Abstract
Flap endonuclease 1 (FEN1) has been shown to remove 5′ overhanging flap intermediates during base excision repair and to process the 5′ ends of Okazaki fragments during lagging-strand DNA replication in vitro. To assess the in vivo role of the mammalian enzyme in repair and replication, we used a gene-targeting approach to generate mice lacking a functional Fen1 gene. Heterozygote animals appear normal, whereas complete depletion of FEN1 causes early embryonic lethality. Fen1−/− blastocysts fail to form inner cell mass during cellular outgrowth, and a complete inactivation of DNA synthesis in giant cells of blastocyst outgrowth was observed. Exposure of Fen1−/− blastocysts to gamma radiation caused extensive apoptosis, implying an essential role for FEN1 in the repair of radiation-induced DNA damage in vivo. Our data thus provide in vivo evidence for an essential function of FEN1 in DNA repair, as well as in DNA replication.
Flap endonuclease 1 (FEN1) is a member of the RAD2 superfamily of nucleases, which play a critical role in DNA replication and repair in both prokaryotes and eukaryotes (16, 17, 24, 26, 55). The main function of FEN1 in replication is proposed to be the removal of displaced RNA-DNA primers synthesized by DNA polymerase α-primase during discontinuous lagging-strand replication. A similar single-stranded DNA flap structure is produced by excessive gap filling by a DNA repair polymerase during long-patch base excision repair (BER). Both biochemical and genetic studies support a role for FEN1 during these cellular processes. Similar 5′-flap intermediates are also formed in nonhomologous DNA end joining of double-strand DNA breaks and DNA recombination (25, 57).
The biochemistry of FEN1 has been studied extensively by several groups (reviewed in reference 32), and the crystal structures of two FEN1 orthologues from archaea have been solved (19, 20). The enzyme employs a unique cleavage mechanism for substrates containing single-stranded 5′ tails or 5′-flap structures. It recognizes the 5′ end, tracking the length of the tail, and cleaves at the junction between double-stranded and single-stranded DNAs (40). Although FEN1 acts less efficiently as an exonuclease than as a flap endonuclease, it is likely that the enzyme employs similar mechanisms for both reactions (16, 34).
FEN1 has been shown to have an important role in the processing of intermediates formed during BER of modified structures in DNA. Recent evidence has indicated that in mammalian cells, BER is mediated through at least two subpathways with different repair patch sizes and different enzymes involved. These pathways have been designated single-nucleotide BER and long-patch BER (12, 27, 28). The choice of subpathways in BER depends on whether the 5′ deoxyribose phosphate intermediate can be efficiently removed by the polymerase β lyase activity to yield a 5′-phosphorylated DNA strand capable of serving as a substrate for DNA ligase (44). When such processing is inefficient, long-patch BER can occur, and FEN1 will remove the overhang formed by repair synthesis, displacing the deoxyribose phosphate-containing strand.
The biological importance of FEN1 is emphasized by characterization of yeast rad27 (a homologue of the mammalian Fen1 gene) mutant strains. Such mutants are conditionally lethal at high temperatures, with a defect in DNA replication, and show sensitivity to UV radiation and alkylating agents and also deficiencies in telomere maintenance (43, 45, 48). These phenotypes are consistent with the participation of Rad27/FEN1 in both DNA replication and repair. As these rad27 deletion strains are strong mutators with destabilized repetitive sequences, it has been suggested that the mammalian enzyme, FEN1, also might be involved in mechanisms through which trinucleotide expansions occur (15, 49). Such expansions have been identified as the basis of >10 hereditary human diseases. In summary, FEN1 is a key enzyme for maintaining genomic stability (30).
Many of the protein components involved in long-patch BER are also components of the DNA replication machinery, providing a mechanistic link between BER and the DNA replication complex (4, 7, 19). The replication proteins PCNA (proliferation cell nuclear antigen) and RPA (replication protein A) have been shown to increase the efficiency of BER reconstituted in vitro (8, 12, 14). In addition to its role in stimulating DNA polymerase δ, PCNA has been shown to directly interact with both FEN1 and DNA ligase I and to stimulate their activities (21, 26, 38, 53, 56). The PCNA-mediated stimulation of these proteins has been shown to be caused by increases in enzyme-substrate binding (53).
This study describes the generation via homologous recombination of mouse blastocysts lacking FEN1. Deletion of both Fen1 alleles causes death early during embryogenesis, indicating an essential role for FEN1 during development. Mice that are heterozygous for the mutation are viable, with no phenotype. Kucherlapati et al. recently demonstrated haploinsufficiency of Fen1 in combination with a mutation in the adenomatous polyposis coli (Apc) gene, resulting in a mild tumor predisposition phenotype (29). We have examined the characteristics of Fen1 null blastocysts. Hypersensitivity to gamma radiation and the absence of DNA synthesis in cellular outgrowth indicate an essential role for FEN1 in DNA repair and replication. No alternative activity for FEN1 could be detected.
MATERIALS AND METHODS
Construction of Fen1 targeting vector.
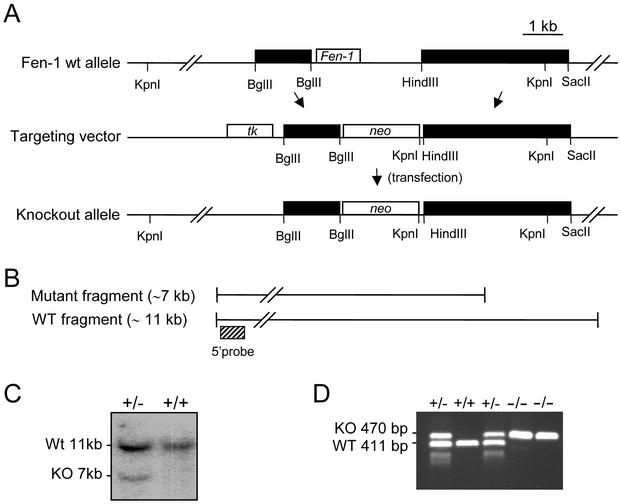
A specific murine probe spanning 600 bp of the FEN1 coding sequence was amplified from murine genomic DNA by PCR and used to screen a mouse 129/SvJ lambda genomic library (Stratagene), using standard hybridization techniques. One genomic clone of ∼19 kb spanning the whole coding region of the Fen1 gene was used to produce the targeting construct. A 3-kb DNA fragment containing Fen1 was replaced by the neomycin resistance gene (neo), as illustrated in Fig. 1A. The resulting construct possesses homologous arms of 1.2 and 3.4 kb flanking the neo gene. The herpes simplex virus thymidine kinase (TK) gene was positioned upstream of the shortest arm. The accuracy of the targeting construct was verified by DNA sequencing and restriction analysis.
FIG. 1.
Targeted disruption of the murine Fen1 locus. (A) Physical map of the genomic DNA containing the Fen1 gene. Exon 2, containing the entire ORF of the Fen1 gene, is boxed. Genomic sequences, represented by solid boxes, were subcloned on either side of a neo gene, generating a targeting construct that deleted the entire Fen1 coding sequence. Murine genomic DNA is represented by solid boxes, vector sequences by a line, and Neo and Tk genes (positive and negative selection, respectively) by open boxes. (B) Restriction digest of genomic DNA by KpnI giving rise to an ∼11-kb fragment, using a 5′ flanking probe as indicated. A gene-targeted locus was detected by hybridization with an ∼7-kb fragment. (C) Southern blot analysis of ES cell lines. A positive, gene-targeted ES-cell line yields a 7-kb fragment in addition to the 11-kb wild-type (WT) fragment. KO, knockout. (D) PCR genotyping of DNA isolated from E3.5 blastocysts resulting from crossing of twoFen1+/− mice. A 411-bp wild-type band and a 470-bp mutant band were amplified with primer pairs P1-P2 and P1-P3. The locations and sequences for PCR primers are as described in Materials and Methods.
Disruption of the mouse Fen1 gene.
The targeting vector was propagated in the SURE Escherichia coli strain (Stratagene), linearized with NotI, and introduced into embryonic stem (ES) cells by electroporation. Colonies doubly resistant to active G418 (250 μg/ml) and ganciclovir (2 μM) were isolated and screened for disruption of the Fen1 gene by Southern hybridization analysis following complete digestion of genomic DNA with KpnI. A 560-bp DNA fragment (illustrated in Fig. 1A) was used as a probe. Two independent ES clones carrying one copy of the disrupted allele of the Fen1 gene were expanded and injected into blastocysts from C57BL/6J mice to generate chimeras. The resultant chimeras were mated with strain C57BL/6 mice, and F1 animals were screened by tail biopsy for Fen1 heterozygosity. Mice heterozygous for the Fen1 allele (Fen1+/−) were interbred, and F2 blastocysts, embryos, and progeny were genotyped as described above and below (Table 1).
TABLE 1.
Genotyping of Fen1+/− intercrosses
| Age | No. with genotype:
|
Total no. of mice | % Fen1−/− resorbeda | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| E2.5 | 5 | 8 | 3 | 16 | 31 |
| E3.5 | 19 | 34 | 6 | 59 | 66 |
| E9.5 | 6 | 13 | 0 | 19 | 100 |
| E18.5 | 3 | 2 | 0 | 5 | 100 |
| 1 wk | 32 | 62 | 0 | 94 | 100 |
Theoretical value of resorbed Fen1−/− blastocysts calculated from the numbers of Fen1+/+ and Fen1+/− blastocysts.
PCR genotyping of the Fen1 locus in mice.
PCR genotyping of mice was performed on tail biopsy specimens. Genomic DNA was isolated from mouse tails (0.3 cm) and incubated overnight at 55°C in 500 μl of lysis buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.5], 0.1 mg of gelatin/ml, and 10 μg of proteinase K/ml). The sample was diluted 1:10, and 1 μl was used in standard PCR with Pfu turbo polymerase (Stratagene). A mixture of three primers was used to detect wild-type and mutant alleles: P1 (forward) (5′-CATAGTCATATTCGCATAGCT-3′), P2 (wild type) (5′-AGGCCTCCTGACAATCAGAAG-3′), and P3 (neo) (5′-TATGGCTTCTGAGGCGGAAAGAACCA-3′). Primers P1 and P2 generate a 411-bp wild-type allele, whereas P1 and P3 generate a 470-bp knockout allele. The PCR profile was as follows: 1 cycle of 94°C for 3 min, followed by 35 cycles of 93°C for 1 min, 59°C for 45 s, and 72°C for 2 min. The PCR products were analyzed by standard 2% agarose gel electrophoresis.
Histological sections.
Embryos at embryonic day 18.5 (E18.5) resulting from heterozygous intercrosses were collected and fixed in 4% (wt/vol) paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4°C, processed, and embedded in paraffin following standard procedures.
Timed pregnancies.
Heterozygous male and female mutant mice were bred to obtain wild-type (Fen1w/t), heterozygous (Fen1+/−), and homozygous (Fen1−/−) mutant embryos. To generate timed pregnancies, Fen1+/− females were injected intaperitoneally with pregnant mare's serum gonadotropin (5 IU per animal; Sigma-Aldrich Co.), followed by injection with human chorionic gonadotropin (5 IU per animal; Sigma-Aldrich Co.) 47 h later and mating overnight with Fen1+/− males. The next morning, the males were removed and the females were examined for the presence of a vaginal plug. The morning on which a vaginal plug was detected was designated E0.5.
Genotyping of preimplantation embryos.
Individual embryos and cells from outgrowths in culture were lysed by incubation at 55°C for 3 h in 20 μl of PCR lysis buffer (10 mM Tris-HCl, pH 8.4, 50 mM KCl, 2 mM MgCl, 0.45% [vol/vol] NP-40, 0.45% [vol/vol] Tween 20, and 60 μg of proteinase K/ml). After being boiled for 10 min, a portion (8 μl) of the lysates was subjected to PCR amplification under the same conditions as for the mouse biopsy specimens described above, except that 45 cycles were used.
In vitro culture of preimplantation embryos.
Heterozygous female mice were superovulated by treatment with gonadotropin as described above. Embryos at different stages of development (E2.5 to E4.5) were collected by flushing the oviducts or the uterus with HEPES-buffered medium 2 (M2; Sigma). For staining of DNA with DAPI (4′,6′-diamidino-2-phenylindole), blastocysts were fixed immediately for 20 min at 4°C with PBS containing 4% PFA, and their nuclei were stained for 10 min with DAPI (Vectashield; Vector). For in vitro culture, blastocysts were cultured for 7 days in 96-well tissue culture plates containing cES medium (Dulbecco’s modified Eagle’s medium with 4.5g of glucose/liter, 15% fetal calf serum, 8μg of 2-mercaptoethanol/ml, 100 U of penicillin/ml, 100 μg of streptomycin, and 1× nonessential amino acids; BioWhittaker) without leukemia factor. Outgrowths were inspected daily and photographed to monitor their development. DAPI staining of outgrowths on day 7 was done as described above. Blastocysts and outgrowths were examined on a Leica DMRB fluorescence microscope. Images were obtained using a Zeiss AxioCam HRC camera and then processed for publication using Axiovision version 3.1 software.
TUNEL assays.
E3.5 blastocysts were collected as described above and fixed with 4% paraformaldehyde in PBS for 20 min at 4°C. The fixed blastocysts were permeabilized for 20 min at room temperature with PBS containing 0.3% (vol/vol) Triton X-100 and 1.5% (wt/vol) bovine serum albumin. DNA fragmentation associated with apoptosis was detected with an in situ cell death detection kit (TMR red; Roche Molecular Biochemicals). In brief, permeabilized embryos were labeled with the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction mixture for 60 min at 37°C. In the last 30 min of incubation, cells were stained with DAPI (1μg/ml; Roche). Fluorescein-labeled DNA was analyzed using a fluorescence microscope.
Gamma irradiation.
E3.5 blastocysts were incubated in M2 and gamma irradiated at 2 Gy (Gammacell 3000 Elan; 5.35 Gy/min). After incubation in M2 for 24 h, the blastocysts were stained with DAPI and TUNEL assayed as described above. The blastocysts were examined by fluorescence microscopy as described above.
Proliferation of blastocyst outgrowths.
DNA synthesis in blastocyst outgrowths was detected using the in situ cell proliferation kit FLUOS (Roche Molecular Biochemicals). E3.5 blastocysts were collected as described above and cultured in cES medium (without leukemia factor) for 3 days before 10 μM BrdU was added for 48 h. The blastocyst outgrowths were fixed and permeabilized with 4% PFA and 0.5% Triton X-100 for 45 min, and the DNA was denatured with 0.5 M HCl for 30 min. Nonspecific binding of anti-BrdU-FLUOS antibody was blocked with incubation buffer for 10 min, and the preparation was covered with anti-BrdU-FLUOS antibody working solution and incubated for 45 min at 37°C in a humid chamber. BrdU-labeled blastocyst outgrowths were detected by fluorescence microscopy as described above.
RESULTS
Targeted disruption of the Fen1 gene.
One genomic clone, ∼19 kb, comprising the complete Fen1 gene (GenBank accession number AC026761) was isolated from a lambda 129/SvJ genomic library. The Fen1 gene is composed of two exons, and the entire open reading frame (ORF) for Fen1 resides in exon 2 in both the human and mouse genomes (22). In the targeting construct, the entire ORF of the Fen1 gene was deleted, and the neomycin resistance marker was inserted in the same transcriptional orientation as Fen1. By this strategy, sequences upstream of the ORF, including exon 1, remain intact after the gene-targeting event. The herpes simplex virus TK gene was used as a negative selection marker (Fig. 1A). The targeting construct was introduced into ES cells by electroporation, and cells that had undergone homologous recombination were identified by Southern hybridization (Fig. 1B). Eight hundred neomycin- and ganciclovir-resistant clones were isolated, and eight correctly targeted clones were identified. ES cells from two of these were microinjected into C57BL/6 blastocysts and transplanted into pseudopregnant C57BL/6 females. Seven chimeric mice were obtained, and one subsequently transmitted the mutated Fen1 allele to its progeny. Male and female Fen1 heterozygote mice appeared phenotypically normal and fertile and were indistinguishable in size and growth from their littermates. Five newborn mice, three heterozygous and two wild type, were analyzed for morphological and histological changes without any significant effects of heterozygosity for Fen1 being found (data not shown). Interbreeding of Fen1+/− mice resulted in Mendelian segregation of wild-type and heterozygous animals in F1 (Table 1). At 12 to 15 months of age, no altered phenotype was present in the population of Fen1+/− mice.
An ORF, C11orf10, is located immediately upstream of the Fen1 gene in the reverse orientation (1). In the human genome, the mRNA of Fen1 and the C11orf10 locus overlap by 14 bp at their 5′ ends, whereas their transcripts are separated by ∼500 bp in mice. The C11orf10 gene is located outside our Fen1 gene-targeting construct, and it seems unlikely that the targeting construct could affect the regulation of C11orf10 or any other genes.
Absence of homozygous Fen1 mutant progeny.
Genotyping of mice arising from Fen1+/− intercrosses failed to produce homozygous Fen1 null mice, indicating prenatal lethality (Fig. 1C), although Mendelian segregation of the wild type and heterozygotes was identified (Table 1). To precisely pinpoint the time of embryonic death, embryos from heterozygous intercrosses were dissected at different stages of gestation and genotyped by PCR. As shown in Table 1, the frequency of Fen1−/− embryos decreased from E2.5 to E3.5 (see Materials and Methods), and they were not identified at later stages. These data indicate that FEN1-deficient embryos die before gastrulation.
FEN1 is required for the expansion of the ICM in vitro.
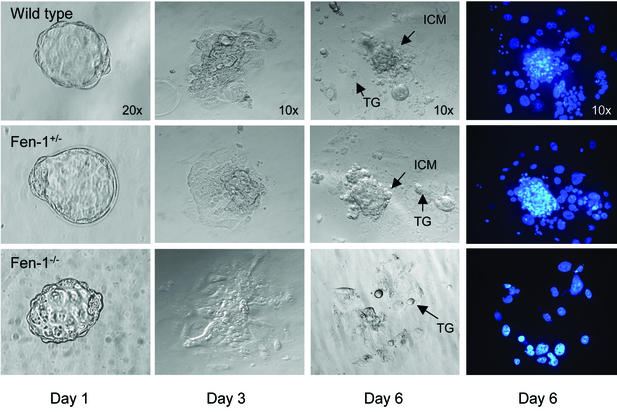
To address whether the abortive development of Fen1-deficient embryos was due to a general cell growth defect or lineage-specific defects, blastocysts isolated at E3.5 were individually cultured in vitro for 6 days and then genotyped. The cultures were examined daily, and photographs were taken. During the early phases of these cultures, most blastocysts, obtained from four separate litters (10 wild type, 14 heterozygous, and 3 null), attached to the substratum, hatched from the zona pellucida, and started to expand their inner cell masses (ICMs) and to form outgrowths having the classical appearance of migrating trophoblastic cells (Fig. 2). However, by day 3, the ICMs of the three FEN1 null blastocysts stopped proliferating, and they degenerated soon after (Fig. 2). This phenotype was fully manifested over the next 2 days, and only FEN1-deficient trophoblastic giant cells persisted in long-term cultures (Fig. 2).
FIG. 2.
Defective growth of Fen1−/− blastocysts in vitro. Heterozygous Fen1+/− mice were intercrossed, and Fen1wt (top), Fen1+/− (middle), and Fen1−/− (bottom) blastocysts were collected by flushing the uterus at E3.5. The genotypes indicated were determined by PCR. Blastocysts were individually cultured for 6 days in cES medium, during which time they developed outgrowths. The outgrowths were inspected daily and photographed to monitor their development. Nuclei of blastocyst outgrowths were visualized by DAPI staining on day 6 (right). The ICM grew normally and was surrounded by trophoblast giant cells (TG) in Fen1wt and Fen1+/− blastocysts. In contrast, the ICM was completely absent in the Fen1−/− blastocysts. Magnifications are indicated on the top row of images.
Loss of FEN1 does not result in spontaneous apoptosis.
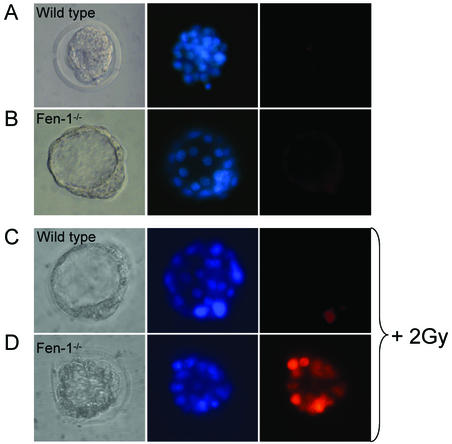
The in vitro growth deficits of Fen1 null mutant ICMs could be due to severe defects in replication and/or DNA repair. From in vitro data, it has been concluded that FEN1 is required for lagging-strand DNA replication and long-patch BER (26, 55). DNA double-strand breaks can arise spontaneously during the processing of DNA adducts, or single-strand breaks can arise through DNA repair or replication processes (2). To address whether the lack of FEN1 resulted in apoptosis, a TUNEL assay was performed on untreated E3.5 blastocysts isolated from three litters (n = 16). No striking difference was observed in any of the embryos analyzed, and Fen1-deficient blastocysts (n = 2) contained numbers of mitotic cells similar to those of their wild-type littermates (Fig. 3). Although we cannot exclude the possibility that apoptosis will occur in Fen1−/− mice after this stage of development, these data suggest that E3.5 Fen1 null embryos do not die from cellular apoptosis.
FIG. 3.
Fen1−/− embryonic cells are hypersensitive to gamma radiation. E3.5 blastocysts were collected as described in Materials and Methods; and placed in M2. The genotypes indicated were determined by PCR. Blastocysts were untreated or gamma irradiated at 2 Gy (Gammacell 3000 Elan; 5.35 Gy/min) prior to incubation for 24 h and double staining with TUNEL reagents (red) and DAPI (blue). Representative photographs of blastocysts following either no radiation (Fen1wt [A] and Fen1−/− [B]) or gamma radiation (2 Gy) (Fen1wt [C] and Fen1−/− [D]). Extensive cell death in Fen1wt E3.5 blastocysts is shown by TUNEL staining (red).
Induced oxidative stress leads to apoptosis in Fen1 mutant blastocysts.
Exposure of DNA to reactive oxygen species and ionizing radiation causes the formation of several different base lesions and apurinic-apyrimidinic (AP) sites that require BER (9, 26). FEN1 participates in long-patch BER. To investigate whether the Fen1 null blastocysts were particularly sensitive to DNA damage, we assayed the ability of E3.5 blastocysts to recover from gamma irradiation. E3.5 blastocysts isolated from two independent litters (n = 10) were irradiated at 2 Gy and allowed to repair DNA damage for 24 h prior to TUNEL assay as described in Materials and Methods. We observed extensive apoptosis in the Fen1−/− blastocysts. When the dose was increased to 4 Gy, apoptosis was observed in both wild-type and Fen1 mutant blastocysts (data not shown). We conclude that FEN1-depleted embryos manifest a pronounced repair deficiency for DNA damage induced by ionizing radiation.
FEN1 is required for S phase entry in trophoblast giant cells.
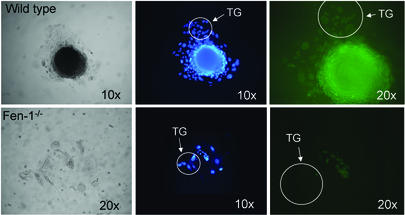
DNA synthesis in trophoblast giant cells occurs by endoreduplication, whereby successive rounds of G and S phases proceed without intervening mitosis (5, 13). As a result, endocycling giant cells acquire vast quantities of DNA in their nuclei, thus becoming polyploid (54). The number of trophoblast giant cells is greatly reduced in cellular outgrowth of Fen1−/− blastocysts compared to the outgrowth of wild-type blastocysts, 10 to 20 or 50 to 150 cells, respectively (Fig. 2 and results not shown). We reasoned that this could reflect the fact that the endocycle was progressing more slowly in Fen−/− blastocysts than in their Fen1wt counterparts, or it could reflect an arrest of the endocycle. To differentiate between these possibilities, E3.5 blastocysts and blastocyst outgrowths (day 3 in culture, equivalent to 6 days postcoitum) were cultured in the presence of bromodeoxyuridine (BrdU) for 24 and 48 h, respectively, and then immunostained with an antibody specific for BrdU to identify cells that had undergone DNA synthesis during the labeling period (Fig. 4). Incorporation of BrdU into blastocyst stage nuclei revealed no difference between the Fen1−/− and Fen1wt embryos, both of which showed extensive labeling (data not shown). In contrast, while the vast majority of giant cells incorporated BrdU in control outgrowth, no BrdU-positive giant cells were detected in the Fen1−/− outgrowth (Fig. 4). These results demonstrate that the Fen1−/− cells were unable to enter S phase and were arrested in the endocycle.
FIG. 4.
Replication and cellular proliferation of Fen1−/− blastocyst outgrowths. Wild-type (top) and Fen1−/− (bottom) embryos were isolated at E3.5 and cultured for 3 days in cES medium (equivalent to 6 days postcoitum). The genotypes indicated were determined by PCR. Cellular outgrowth was extended for 48 h in the presence of BrdU, and the cells were then immunostained with an antibody specific for BrdU to identify cells that had undergone DNA synthesis during the labeling period. The outgrowths were photographed to monitor their development (left). Nuclei were visualized by DAPI staining (middle). While the vast majority of trophoblast giant cells (TG) easily incorporated BrdU in control outgrowth, no BrdU-positive cells were detected in the Fen1−/− outgrowth (right, green cells). Magnifications are indicated on the images.
DISCUSSION
Role of BER during embryonic development.
The biochemical role of FEN1 in DNA BER has been studied extensively in vitro (24, 26). FEN1 is a key enzyme for the removal of 5′ single-stranded DNA flap structures produced by excessive gap-filling reactions during BER. In this study, the biological significance of this activity was addressed by targeted disruption of Fen1 from the mouse genome.
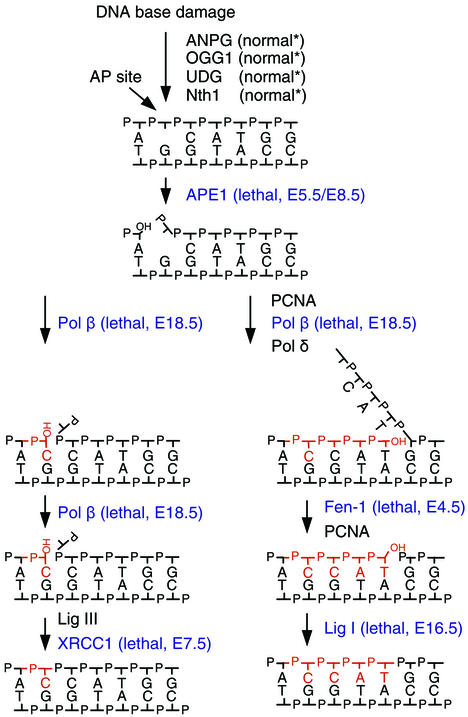
Several proteins required for the removal of BER intermediates are essential for viability, as judged by the embryonic lethality in gene-targeted mice (Fig. 5). This phenotype is produced regardless of whether proteins from the 1-nucleotide BER or the long-patch BER are inactivated (Fig. 5, left and right branches, respectively). However, the causes and stages of lethality vary. Whereas the XRCC1 (X-ray cross-complementing group 1), APE1 (AP endonuclease I), and FEN1 homozygous knockout mice die early during embryonic development (references 35, 52, and 59 and this work), DNA ligase I and DNA polymerase β null mutants die at late embryonic stages or immediately after birth, respectively (6, 50). The cause of this variation might reflect additional roles of the deleted DNA repair enzyme (32, 58) or the existence of backup activities that would alleviate some of the effects of the phenotype (discussed below). It is therefore unlikely that the individual phenotype reflects solely the detrimental effects of the accumulation of specific BER intermediates, such as AP sites, single-strand breaks, gaps, or flap structures.
FIG. 5.
Summary of gene-targeted knockout mice in BER. A simplified view of the branched pathways for BER is presented. In the left pathway, a single nucleotide is replaced, whereas several nucleotides are replaced in the right pathway. Regular AP sites, resulting from spontaneous or glycosylase-initiated removal of the base, can be repaired via the single-nucleotide pathway, although a minor fraction are repaired via the right pathway. Oxidized or reduced AP sites are removed via the right pathway. BER initiated by a DNA glycosylase with associated AP lyase activity, OGG1 or Nth1, is repaired slightly differently from the pathways illustrated. Gene-targeted knockout mice of DNA glycosylases, ANPG, OGG1, UDG, and Nth1, have a mild or no phenotype. Targeted disruptions of enzymes for processing of key BER intermediates, highlighted in blue, all have a severe phenotype and are embryonic lethal. UDG, uracil-DNA glycosylase; Nth1, (mammalian) endonuclease III homolog 1; Pol β, DNA polymerase β; Pol δ, DNA polymerase δ; Lig I, DNA ligase I; Lig III, DNA ligase III.
So far, no obvious phenotypes have been produced by targeted disruption of DNA glycosylases of the murine genome (10, 27, 37, 41, 51). However, all of the studies have been followed by discoveries of alternative DNA repair pathways for specific DNA lesions. For example, although ANPG (alkyl-N-purine DNA glycosylase) is the major DNA glycosylase for 3-methyladenine repair, 3-methyladenine disappears faster from the genome of Anpg−/− mice than would be expected from spontaneous depurination alone (47). Alternative repair of uracil residues in DNA is accomplished by the SMUG enzyme (18, 41). In mice depleted of OGG1 (8-oxoguanine [8-oxoG] DNA glycosylase), the enzyme initiating BER of the highly mutagenic 8-oxoG, a 10-fold-elevated frequency of G-to-T transversions has been reported (27). However, a backup activity for 8-oxoG repair, which might have priority for transcribed sequences, removes 8-oxoG lesions in vivo at significant rates (31, 42). Finally, a novel nuclear and mitochondrial glycosylase for thymine glycols was revealed by disruption of the mouse Nth1 gene (51).
Repair and replication failure of Fen1−/− blastocysts.
Extensive apoptosis was observed in cells from Fen1−/− blastocysts exposed to gamma radiation (Fig. 3), providing new evidence for an essential role for FEN1, and long-patch BER, in the repair of radiation-damaged DNA (23). Haploinsufficiency of FEN1, combined with a mutation in the Apc gene, causes increased numbers of adenocarcinomas and decreased survival (29). The tumors from these mice show microsatellite instability. These results, taken together, emphasize the importance of FEN1 for the maintenance of genomic stability during normal life and following exposure to stress conditions. Oxidation of DNA occurs at significant rates in vivo, and the decay of DNA is likely to be a major factor in carcinogenesis (33). These data indicate that endogenously induced DNA damage, e.g., caused by reactive oxygen species, triggers the long-patch BER for which FEN1 is required.
Inactivation of FEN1 caused a complete failure of DNA synthesis in giant cells of blastocyst outgrowth (Fig. 4). One might expect that blockage of replication should cause immediate cell death. However, the mouse embryo develops very slowly, reaching the two-cell stage 24 h after fertilization, and will continue to divide slowly without any increase in mass as it moves along the oviduct to implantation 4.5 days after fertilization. Fen1 null embryos were able to hatch from the zona pellucida, but the ICM disappeared shortly thereafter. We suggest that Fen1−/− embryos develop normally until the maternally supplied Fen1 mRNA or gene product is exhausted. The transfer of maternal wild-type products from oocytes will allow survival through the initial cell divisions (e.g., inactivation of the survival motor neuron [SMN] gene [46]), and it seems likely that this may occur with several essential genes. Genetic analysis of Saccharomyces cerevisiae has revealed that deletion of the yeast homologue of Fen1, rad27, does not confer lethality and that Dna2 and Exo1 have redundant functions contributing to lagging-strand synthesis (3, 39). Insect cells lacking FEN1 endonuclease are viable but hypersensitive to DNA-damaging agents (36). No redundant activity for FEN1 has been identified in human or mouse cells.
Finally, deletion of rad27 in S. cerevisiae causes expansion and length-dependent fragility of CTG repeats (11). A number of genetic diseases, such as myotonic dystrophy, Huntington's disease, fragile-X syndrome, and cancer, result from genomic instability, indicating a central role for the FEN1 enzyme. The design of new mammalian Fen1 mutant models (e.g., gene targeting with site-specific mutations) should address these questions. Such models could also help elucidate the role of FEN1 interaction with PCNA and its implication for genetic risk (14).
Acknowledgments
We thank Jeanette Ringvoll, Torbjørn Rognes, Lars Eide, and Pål Falnes for their contributions to different aspects of this work, and the Centre for Comparative studies, the National Hospital in Oslo, and the animal facility of Umeå University for animal care and ES cell microinjection. We are grateful to Jeffrey R. Needham, FIBMS, United Kingdom, for histology reports.
This work was supported by the Norwegian Cancer Society, the Research Council of Norway, and the DNage Collaborative Project, EU. A.K. acknowledges the economic support of the Norwegian Advanced Research Program.
REFERENCES
- 1.Adachi, N., Z. E. Karanjawala, Y. Matsuzaki, H. Koyama, and M. R. Lieber. 2002. Two overlapping divergent transcription units in the human genome: the FEN1/C11orf10 locus. OMICS 6:273-279. [DOI] [PubMed] [Google Scholar]
- 2.Akyuz, N., G. S. Boehden, S. Susse, A. Rimek, U. Preuss, K. H. Scheidtmann, and L. Wiesmuller. 2002. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol. Cell. Biol. 22:6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, S. H., and Y. S. Seo. 2000. Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 275:38022-38031. [DOI] [PubMed] [Google Scholar]
- 4.Bambara, R. A., R. S. Murante, and L. A. Henricksen. 1997. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 272:4647-4650. [DOI] [PubMed] [Google Scholar]
- 5.Barlow, P. W., and M. I. Sherman. 1972. The biochemistry of differentiation of mouse trophoblast: studies on polyploidy. J. Embryol. Exp. Morphol. 27:447-465. [PubMed] [Google Scholar]
- 6.Bentley, D., J. Selfridge, J. K. Millar, K. Samuel, N. Hole, J. D. Ansell, and D. W. Melton. 1996. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 13:489-491. [DOI] [PubMed] [Google Scholar]
- 7.DeMott, M. S., B. Shen, M. S. Park, R. A. Bambara, and S. Zigman. 1996. Human RAD2 homolog 1 5′-to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J. Biol. Chem. 271:30068-30076. [DOI] [PubMed] [Google Scholar]
- 8.DeMott, M. S., S. Zigman, and R. A. Bambara. 1998. Replication protein A stimulates long patch DNA base excision repair. J. Biol. Chem. 273:27492-27498. [DOI] [PubMed] [Google Scholar]
- 9.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 10.Engelward, B. P., A. Dreslin, J. Christensen, D. Huszar, C. Kurahara, and L. Samson. 1996. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 15:945-952. [PMC free article] [PubMed] [Google Scholar]
- 11.Freudenreich, C. H., S. M. Kantrow, and V. A. Zakian. 1998. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279:853-856. [DOI] [PubMed] [Google Scholar]
- 12.Frosina, G., P. Fortini, O. Rossi, F. Carrozzino, G. Raspaglio, L. S. Cox, D. P. Lane, A. Abbondandolo, and E. Dogliotti. 1996. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 271:9573-9578. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, R. L. 1983. Origin and differentiation of extraembryonic tissues in the mouse. Int. Rev. Exp. Pathol. 24:63-133. [PubMed] [Google Scholar]
- 14.Gary, R., K. Kim, H. L. Cornelius, M. S. Park, and Y. Matsumoto. 1999. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem. 274:4354-4363. [DOI] [PubMed] [Google Scholar]
- 15.Gordenin, D. A., T. A. Kunkel, and M. A. Resnick. 1997. Repeat expansion—all in a flap? Nat. Genet. 16:116-118. [DOI] [PubMed] [Google Scholar]
- 16.Harrington, J. J., and M. R. Lieber. 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13:1235-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrington, J. J., and M. R. Lieber. 1994. Functional domains within FEN1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 8:1344-1355. [DOI] [PubMed] [Google Scholar]
- 18.Haushalter, K. A., M. W. Todd Stukenberg, M. W. Kirschner, and G. L. Verdine. 1999. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 9:174-185. [DOI] [PubMed] [Google Scholar]
- 19.Hosfield, D. J., G. Frank, Y. Weng, Y. A. Tainer, and B. Shen. 1998. Newly discovered archaebacterial flap endonucleases show a structure-specific mechanism for DNA substrate binding and catalysis resembling human flap endonuclease-1. J. Biol. Chem. 273:27154-27161. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, K. Y., K. Baek, H. Y. Kim, and Y. Cho. 1998. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 5:707-713. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson, Z. O., R. Hindges, and U. Hubscher. 1998. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 17:2412-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karanjawala, Z. E., X. Shi, C. L. Hsieh, and M. R. Lieber. 2000. The mammalian FEN-1 locus: structure and conserved sequence features. Microb. Comp. Genomics 5:173-177. [DOI] [PubMed] [Google Scholar]
- 23.Karmakar, P., A. S. Balajee, and A. T. Natarajan. 2001. Analysis of repair and PCNA complex formation induced by ionizing radiation in human fibroblast cell lines. Mutagenesis 16:225-232. [DOI] [PubMed] [Google Scholar]
- 24.Kim, C. Y., B. Shen, M. S. Park, and G. A. Olah. 1999. Structural changes measured by X-ray scattering from human flap endonuclease-1 complexed with Mg2+ and flap DNA substrate. J. Biol. Chem. 274:1233-1239. [DOI] [PubMed] [Google Scholar]
- 25.Kim, K., S. Biade, and Y. Matsumoto. 1998. Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem. 273:8842-8848. [DOI] [PubMed] [Google Scholar]
- 26.Klungland, A., and T. Lindahl. 1997. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J. 16:3341-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klungland, A., I. Rosewell, S. Hollenbach, E. Larsen, G. Daly, B. Epe, E. Seeberg, T. Lindahl, and D. E. Barnes. 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA 96:13300-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubota, Y., R. A. Nash, A. Klungland, P. Schar, D. E. Barnes, and T. Lindahl. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 29.Kucherlapati, M., K. Yang, M. Kuraguchi, J. Zhao, M. Lia, J. Heyer, M. F. Kane, K. Fan, R. Russell, A. M. Brown, B. Kneitz, W. Edelmann, R. D. Kolodner, M. Lipkin, and R. Kucherlapati. 2002. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA 99:9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunkel, T. A., M. A. Resnick, and D. A. Gordenin. 1997. Mutator specificity and disease: looking over the FENce. Cell 88:155-158. [DOI] [PubMed] [Google Scholar]
- 31.Le Page, F., A. Klungland, D. E. Barnes, A. Sarasin, and S. Boiteux. 2000. Transcription coupled repair of 8-oxoguanine in murine cells: the ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl. Acad. Sci. USA 97:8397-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber, M. R. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19:233-240. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 34.Lindahl, T., J. A. Gally, and G. M. Edelman. 1969. Deoxyribonuclease IV: a new exonuclease from mammalian tissues. Proc. Natl. Acad. Sci. USA 62:597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludwig, D. L., M. A. MacInnes, Y. Takiguchi, P. E. Purtymun, M. Henrie, M. Flannery, J. Meneses, R. A. Pedersen, and D. J. Chen. 1998. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res. 409:17-29. [DOI] [PubMed] [Google Scholar]
- 36.Matsuzaki, Y., N. Adachi, and H. Koyama. 2002. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 30:3273-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minowa, O., T. Arai, M. Hirano, Y. Monden, S. Nakai, M. Fukuda, M. Itoh, H. Takano, Y. Hippou, H. Aburatani, K. Masumura, T. Nohmi, S. Nishimura, and T. Noda. 2000. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. USA 97:4156-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montecucco, A., R. Rossi, D. S. Levin, R. Gary, M. S. Park, T. A. Motycka, G. Ciarrocchi, A. Villa, G. Biamonti, and A. E. Tomkinson. 1998. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 17:3786-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murante, R. S., L. Rust, and R. A. Bambara. 1995. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 270:30377-30383. [DOI] [PubMed] [Google Scholar]
- 41.Nilsen, H., I. Rosewell, P. Robins, C. F. Skjelbred, S. Andersen, G. Slupphaug, G. Daly, H. E. Krokan, T. Lindahl, and D. E. Barnes. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell 5:1059-1065. [DOI] [PubMed] [Google Scholar]
- 42.Osterod, M., E. Larsen, F. Le Page, J. G. Hengstler, G. T. Van Der Horst, S. Boiteux, A. Klungland, and B. Epe. 2002. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene 21:8232-8239. [DOI] [PubMed] [Google Scholar]
- 43.Parenteau, J., and R. J. Wellinger. 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podlutsky, A. J., I. I. Dianova, V. N. Podust, V. A. Bohr, and G. L. Dianov. 2001. Human DNA polymerase beta initiates DNA synthesis during long-patch repair of reduced AP sites in DNA. EMBO J. 20:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrank, B., R. Gotz, J. M. Gunnersen, J. M. Ure, K. V. Toyka, A. G. Smith, and M. Sendtner. 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA 94:9920-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, S. A., and B. P. Engelward. 2000. In vivo repair of methylation damage in Aag 3-methyladenine DNA glycosylase null mouse cells. Nucleic Acids Res. 28:3294-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash, and L. Prakash. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′-to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270:4193-4196. [DOI] [PubMed] [Google Scholar]
- 49.Spiro, C., R. Pelletier, M. L. Rolfsmeier, M. J. Dixon, R. S. Lahue, G. Gupta, M. S. Park, X. Chen, S. V. Mariappan, and C. T. McMurray. 1999. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell 4:1079-1085. [DOI] [PubMed] [Google Scholar]
- 50.Sugo, N., Y. Aratani, Y. Nagashima, Y. Kubota, and H. Koyama. 2000. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 19:1397-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takao, M., S. Kanno, T. Shiromoto, R. Hasegawa, H. Ide, S. Ikeda, A. H. Sarker, S. Seki, J. Z. Xing, X. C. Le, M. Weinfeld, K. Kobayashi, J. Miyazaki, M. Muijtjens, J. H. J. Hoeijmakers, G. van der Horst, A. Yasui, and A. H. Sarker. 2002. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 21:3486-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tebbs, R. S., M. L. Flannery, J. J. Meneses, A. Hartmann, J. D. Tucker, L. H. Thompson, J. E. Cleaver, and R. A. Pedersen. 1999. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 208:513-529. [DOI] [PubMed] [Google Scholar]
- 53.Tom, S., L. A. Henricksen, M. S. Park, and R. A. Bambara. 2001. DNA ligase I and proliferating cell nuclear antigen form a functional complex. J. Biol. Chem. 276:24817-24825. [DOI] [PubMed] [Google Scholar]
- 54.Varmuza, S., V. Prideaux, R. Kothary, and J. Rossant. 1988. Polytene chromosomes in mouse trophoblast giant cells. Development 102:127-134. [DOI] [PubMed] [Google Scholar]
- 55.Waga, S., and B. Stillman. 1994. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 369:207-212. [DOI] [PubMed] [Google Scholar]
- 56.Wu, X., J. Li, X. Li, C. L. Hsieh, P. M. Burgers, and M. R. Lieber. 1996. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 24:2036-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, X., T. E. Wilson, and M. R. Lieber. 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xanthoudakis, S., and T. Curran. 1992. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 11:653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xanthoudakis, S., R. J. Smeyne, J. D. Wallace, and T. Curran. 1996. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl. Acad. Sci. USA 93:8919-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]