Abstract
Transcription of the Escherichia coli osmC gene is induced by several stress conditions. osmC is expressed from two overlapping promoters, osmCp1 and osmCp2. The proximal promoter, osmCp2, is transcribed at the entry into the stationary phase by the σs sigma factor. The distal promoter, osmCp1, is activated by NhaR and RcsB. NhaR is a positive regulator of the LysR family and is known to be an activator of the nhaA gene encoding an Na+/H+ antiporter. RcsB is the response regulator of the RcsCDB His-Asp phosphorelay signal transduction system. Genetic data indicated that activation of osmCp1 by both NhaR and RcsB requires the same short sequences upstream of the −35 region of the promoter. Accordingly, DNase I footprint analysis indicated that both activators protect an overlapping region close to the −35 box of the promoter and suggested that the regulatory effect is direct. Despite the overlap of the binding sites, each activator acts independent of the other and is specific for a particular stress. NhaR can stimulate osmCp1 in response to an osmotic signal even in the absence of RcsB. RcsB is responsible for the induction of osmCp1 by alteration of the cell envelope, even in the absence of NhaR. osmCp1 as an example of multiple-stress-responsive promoter is discussed in light of a comparison of the NhaR and RcsB target regions in the Enterobacteriaceae.
When subjected to an environmental stress, bacterial cells induce specific families of genes that contribute to better survival under adverse conditions. Many of these genes are induced by several stresses through the action of a number of transcriptional regulators that bind to specific sites in the vicinity of stress-inducible promoters. In Escherichia coli, osmC is an example of a multiple-stress-responsive gene, and we are analyzing the regulation of this gene in order to better understand the complex interplay of regulators resulting in the multiple stress response. osmC belongs to a family of genes that are highly conserved in gram-positive and -negative bacteria that code for putative envelope proteins required for resistance to organic peroxides and long-term survival in the stationary phase (2, 7, 22, 34). In E. coli, osmC is induced by high osmolarity during the transition to the stationary phase and by the weak acids acetate and salicylate (1, 12, 15, 27). It is transcribed from two overlapping promoters, osmCp1 and osmCp2 (12, 15). The proximal promoter, osmCp2, is mainly recognized by σS and is responsible for growth phase regulation of the gene. In minimal medium, this promoter is also activated by the leucine-responsive protein and is repressed by the nucleoid-associated protein H-NS. Transcription from the distal promoter, osmCp1, is independent of σS and is repressed by both the leucine-responsive protein and H-NS in minimal medium (4). Recently, the following two positive regulators of osmCp1 transcription have been identified: NhaR and RcsB (8, 33). NhaR is a positive regulator belonging to the LysR family and was previously identified as an activator of nhaA, a gene encoding an Na+/H+ antiporter (6, 9, 23, 28). NhaR is necessary for the osmotic induction of osmCp1 by NaCl, LiCl, and, to a lesser extent, the nonionic solute sucrose (33). RcsB is the response regulator of the RcsCDB His-Asp phosphorelay system, which was initially identified as a regulator of the synthesis of the capsular polysaccharide in E. coli (13). RcsB has been shown to be necessary for induction of osmCp1 by the cationic amphipathic molecule chlorpromazine (CPZ) (7). Deletion analysis indicated that a short region upstream from the −35 box was sufficient for stimulation of osmCp1 by overproduction of NhaR or RcsB (8, 33). RcsB directly activated osmCp1 in in vitro transcription experiments and stimulated the binding of RNA polymerase to the promoter, but direct binding of RcsB to its DNA target has not been demonstrated so far (8). In this paper we show that RcsB and probably also NhaR are able to directly bind to overlapping sites next to osmCp1. We also show that the two activators act independent of each other to induce osmCp1 in response to specific signals.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. All of the strains were derived from E. coli K-12 wild-type strain MG1655 (3).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| OR100 | melBLid ΔlacZY nhaR1::kan | 28 |
| MZ60 | φ(cps-lacZ) rcsB::tet | 36 |
| CF6343 | MG1655 ΔlacIZ(MluI) | M. Cashel |
| CLG723 | CF6343 φ(malP-lac)a | Laboratory collection |
| CLG771 | CF6343 φ[osmCp1−p2+(7E)-φ(malP-lac)]b | This study |
| CLG772 | CF6343 φ[osmCp1+p2−(7E)-φ(malP-lac)]c | This study |
| CLG806 | CLG772 nhaR1::kan | This study |
| CLG805 | CLG772 resB::tet | This study |
| CLG809 | CLG772 nhaR1::kan rcsB::tet | This study |
| Plasmids | ||
| pCG302 | pBR322 carrying a 1-kbp osmC+ fragment | 15 |
| pAPT110 | p15A derivative carrying lacUV5 promoter, Kanr/Sprd | 26 |
| pAPTnhaR | pAPT110 derivative carrying nhaR | 33 |
| pAPT-6His-nhaR | pAPT110 derivative carrying nhaR with an N-terminal six-histidine tag | 33 |
| pHRcsB | pFAB1, pAPT110 derivative carrying rcsB with an N-terminal six-histidine tag | 5 |
| pHRcsBD56E | pAPT110 derivative carrying rcsBD56E with an N-terminal six-histidine tag | 8 |
The notation indicates a transcriptional fusion between the intact lacZ and lacY genes and the malP gene. In CLG723, this fusion is transcribed under control of the malP promoter.
In this transcriptional fusion, the malP-lac hybrid operon is transcribed under control of a 138-bp DNA fragment carrying the osmC promoter region with the osmCp11 mutation, which inactivates osmCp1 (12). 7E refers to utilization of oligonucleotide OsmC7E (see Materials and Methods) to obtain the osmC promoter DNA fragment by PCR amplification.
In this fusion, the transcription is directed by the same DNA fragment, but it carries the osmCp21 mutation, which inactivates osmCp2 (12).
Kanr, kanamycin resistant; Spr, spectinomycin resistant.
Culture conditions and enzyme assay.
Cells were grown aerobically at 37°C in Luria-Bertani broth with 0.17 M NaCl (5 g/liter) (LB170) or without NaCl. β-Galactosidase activities were assayed as described by Miller (20).
Genetic procedures.
Strains carrying an osmCp1-lac or osmCp2-lac transcriptional fusion were constructed in strain CLG723, as described previously (33). The 138-bp osmCp DNA fragment controlling transcription of these fusions was obtained by PCR amplification by using the osmC7E sense primer (5′-GGGGGAATTCCCGGTAATCTATTGTGGG-3′) and the osmC3E antisense primer (5′-GGGAATTCGTTGCTCTCCTGTGGGC-3′) along with plasmid templates derived from pCG302 (Table 1) carrying the osmCp11 mutation (osmCp1 osmCp2+) that inactivates osmCp1 or the osmCp21 mutation (osmCp1+ osmCp2) that inactivates osmCp2 (12) (see Fig. 2). Bacterial strains carrying nhaR::kan and strains carrying rcsB::tet were constructed by P1vir transduction as described by Silhavy et al. (31) by using OR100 and MZ60 as donors and selecting for resistance to kanamycin (40 μg/ml) and tetracycline (10 μg/ml), respectively.
FIG. 2.
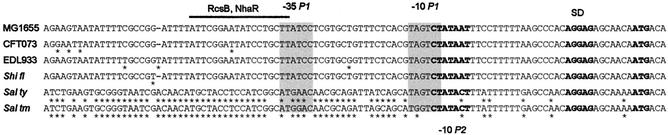
Sequences of the RcsB and NhaR binding sites upstream of osmCp1. The sequence of the osmC promoter region is shown. Solid boxes indicate the −35 and −10 hexanucleotides of osmCp1. The dashed box indicates the −10 region of the proximal promoter osmCp2. Arrows indicate the transcription start sites. The osmCp11 and osmCp21 mutations, eliminating activity of osmCp1 and osmCp2, respectively, are indicated. The dots and arrowheads indicate the sites protected from and hypersensitive to cleavage by DNase I, respectively. The consensus sequence for RcsA-independent RcsB sites is shown above the RcsB sequence.
Preparation of E. coli crude extracts.
Bacterial cells transformed with the vector pAPT110 or derivatives of this vector (pAPTnhaR, pAPT-6His-nhaR, or pHRcsB) were grown at 37°C in LB170 until the optical density at 600 nm was 0.5, and then they were induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h. Cells were harvested by centrifugation, and the pellets were washed in Tris-NaCl buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl) and stored at −70°C. The cells were then washed in B buffer (20 mM HEPES [pH 8.0], 1 mM EDTA, 150 mM NaCl, 7 mM β-mercaptoethanol, 10% [vol/vol] glycerol) and lysed by sonication. The lysates were centrifuged for 1 h at 12,000 × g at 4°C, and the supernatants were each mixed with an equal volume of saturated ammonium sulfate and incubated for 30 min at 4°C. After centrifugation at 12,000 × g at 4°C, the pellets were resuspended in B buffer, and the protein concentrations were adjusted to 1 μg/μl after the total protein contents were assayed with a Bradford protein assay kit (Bio-Rad).
Purification of His6-RcsB and His6-RcsBD56E proteins.
The six-histidine-tagged RcsB and RcsBD56E proteins were purified as described by Carballes et al. (5).
DNase I footprint analysis.
DNase I footprint experiments were performed as described previously (10, 21), with the following modifications. The 188-bp osmC DNA fragment used for footprint analysis was generated by PCR with primers OsmC7 (5′-CCGGTAATCTATTGTGGG-3′) and OsmC14 (5′-GTTTGATATCGCCTTCCC-3′) and contained 49 bp of the osmC coding sequence, as well as 139 bp upstream of the osmC translation start. For DNase I footprint experiments the 32P-end-labeled probe (5 × 104 cpm) was incubated with crude extract or purified protein as indicated below in a 16-μl solution containing 20 mM HEPES (pH 8), 1 mM EDTA, 150 mM NaCl, 7 mM β-mercaptoethanol, 10% (vol/vol) glycerol, and 1 μg of poly(dI-dC)/poly(dI-dC) (Pharmacia). After 5 min of incubation, DNase I (Appligene) was added at a final concentration of 1.25 μg/ml. The digestion was stopped after 3 min by adding 4 μl of a solution containing 1.5 M COONH4, 0.25 M EDTA, and 125 μl of tRNA per ml. After ethanol precipitation, chloroform protein extraction, and another ethanol precipitation, the pellet was resuspended in 4 μl of loading buffer. Digests and their corresponding sequences (obtained with the same labeled primer) were analyzed on 6% denaturing polyacrylamide gels.
Comparison of protein and DNA sequences.
Sequence analysis was performed by using the facilities of the National Center for Biotechnology Information microbial genome database (http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/bact.html) and of the Penn State Computational Molecular Biology Group (http://globin.cse.psu.edu/enterix/) (11).
RESULTS
RcsB and NhaR have overlapping binding sequences.
In order to demonstrate that RcsB and NhaR directly bind to the osmCp1 region and to identify their binding sites, we performed DNase I footprint experiments. To do this, we used a 188-bp DNA fragment that contained the upstream region of osmCp1 and was sufficient for activation by RcsB and NhaR (8, 33).
Neither purified wild-type RcsB (Fig. 1A, lane 2) nor crude extract enriched in RcsB (Fig. 1C, lane 3) resulted in protection against cleavage by DNase I. We then used purified RcsBD56E, a mutant form of the protein in which the conserved aspartate residue at position 56 was replaced by glutamate. This mutation makes the protein more active and is thought to mimic the phosphorylated state of the response regulator (5, 14). RcsBD56E gave a footprint on both DNA strands of the osmCp1 region. The protected sequences extended from position −52 to position −40 on the template strand (Fig. 1A, lanes 3 and 4) and from position −52 to position −37 on the nontemplate strand (Fig. 1C, lane 4). Two sites on the template strand, at positions −48 and −16, appeared to be hypersensitive to DNase I (Fig. 1A). The protected region is in agreement with the location of the RcsB box proposed previously from genetic data (5, 8). The footprint was obtained with 10 μM RcsBD56E (Fig. 1A, lane 4), a concentration that is not high enough to observe an effect with wild-type RcsB (Fig. 1A, lane 2).
FIG. 1.
DNase I protection footprint of NhaR and RcsB in the osmCp1 region. A 188-bp DNA fragment encompassing the osmC promoter region, end labeled with 32P on the strand indicated at the bottom, was incubated with purified RcsB or RcsBD56E protein or with crude extracts enriched or not enriched in RcsB or NhaR protein, digested with DNase I, and analyzed on denaturing polyacrylamide gels. The results of autoradiography of the dried gels are shown. (A) Lane 1, DNA probe alone; lane 2, DNA probe incubated with purified RcsB protein (10 μM); lanes 3 and 4, DNA probe incubated with 100 μM (lane 3) or 10 μM (lane 4) purified RcsBD56E protein. (B) Lane 1, DNA probe alone; lanes 2 and 3, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pAPTnhaR (lane 2) or pAPT110 (lane 3). (C) Lane 1, DNA probe alone; lane 2, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pAPT110; lane 3, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pHRcsB; lane 4, DNA probe incubated with 100 μM purified RcsBD56E protein; lane 5, DNA probe incubated with crude extract (2 μg of protein) of strain CLG772 transformed with pAPTnhaR. The vertical lines and arrowheads indicate the positions protected from and hypersensitive to cleavage by DNase I, respectively.
When the osmCp1 DNA fragment was incubated with a crude extract from a wild-type strain, no protection against limited DNase I digestion was observed (Fig. 1B, compare lanes 1 and 3). In contrast, when the crude extract was prepared from a strain overexpressing NhaR, a footprint was obtained on both DNA strands (Fig. 1B and C), suggesting that NhaR directly binds to the osmCp1 regulatory region. This footprint extended from position −61 to position −27 on the template strand and from position −57 to position −24 on the nontemplate strand. The protected region was limited at both sides by sites where there was enhanced DNase I cleavage. This region included not only the −35 box of osmCp1 but also the RcsB binding site (Fig. 2). However, crude extracts prepared from a strain carrying an rcsB::tet mutation and containing plasmid pAPTnhaR gave the same footprint (data not shown), indicating that the binding of NhaR does not require RcsB.
NhaR and RcsB can independently activate the osmCp1 promoter.
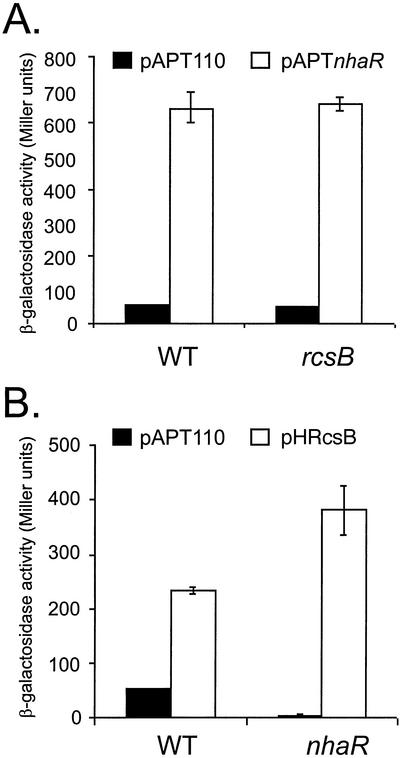
Because RcsB and NhaR act through the same region upstream of osmCp1, we investigated putative functional interference between these two activators in regulating osmCp1. RcsB or NhaR was overproduced in strains with mutations either in nhaR (CLG806) or in rcsB (CLG805), and the expression of a chromosomal osmCp1-lac fusion was monitored (Fig. 3). The 13-fold activation of osmCp1 by overproduction of NhaR was the same in the presence or in the absence of RcsB, indicating that the activity of NhaR is independent of RcsB, in agreement with the footprint data.
FIG. 3.
RcsB and NhaR activate an osmCp1-lacZ transcriptional fusion independent of each other. Strain CLG772 (WT), carrying an osmCp1-lac fusion, and derivatives of this strain with mutations in rcsB (CLG805) (A) and nhaR (CLG806) (B) were transformed with plasmid pAPT110 (vector) (A and B), pAPTnhaR (A), or pHRcsB (B). Overnight cultures of these strains in LB170 containing spectinomycin were diluted 1,000-fold and grown for five generations. They were then diluted 40-fold in prewarmed medium with IPTG (final concentration, 500 μM), and samples were used for β-galactosidase assays after 2 h. The values are the means of the results of three independent experiments.
As shown in Fig. 3B, transcription from osmCp1 was stimulated by overproduction of RcsB both in wild-type and nhaR mutant backgrounds, indicating that RcsB can activate transcription of osmCp1 independent of NhaR. However, we observed approximately twofold-higher expression when RcsB was overproduced in the absence of nhaR. Therefore, NhaR had a negative effect on activation by RcsB, presumably through competition for the same binding site. We also observed that the basal expression was much reduced in an nhaR mutant background, indicating that NhaR is the major regulator of osmCp1 during growth in Luria broth. The combination of a reduced basal level and higher induced expression resulted in an induction rate that increased from 5-fold in the wild type to 100-fold in an nhaR mutant background (Fig. 3B). In control experiments with the isogenic strain CLG771, an osmCp2-lac transcriptional fusion was not affected by overproduction of NhaR or RcsB (data not shown).
NhaR and RcsB are specifically required for induction of osmCp1 by different stress conditions.
In agreement with previous results (33), we observed that NaCl and LiCl induced transcription from osmCp1 and that this induction required nhaR (Table 2). In contrast, the response to NaCl or LiCl was independent of RcsB, because it was identical in rcsB+ and rcsB mutant backgrounds (compare CLG772 and CLG805 in Table 2). The cationic amphipathic molecule CPZ also induces osmCp1, and this activation requires a wild-type rcsB allele (7). In agreement, we observed that induction of osmCp1 by CPZ was essentially eliminated in the absence of RcsB. In contrast, induction by CPZ was still observed in an nhaR mutant background, indicating that the RcsB-dependent response to CPZ does not require NhaR (Table 2). Altogether, these data demonstrate that NhaR and RcsB are able to transduce different regulatory signals to osmCp1 independent of each other. We noted, however, that the activation by CPZ was increased in an nhaR background (92 versus 140 U), indicating that NhaR partially hampers the complete activation of osmCp1 by RcsB.
TABLE 2.
Effects of nhaR and rcsB mutations on osmotic and CPZ-induced stress responses of osmCp1
| Strain (relevant genotype) | β-Galactosidase activity (Miller units)a
|
|||
|---|---|---|---|---|
| No addition | NaCl (0.3 M) | LiCl (0.3 M) | CPZ (0.1 mM) | |
| CLG772 | 18 ± 2 | 70 ± 9 | 77 ± 6 | 92 ± 10 |
| CLG805 (rcsB::tet) | 13 ± 2 | 68 ± 5 | 62 ± 4 | 21 ± 1 |
| CLG806 (nhaR1::kan) | 14 ± 1 | 14 ± 2 | 15 ± 1 | 140 ± 15 |
| CLG809 (rcsB::tet nhaR1::kan) | 5 ± 1 | 7 ± 1 | 5 ± 1 | 5 ± 1 |
Cultures of the strains were grown aerobically in Luria-Bertani broth without NaCl at 37°C. At an optical density at 600 nm of 0.1, NaCl, LiCl, or CPZ was added, and the β-galactosidase activity was assayed after 2 h. The values are the means of the results of two independent experiments for CLG809 and three independent experiments for the other strains.
Organization of RcsB and NhaR target regions in some members of the Enterobacteriaceae.
E. coli strains EDL933 (O157:H7, enterohemorrhagic) (25) and CFT073 (O6:H1, uropathogenic) (35) both have an osmC gene, which encodes a protein that is the same length (143 amino acids) as and is almost identical to the protein of K-12 strain MG1655 (142 of 143 positions are identical). The osmC gene of Shigella flexneri serotype 2a (17) is identical to the osmC gene of MG1655. Salmonella enterica serovar Typhimurium LT2 (19) and S. enterica serovar Typhi CT18 (24) have an identical osmC gene, which encodes a protein with 92% identity to the protein of MG1655 (132 of 143 amino acids are identical). Homologues of NhaR and of the RcsCDB system are also found in all these strains. They exhibit between 85 and 100% identity with the proteins of E. coli MG1655, suggesting that NhaR and RcsB function similarly, if not identically, in these phylogenetically closely related bacteria. In order to see whether regulation of osmC by NhaR and RcsB was also conserved, we compared the osmC promoter regions in these different bacteria (Fig. 4). The three E. coli strains and the S. flexneri serotype 2a strain had very similar sequences in the osmC promoter region, which was indicative of both the conservation of functional osmCp1 and osmCp2 promoters and the conservation of their regulation by NhaR, RcsB, and σS. In contrast, in the two Salmonella strains, despite the conservation of very similar osmC genes, the DNA sequence is completely divergent upstream of the osmCp2 −10 box (Fig. 4), suggesting that osmC regulation in these organisms might be different from that in E. coli and S. flexneri.
FIG. 4.
Alignment of the DNA sequences of the osmCp regions of different members of the Enterobacteriaceae. MG1655, E. coli K-12 strain; CFT073, uropathogenic E. coli O6:H1 strain; EDL933, enterohemorrhagic E. coli O157:H7 strain; Shi fl, S. flexneri serotype 2a; Sal thy, S.enterica serovar Typhi CT18; Sal tm, S.enterica serovar Typhimurium LT2. The stars indicate differences compared with the MG1655 sequence. SD, Shine-Dalgarno sequence.
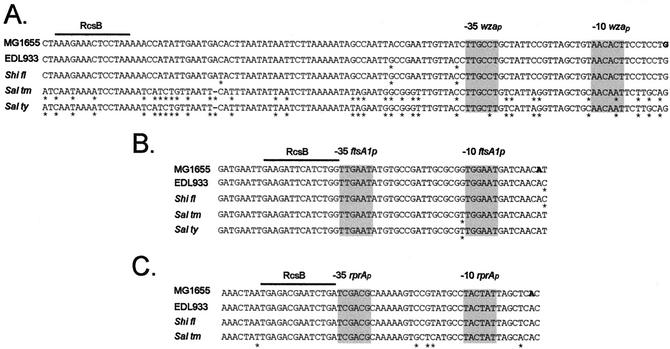
The nhaA promoter is the other known target of NhaR. All the Escherichia, Shigella, and Salmonella strains have closely related nhaA and nhaR genes. A comparison of the nhaA regulatory region (http://globin.cse.psu.edu/enterix; Menteric server) revealed very good conservation of the promoter and of the NhaR binding sites (data not shown). Similarly, comparison of the known RcsB targets at the wza (cps), ftsA1p, and rprAp promoters revealed strong conservation of the putative RcsB binding sites in the Escherichia, Shigella, and Salmonella strains (Fig. 5).
FIG. 5.
Alignment of putative RcsB target sites of different members of the Enterobacteriaceae: regulatory regions of the wza (A), ftsA1p (B), and rprA (C) genes. MG1655, E. coli K-12 strain; EDL933, enterohemorrhagic E. coli O157:H7 strain; Shi fl, S. flexneri serotype 2a; Sal thy, S.enterica serovar Typhi CT18; Sal tm, S.enterica serovar Typhimurium LT2. The stars indicate differences compared with the MG1655 sequence. The −10 and −35 regions of the promoters (shaded background) and the transcription start sites (boldface type) have been described previously (5, 18, 32).
DISCUSSION
Previous work showed that osmCp1, one of the two overlapping promoters of the osmC gene, could be activated by the transcriptional regulators RcsB (8) and NhaR (33). Deletion analysis showed that activation by these two regulators required the same 16-bp DNA sequence upstream of the −35 box of osmCp1, raising the question of possible interference between RcsB and NhaR or even an indirect effect of one regulator through the other. The biochemical and genetic data reported here demonstrate that RcsB and NhaR act independent of each other, probably through direct binding to overlapping sites close to the osmCp1 −35 box.
We previously showed with gel shift assays that RcsBD56E, a constitutively active mutant form of RcsB, potentiated the binding of RNA polymerase to osmCp1, but binding of the activator alone to the RcsB box could not be demonstrated (8). The footprint analysis described here demonstrated that RcsBD56E binds on its own to its target DNA (Fig. 1), and to our knowledge, this is the first time that a footprint has been described with RcsB. It is not clear why binding of RcsBD56E to its target site is not seen in band shift experiments. Possibly, the affinity of binding is too low to obtain a stable complex during the electrophoretic migration. The consensus sequence derived from the comparison of several RcsB target regions was KMRGAWTMWYCTGS (W = A or T; K = G or T; M = A or C; R = A or G; W = A or T; Y = C or T; S = C or G), and this sequence allowed prediction of an RcsB box upstream from osmCp1 (8). The binding site defined here by the DNase I footprint analysis confirms the predicted site; it is centered on position −41/−42, and the protection extends roughly over one turn of the DNA double helix (Fig. 2). We noted that the pattern of protection against DNase I is asymmetric, in agreement with the probable binding of RcsB as a dimer, with each monomer protecting one-half of the site.
A crude extract of wild-type E. coli does not contain enough of any protein to protect the osmCp1 region against cleavage by DNase I in our experimental conditions (Fig. 1). In contrast, an extract enriched in NhaR protects a site that encompasses the −35 box of osmCp1 and includes the region required for regulation by NhaR (33). Although the protected site includes the region protected by RcsB, RcsB is not necessary for the protection observed with extracts enriched in NhaR, and it is likely due to direct binding of NhaR upstream from osmCp1. NhaR belongs to the LysR family, a group of transcription activators characterized by unusually large binding sites (16, 28, 30). Upstream from the nhaA promoter, NhaR protects approximately 90 bp, spanning a compound site composed of three core binding sites separated by positions hypersensitive to DNase I (6). Because almost all the positions in the core binding sites vary from one site to another, no consensus sequence could be defined (6). The site protected at osmCp1 is smaller than that protected at nhaAp, encompassing approximately 30 bp with positions hypersensitive to DNase I on both sides (Fig. 2). The NhaR binding site at osmCp1 is composed of a single core binding site and thus appears to be simpler than the binding site at nhaAp. It is therefore likely that the affinity of NhaR for osmCp1 is weaker than that for nhaAp, explaining why the same His-tagged NhaR variant is able to bind to nhaAp but not to a DNA fragment carrying osmCp1 (6, 8).
osmC is a multiple-stress-responsive gene. Here, we established that induction of this gene by osmotic stress resulting from NaCl or LiCl addition and induction by membrane alteration due to CPZ are accounted for by independent pathways, involving different transcriptional regulators. Activated promoters often result from the association of intrinsically weak promoters and regulatory modules that allow the binding of specific activators that help RNA polymerase initiate transcription (29). During evolution, acquisition of a new module near a promoter should confer the ability to respond to a new specific stress by recruiting a dedicated regulator. However, most activators must be precisely located with respect to RNA polymerase, and this results in a strong constraint on the position of the binding site. In osmCp1, the same short sequence appears to be a bifunctional module that fulfills the requirements for accommodating two different transcriptional activators. Combined with utilization of overlapping alternative promoters, this makes the osmC regulatory region in E. coli K-12 unusually compact and complex. The three E. coli strains and the S. flexneri strain sequenced have almost identical osmC regulatory regions, suggesting that the patterns of regulation of osmC must be similar in all of these strains. In contrast, the Salmonella strains have a very different region (Fig. 4). Analysis of the sequence in Salmonella indicates that there is probably a functional, presumably σS-dependent homologue of osmCp2. In contrast, if there is a functional homologue of osmCp1, it must be located at a different position, because several sequence changes affect positions corresponding to the −10 or −35 regions of osmCp1 and decrease the similarity to the consensus sequence for σ70-dependent promoters. Experimental work is needed to determine the pattern of regulation of osmC in Salmonella, but the analysis of the promoter region suggests that it might be different from that in E. coli. We note that the difference in organization of the target promoters of NhaR or RcsB in E. coli and Salmonella is not a general phenomenon. For instance, comparison of the nhaAp regions revealed very good conservation up to the 5′ limit of the three NhaR binding sites, followed by complete divergence between the Escherichia-Shigella group and the Salmonella strains. Known targets for RcsB also show very good conservation of the RcsB binding sites (Fig. 5), suggesting that again, the regulatory patterns and mechanisms are highly conserved in members of the Enterobacteriaceae. Finally, the very different organizations of the osmC promoter region suggest that acquisition of the osmCp1 promoter and its regulatory cassettes was recent and occurred after separation of the genera Escherichia and Salmonella.
Acknowledgments
We are grateful to G. Duval-Valentin for helpful discussions and to L. Poljak for improvements in the language.
This work was supported in part by a grant from the microbiology program of the Génopôle of Toulouse and benefited from the technical platform of the Institut d'Exploration Fonctionnelle des Génomes (IFR 109).
REFERENCES
- 1.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 4.Bouvier, J., S. Gordia, G. Kampmann, R. Lange, R. Hengge-Aronis, and C. Gutierrez. 1998. Interplay between global regulators of Escherichia coli: effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol. Microbiol. 28:971-980. [DOI] [PubMed] [Google Scholar]
- 5.Carballes, F., C. Bertrand, J. P. Bouche, and K. Cam. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 34:442-450. [DOI] [PubMed] [Google Scholar]
- 6.Carmel, O., O. Rahav-Manor, N. Dover, B. Shaanan, and E. Padan. 1997. The Na+-specific interaction between the LysR-type regulator, NhaR, and the nhaA gene encoding the Na+/H+ antiporter of Escherichia coli. EMBO J. 16:5922-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conter, A., R. Sturny, C. Gutierrez, and K. Cam. 2002. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J. Bacteriol. 184:2850-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davalos-Garcia, M., A. Conter, I. Toesca, C. Gutierrez, and K. Cam. 2001. Regulation of osmC gene expression by the two-component system rcsB-rcsC in Escherichia coli. J. Bacteriol. 183:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dover, N., C. F. Higgins, O. Carmel, A. Rimon, E. Pinner, and E. Padan. 1996. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by NaR and affected by H-NS. J. Bacteriol. 178:6508-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duval-Valentin, G., and R. Ehrlich. 1988. Far upstream sequences of the bla promoter from TN3 are involved in complexation with E. coli RNA-polymerase. Nucleic Acids Res. 16:2031-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florea, L., C. Riemer, S. Schwartz, Z. Zhang, N. Stojanovic, W. Miller, and M. McClelland. 2000. Web-based visualization tools for bacterial genome alignments. Nucleic Acids Res 28:3486-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordia, S., and C. Gutierrez. 1996. Growth-phase-dependent expression of the osmotically inducible gene osmC of Escherichia coli K-12. Mol. Microbiol. 19:729-736. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S., P. Trisler, and A. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupte, G., C. Woodward, and V. Stout. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez, C., and J. C. Devedjian. 1991. Osmotic induction of gene osmC expression in Escherichia coli K12. J. Mol. Biol. 220:959-973. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, J. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Wen, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 19.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1992. A short course in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Mizuno, T. 1987. Static bend of DNA helix at the activator recognition site of the ompF promoter in Escherichia coli. Gene 54:57-64. [DOI] [PubMed] [Google Scholar]
- 22.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padan, E., and S. Schuldiner. 1994. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim. Biophys. Acta 1185:129-151. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 25.Perna, N. T., G. Plunkett 3rd, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 26.Polard, P., and M. Chandler. 1995. An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev. 9:2846-2858. [DOI] [PubMed] [Google Scholar]
- 27.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahav-Manor, O., O. Carmel, R. Karpel, D. Taglicht, G. Glaser, S. Schuldiner, and E. Padan. 1992. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in Escherichia coli. J. Biol. Chem. 267:10433-10438. [PubMed] [Google Scholar]
- 29.Raibaud, O., and M. Schwartz. 1984. Positive control of transcription initiation in bacteria. Annu. Rev. Genet. 18:173-206. [DOI] [PubMed] [Google Scholar]
- 30.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 31.Silhavy, T., M. Berman, and L. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Stout, V. 1996. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J. Bacteriol. 178:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toesca, I., C. Perard, J. Bouvier, C. Gutierrez, and A. Conter. 2001. The transcriptional activator NhaR is responsible for the osmotic induction of osmCp1, a promoter of the stress-inducible gene osmC in Escherichia coli. Microbiology 147:2795-2803. [DOI] [PubMed] [Google Scholar]
- 34.Volker, U., K. K. Andersen, H. Antelmann, K. M. Devine, and M. Hecker. 1998. One of two osmC homologs in Bacillus subtilis is part of the σB-dependent general stress regulon. J. Bacteriol. 180:4212-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch, R. A., V. Burland, G. Plunkett 3rd, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuber, M., T. A. Hoover, and D. L. Court. 1995. Analysis of a Coxiella burnetti gene product that activates capsule synthesis in Escherichia coli: requirement for the heat shock chaperone DnaK and the two-component regulator RcsC. J. Bacteriol. 177:4238-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]