocytosis and degradation rates. Wild-type hepatocytes and hepatocytes derived from vti1b-deficient mice of normal size degraded asialofetuin at similar rates (Fig. 8). Wild-type hepatocytes degraded 66.7% ± 6.3% of the endocytosed 125I-asialofetuin after a chase period of 120 min; hepatocytes from normal-size vti1b-deficient mice degraded 68.4% ± 0.5%. By contrast, less 125I-asialofetuin was degraded by hepatocytes derived from small vti1b-deficient mice after all chase periods, for example, 49.4% ± 4.3% after a 120-min chase. These data indicate that hepatocytes derived from small mice had a slight delay in lysosomal delivery and degradation of an endocytosed protein.
FIG. 8.
Lysosomal degradation of endocytosed 125I-asialofetuin was slower in hepatocytes derived from small vti1b-deficient mice. No difference in degradation of 125I-asialofetuin was observed in hepatocytes from wild-type (+/+) and vti1b-deficient (−/−) mice of normal weight. Hepatocytes derived from wild-type or vti1b-deficient mice of normal or smaller size were cultured for 1 day. They were incubated with 125I-asialofetuin for 20 min at 37°C and chased for the indicated periods after removal of the radioactive medium. Degraded (soluble in TCA) and intact 125I-asialofetuins were separated by TCA precipitation of the medium and cells and quantified. About 8,000 to 18,000 cpm was endocytosed by the hepatocytes, depending on cell density but independent of the genotype. Three individual experiments were performed with double values. The error bars indicate standard deviations.
Lysosomal function is also required for degradation of proteins via autophagocytosis (22). Cytosol and organelles are wrapped by double membranes, which form autophagosomes. The outer membranes of these autophagosomes fuse with lysosomal membranes, and the inner membranes and the contents are degraded. Degradation of long-lived proteins occurs mainly via autophagy in hepatocytes (38). Long-lived proteins were degraded at wild-type rates in vti1b-deficient hepatocytes, suggesting that autophagic protein degradation was not impaired (data not shown).
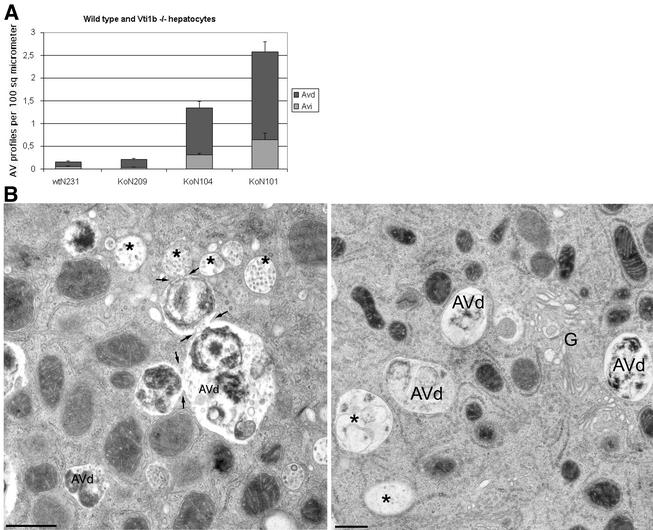
Hepatocytes from a wild-type mouse and three smaller vti1b-deficient mice were used to quantify the accumulation of autophagic vacuoles on ultrathin Epon sections. In hepatocytes derived from two of the small vti1b−/− mice, a strong accumulation of both early (Avi) and late (Avd) autophagic vacuoles was observed (Fig. 9A). An increased proportion of these autophagic vacuoles were connected to each other, suggesting that they might have been caught in the process of fusion (Fig. 9B). In addition, more multivesicular bodies were present. However, autophagic vacuoles did not accumulate in hepatocytes from the third small vti1b-deficient mouse, further emphasizing the variability in phenotypes.
FIG. 9.
Hepatocytes of some smaller vti1b-deficient mice accumulated autophagic vacuoles and multivesicular bodies. (A) The numbers of early (Avi) and late (Avd) autophagic vacuole profiles per 100 μm2 of cell area on thin sections were determined for hepatocytes isolated from wild-type (wt) and three different smaller vti1b-deficient (Ko) mice. The error bars represent standard errors of the mean. (B) Fusion profiles of autophagic vacuoles (arrows), as well as multivesicular bodies (∗), were more prominent in hepatocytes from vti1b−/− mouse 101 (left) than in wild-type (right) hepatocytes. G, Golgi. Bar, 500 nm.
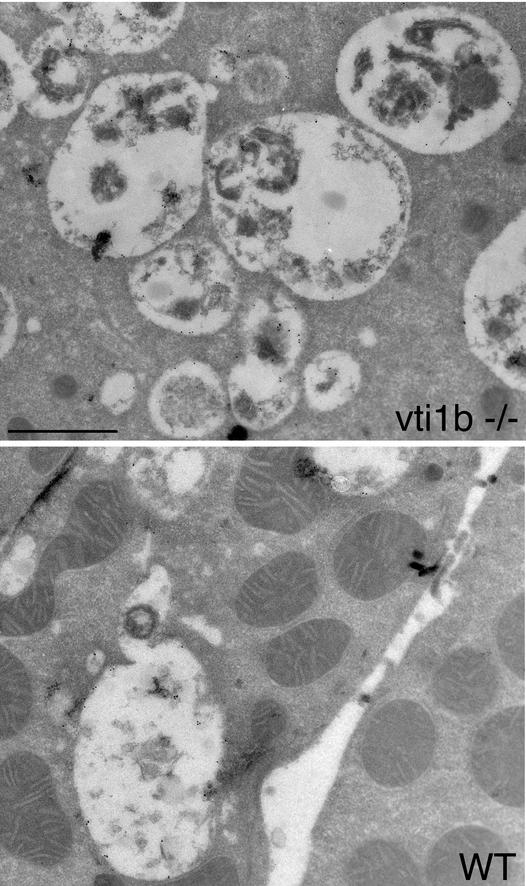
Immunoelectron microscopy was used to investigate the morphologies of late endosomes and lysosomes in hepatocytes derived from wild-type and the smaller vti1b-deficient mice. Ultrathin cryosections were stained with antibodies specific for LAMP-1, followed by detection with 10-nm-diameter protein A-gold (Fig. 10). LAMP-1 is a lysosomal membrane protein which is also present in late endosomes and autophagic vacuoles. The overall morphologies of LAMP-1-stained structures were similar in wild-type and vti1b-deficient hepatocytes. However, the vti1b−/− cells seemed to contain more LAMP-1-positive organelles, which were sometimes clustered or appeared to be in the process of fusion.
FIG.10.
LAMP-1-positive structures had similar appearances in wild-type (WT) and vti1b-deficient hepatocytes. Ultrathin cryosections were obtained from hepatocytes of wild-type and smaller vti1b-deficient mice and were decorated with antibodies specific for LAMP-1 and 10-nm-diameter protein A-gold. LAMP-1 is found in lysosomes, late endosomes, and late autophagic vacuoles. vti1b−/− cells seemed to contain more LAMP-1-positive organelles, which were sometimes clustered. Bar, 1 μm.
In summary, the absence of vti1b did not result in lysosomal defects in most mice. In some smaller mice, lysosomal degradation was delayed and autophagic vacuoles, as well as multivesicular bodies, accumulated.
DISCUSSION
Loss of syntaxin 8 protein.
The syntaxin 8 protein was found in reduced amounts in different tissues and embryonic fibroblasts obtained from vti1b-deficient mice, despite the presence of mRNA encoding syntaxin 8. Syntaxin 8 was degraded in lysosomes because incubation with the lysosomal protease inhibitor leupeptin stabilized the protein somewhat. The proteasomal inhibitor lactacystin did not stabilize syntaxin 8 within 16 h. However, we cannot exclude the possibility that part of syntaxin 8 is degraded via proteasomes, because apoptosis may have been induced under these conditions (10). Syntaxin 8 forms a SNARE complex with vti1b, endobrevin/VAMP-8, and syntaxin 7 (4). The amounts of endobrevin and syntaxin 7 were not reduced in vti1b-deficient tissues. These data indicate that vti1b is specifically required for the stability of syntaxin 8. vti1b may control the localization of syntaxin 8; the subcellular distribution of syntaxin 8 is similar to that of vti1b (25). In PC12 cells syntaxin 8 is predominantly localized to the TGN, vesicular structures, late endosomes, and lysosomes (35). Syntaxin 8 may be stable on lysosomal membranes, because the luminal domain consists of only three amino acid residues. Syntaxin 8 is found on the limiting membrane but not on internal vesicles of multivesicular bodies which are destined for lysosomal degradation (32). Syntaxin 8 may be mislocalized to these internal vesicles in the absence of vti1b. Alternatively, syntaxin 8 may be misfolded in the absence of vti1b, resulting in reduced stability. In both cases, extensive interactions must occur between vti1b and syntaxin 8. Interestingly, vti1b and syntaxin 8 take the position of the N-terminal and C-terminal SNARE motifs of SNAP-25 in the SNARE complex (3). In vitro SNARE complex assembly also points to a tight connection between vti1b and syntaxin 8. Recombinant SNARE motifs of neuronal and late-endosomal SNAREs were mixed and tested for the ability to form stable complexes (4). vti1b, together with syntaxin 8, can substitute for both SNARE motifs of SNAP-25, and vice versa. By contrast, exchange of just one of these helices does not result in stable complexes. This indicates that the Qb- and Qc-SNARE helices can be provided either by a single protein, like SNAP-25, or by two proteins with close functional connections.
Defects and phenotypic variability in vti1b-deficient mice.
Twenty percent of the vti1b-deficient mice had lower body weights than their littermates, especially around the time of weaning, at an age of 3 to 5 weeks. The mortality of the smaller mice increased during this period. Anatomical abnormalities were detected in the liver and gall bladder. Eighteen percent of smaller vti1b−/− mice had enlarged gall bladders. Multiple liver cysts were found in a third of normal-size vti1b−/− mice older than 15 months. Therefore, hepatocytes were used to study lysosomal function and morphology. Lysosomal degradation of an endocytosed protein, asialofetuin, was slightly delayed in hepatocytes isolated from smaller vti1b-deficient mice but was not affected in hepatocytes derived from normal-size vti1b-deficient mice. These data fit with electron microscopical analysis of hepatocytes. Hepatocytes derived from normal-size vti1b-deficient mice were indistinguishable from wild-type hepatocytes. An accumulation of multivesicular endosomes and autophagic vacuoles was observed in hepatocytes isolated from some of the smaller vti1b-deficient mice, but this finding was not consistent. In addition, more multivesicular endosomes and autophagic vacuoles were observed in close contact with each other or seemed to be in the process of fusion. These data indicate that fusion of these organelles may be slowed down. However, the effect was not strong enough to severely impact lysosomal function. Mice lacking the lysosomal membrane protein LAMP-2 accumulate autophagic vacuoles in hepatocytes (41) due to the prolonged half-life of early and late autophagic vacuoles (13). However, they do not accumulate multivesicular endosomes or fusion profiles, indicating that LAMP-2 and vti1b function in distinct molecular steps.
The phenotypic variability of vti1b-deficient mice may be due to the mixed genetic background of these mice. The embryonic stem cells were derived from mouse strain 129/SvJ. Chimeric animals were crossed with C57BL/6J mice. The offspring of this cross were bred further to obtain the animals used in this study. Variations in phenotype are also observed in other knockout mice. About 50% of mice deficient for the lysosomal membrane protein LAMP-2 die between postnatal days 20 and 40, while the rest are long-term survivors (41).
Compensation for vti1b deficiency.
The mild phenotypes suggest that the loss of vti1b is compensated for. As vti1b is a Qb-SNARE, another Qb-SNARE is needed to replace it. The best candidate is vti1a, because it is the closest relative of vti1b, even though the proteins share only 30% of their amino acid residues.
Mice deficient for the R-SNARE VAMP-3/cellubrevin are the only examples of compensation for the loss of a SNARE (51). VAMP-2/synaptobrevin 2 probably substitutes for VAMP-3, as the two proteins share 74% of their amino acid residues, differ only in a single residue in the SNARE motif, and are coexpressed in nonneuronal tissues (36). By contrast, the loss of syntaxin 4 results in embryonic lethality (50), even though syntaxin 2 and syntaxin 3 share 44 and 41% of their amino acid residues, respectively, with syntaxin 4. These three proteins are coexpressed and are predominantly present on the plasma membrane but differ in their distributions between apical and basolateral domains in polarized cells (26).
According to sequencing projects, the only additional known mammalian Qb-SNAREs with a single SNARE motif are membrin and GOS-28, which are involved in endoplasmic reticulum-to-Golgi and intra-Golgi traffic (18, 31). They are unlikely candidates for replacing vti1b because of their subcellular localization and low sequence homology.
The compensatory protein did not stabilize the Qc-SNARE syntaxin 8, since amounts of syntaxin 8 were strongly reduced in all vti1b-deficient mice. vti1a forms a SNARE complex including the Qc-SNARE syntaxin 6. As the closest relative, syntaxin 6 may compensate for the loss of syntaxin 8, since both proteins occupy the same position in a SNARE complex. However, syntaxin 6 and syntaxin 8 share only 23% of their amino acid residues (39). A candidate for substituting for both vti1b and syntaxin 8 is SNAP-29, which contains a Qb- and a Qc-SNARE motif. SNAP-29 has been localized to the Golgi and binds several syntaxins, preferentially syntaxin 6 (39, 47, 48), but its function is not known. A complex containing endobrevin, syntaxin 6, and SNAP-29 was immunoprecipitated upon overexpression of all components (19). This complex contains an R-SNARE, a Qb-SNARE, and two Qc-SNARE motifs, but no Qa-SNARE, distinguishing it from all well-characterized SNARE complexes found in vivo under endogenous expression levels.
Another reason for the lack of phenotype in vti1b-deficient mice could be the existence of a redundant SNARE complex. The R-SNARE VAMP-7 functions in fusion with the lysosome (2, 46). VAMP-7 forms a complex with syntaxin 7 (45). The Qb- and Qc-SNAREs of this complex are not known, but vti1b plus syntaxin 8 or SNAP-29 is the best candidate among the known SNAREs.
In summary, three different compensatory mechanisms are most likely: the presence of a distinct SNARE complex containing VAMP-7 or replacement of vti1b and syntaxin 8 in an endobrevin- and syntaxin 7-containing SNARE complex either by vti1a and syntaxin 6 or by SNAP-29.
Acknowledgments
We thank Paul Saftig (Universität Kiel) for invaluable help with generating targeted embryonic stem cells and isolation of primary mouse hepatocytes, K. von Figura for support, and Beate Veith for excellent technical assistance. We are indebted to Monika Schindler and H. Riedesel (Max-Planck-Institut für experimentelle Medizin, Göttingen) for blastocyst injections. We acknowledge Peter Schu (Universität Göttingen) for stimulating discussions and for critical reading of the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 523 and TP B7 and Z2) and from the Volkswagen Stiftung (Nachwuchsgruppen an Universitäten program).
REFERENCES
- 1.Advani, R. J., H. R. Bae, J. B. Bock, D. S. Chao, Y. C. Doung, R. Prekeris, J. S. Yoo, and R. H. Scheller. 1998. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem. 273:10317-10324. [DOI] [PubMed] [Google Scholar]
- 2.Advani, R. J., B. Yang, R. Prekeris, K. C. Lee, J. Klumperman, and R. H. Scheller. 1999. VAMP-7 mediates vesicular transport from endosomes to lysosomes. J. Cell Biol. 146:765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonin, W., D. Fasshauer, S. Becker, R. Jahn, and T. Schneider. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9:107-111. [DOI] [PubMed] [Google Scholar]
- 4.Antonin, W., C. Holroyd, D. Fasshauer, S. Pabst, G. Fischer von Mollard, and R. Jahn. 2000. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonin, W., C. Holroyd, R. Tikkanen, S. Höning, and R. Jahn. 2000. The R-SNARE endobrevin/VAMP-8 mediates homotypic fusion of early and late endosomes. Mol. Biol. Cell 11:3289-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonin, W., D. Riedel, and G. Fischer von Mollard. 2000. The SNARE Vti1a-β is localized to small synaptic vesicles and participates in a novel SNARE complex. J. Neurosci. 20:5724-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock, J. B., H. T. Matern, A. A. Peden, and R. H. Scheller. 2001. A genomic perspective on membrane compartment organization. Nature 409:839-841. [DOI] [PubMed] [Google Scholar]
- 8.Bordier, C. 1981. Phase separation of internal membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 9.Brickner, J., J. Blanchette, G. Sipos, and R. Fuller. 2001. The Tlg SNARE complex is required for TGN homotypic fusion. J. Cell Biol. 155:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, F., D. Chang, M. Goh, S. Klibanov, and M. Ljungman. 2000. Role of p53 in cell cycle regulation and apoptosis following exposure to proteasome inhibitors. Cell Growth Differ. 11:239-246. [PubMed] [Google Scholar]
- 11.Chen, Y. A., and R. H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2:98-106. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, W. 1990. Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J. Cell Biol. 110:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eskelinen, E., A. Illert, Y. Tanaka, G. Schwarzmann, J. Blanz, K. von Figura, and P. Saftig. 2002. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol. Biol. Cell 13:3355-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fasshauer, D., W. Antonin, M. Margittai, S. Pabst, and R. Jahn. 1999. Mixed and non-cognate SNARE complexes. J. Biol. Chem. 274:15440-15446. [DOI] [PubMed] [Google Scholar]
- 15.Fischer von Mollard, G., S. F. Nothwehr, and T. H. Stevens. 1997. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol. 137:1511-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer von Mollard, G., and T. H. Stevens. 1998. A human homolog can functionally replace the yeast v-SNARE Vti1p in two vesicle transport pathways. J. Biol. Chem. 273:2624-2630. [DOI] [PubMed] [Google Scholar]
- 17.Fischer von Mollard, G., and T. H. Stevens. 1999. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell 10:1719-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay, J., D. Chao, C. Kuo, and R. Scheller. 1997. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell 89:149-158. [DOI] [PubMed] [Google Scholar]
- 19.Hohenstein, A., and P. Roche. 2001. SNAP-29 is a promiscuous syntaxin-binding SNARE. Biochem. Biophys. Res. Commun. 285:167-171. [DOI] [PubMed] [Google Scholar]
- 20.Hunziker, W., and H. Geuze. 1996. Intracellular trafficking of lysosomal membrane proteins. Bioessays 18:379-389. [DOI] [PubMed] [Google Scholar]
- 21.Jahn, R., and T. Sudhof. 1999. Membrane fusion and exocytosis. Annu. Rev. Biochem. 68:863-911. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky, D., and S. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornfeld, S., and I. Mellman. 1989. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5:483-525. [DOI] [PubMed] [Google Scholar]
- 24.Köster, A., P. Saftig, U. Matzner, K. von Figura, C. Peters, and R. Pohlmann. 1993. Targeted disruption of the M(r) 46,000 mannose 6-phosphate receptor gene in mice results in misrouting of lysosomal proteins. EMBO J. 12:5219-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreykenbohm, V., D. Wenzel, W. Antonin, V. Atlachkine, and G. Fischer von Mollard. 2002. The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell. Biol. 81:273-280. [DOI] [PubMed] [Google Scholar]
- 26.Low, S. H., S. Chapin, C. Wimmer, S. Whiteheart, L. Komuves, K. Mostov, and T. Weimbs. 1998. The SNARE machinery is involved in apical plasma membrane trafficking in MDCK cells. Mol. Biol. Cell 141:1503-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupashin, V. V., I. D. Pokrovskaya, J. A. McNew, and M. G. Waters. 1997. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol. Biol. Cell 8:2659-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallard, F., B. Tang, T. Galli, D. Tenza, A. Saint-Pol, X. Yue, C. Antony, W. Hong, B. Goud, and L. Johannes. 2002. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156:653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meredith, M. 1988. Rat hepatocytes prepared without collagenase: prolonged retention of differentiated characteristics in culture. Cell Biol. Toxicol. 4:405-425. [DOI] [PubMed] [Google Scholar]
- 30.Mullins, C., and J. Bonifacino. 2001. The molecular machinery for lysosome biogenesis. Bioessays 23:333-343. [DOI] [PubMed] [Google Scholar]
- 31.Nagahama, M., L. Orci, M. Ravazzola, M. Amherdt, L. Lacomis, J. Rothman, and T. Sollner. 1996. A v-SNARE implicated in intra-Golgi transport. J. Cell Biol. 133:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piper, R., and J. Luzio. 2001. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic 2:612-621. [DOI] [PubMed] [Google Scholar]
- 33.Pohlmann, R., M. Wendland, C. Boeker, and K. von Figura. 1995. The two mannose 6-phosphate receptors transport distinct complements of lysosomal proteins. J. Biol. Chem. 27311-27318. [DOI] [PubMed]
- 34.Porter, M., A. Fluharty, and H. Kihara. 1969. Metachromatic leukodystrophy: arylsulfatase-A deficiency in skin fibroblast cultures. Proc. Natl. Acad. Sci. USA 62:887-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prekeris, R., B. Yang, V. Oorschot, J. Klumperman, and R. H. Scheller. 1999. Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol. Biol. Cell 10:3891-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralston, E., S. Beushausen, and T. Ploug. 1994. Expression of the synaptic vesicle proteins VAMPs/synaptobrevins 1 and 2 in non-neural tissues. J. Biol. Chem. 269:15403-15406. [PubMed] [Google Scholar]
- 37.Sachse, M., G. Ramm, G. Strous, and J. Klumperman. 2002. Endosomes: multipurpose designs for integrating housekeeping and specialized tasks. Histochem. Cell Biol. 117:91-104. [DOI] [PubMed] [Google Scholar]
- 38.Seglen, P., and P. Gordon. 1984. Amino acid control of autophagic sequestration and protein degradation in isolated rat hepatocytes. J. Cell Biol. 99:435-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steegmaier, M., B. Yang, J. Yoo, B. Huang, M. Shen, S. Yu, Y. Luo, and R. H. Scheller. 1998. Three novel proteins of the syntaxin/SNAP-25 family. J. Biol. Chem. 273:34171-34179. [DOI] [PubMed] [Google Scholar]
- 40.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395:347-353. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, Y., G. Guhde, A. Suter, E. Eskelinen, D. Hartmann, R. Lullmann-Rauch, P. M. Janssen, J. Blanz, K. von Figura, and P. Saftig. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406:902-906. [DOI] [PubMed] [Google Scholar]
- 42.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryos in culture and their development into established cell lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tokuyasu, K. 1973. A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tolleshaug, H., M. Berg, M. Nilssom, and K. Norum. 1977. Uptake and degradation of 125I-labelled asialofetuin by isolated rat hepatocytes. Biochem. Biophys. Acta 499:73-84. [DOI] [PubMed] [Google Scholar]
- 45.Wade, N., N. J. Bryant, L. M. Connolly, R. J. Simpson, J. P. Luzio, R. C. Piper, and D. E. James. 2001. Syntaxin 7 complexes with mVti1b, syntaxin 6, VAMP8, and VAMP7 in b16 melanoma cells. J. Biol. Chem. 276:19820-19827. [DOI] [PubMed] [Google Scholar]
- 46.Ward, D., J. Pevsner, M. A. Scullion, M. Vaughn, and J. Kaplan. 2000. Syntaxin 7 and VAMP-7 are SNAREs required for late endosome-lysosome and homotypic lysosome fusion in alveolar macrophages. Mol. Biol. Cell 11:2327-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wendler, F., L. Page, S. Urbe, and S. A. Tooze. 2001. Homotypic fusion of immature secretory granules during maturation requires syntaxin 6. Mol. Biol. Cell 12:1699-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong, S. H., Y. Xu, T. Zhang, G. Griffiths, L. Lowe, V. N. Subramaniam, K. Seow, and W. Hong. 1999. GS32, a novel Golgi SNARE of 32 kDa, interacts preferentially with syntaxin 6. Mol. Biol. Cell 10:119-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, Y., S. H. Wong, B. L. Tang, V. N. Subramaniam, T. Zhang, and W. Hong. 1998. A 29-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Vti1-rp2) implicated in protein trafficking in the secretory pathway. J. Biol. Chem. 273:21783-21789. [DOI] [PubMed] [Google Scholar]
- 50.Yang, C., K. Coker, J. Kim, S. Mora, D. Thurmond, A. Davis, B. Yang, R. Williamson, G. Shulman, and J. E. Pessin. 2001. Syntaxin 4 heterozygous knockout mice develop muscle insulin resistance. J. Clin. Investig. 107:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, C., S. Mora, J. Ryder, K. Coker, P. Hansen, L. Allen, and J. Pessin. 2001. VAMP3 null mice display normal constitutive, insulin- and exercise-regulated vesicle trafficking. Mol. Cell. Biol. 21:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]