Abstract
Analyses of the F420s present in Methanococcus jannaschii have shown that these cells contain a series of γ-glutamyl-linked F420s capped with a single, terminal α-linked l-glutamate. The predominant form of F420 was designated as α-F420-3 and represented 86% of the F420s in these cells. Analyses of Methanosarcina thermophila, Methanosarcina barkeri, Methanobacterium thermoautotrophicum, Archaeoglobus fulgidus, and Mycobacterium smegmatis showed that they contained only γ-glutamyl-linked F420s.
Coenzyme F420 (Fig. 1) is a name given to a group of redox active cofactors that are presently known to have only a limited distribution among the archaea and high G+C gram-positive bacteria (3). Although playing a crucial role in methanoarchaeal metabolism (4), coenzyme F420 has also been found in various eubacteria such as Streptomyces, Rhodococcus, Nocardioides, and Mycobacterium spp. and their relatives (3). The coenzyme, in fact, was first isolated in 1960 as a cofactor involved in the biosynthesis of chlortetracycline in Streptomyces aureofaciens (18). The coenzyme is presently known to be involved in the biosynthesis of a number of secondary metabolites (20), the degradation of nitroaromatics (5), and activation of nitroimidazofurans (23). Mycobacterium and Nocardia spp. contain an F420-dependent glucose-6-phosphate dehydrogenase (21, 22), and an F420 containing photolyase functions in DNA repair mechanisms in a number of microorganisms (7).
FIG. 1.
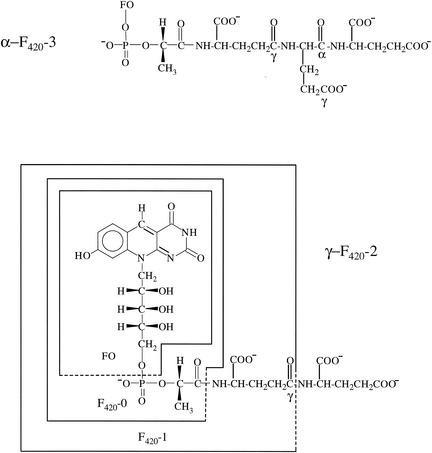
The chemical structures of the F420 coenzyme analogs and their biosynthetic precursors. FO is defined in the text and is the fluorescent chromophore in F420.
The first F420 whose structure was completely characterized was the γ-F420-2 [N-(N-l-lactyl-γ-l-glutamyl)-l-glutamic acid phosphodiester of 7,8-didemethyl-8-hydroxy-5-deazariboflavin] isolated from Methanobacterium thermoautotrophicum (γ-F420-2 in Fig. 1) (6). This F420 contained two glutamic acids, with the terminal glutamate being bound through an amide bond to the γ position of the other glutamate, as is found in most folates and glutathione. This structure will be designated here as γ-F420-2 to indicate the γ attachment of this terminal glutamate to the core structure of F420-1 (N-l-lactyl-γ-l-glutamic acid phosphodiester of 7,8-didemethyl-8-hydroxy-5-deazariboflavin) (Fig. 1). Subsequent work, however, has revealed that most organisms contain a series of γ-polyglutamated F420 cofactors with up to a total of seven γ-linked glutamates (1, 10-12, 17, 19).
The assembly of F420-0 and its polyglutamate derivatives from 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) (Fig. 1), pyruvate, and glutamate requires at least six steps (14, 15). The fourth step in this sequence of reactions is the reaction of lactyl (2) diphospho-(5′) guanosine (LPPG) with FO to form F420-0 (F420 with no glutamic acid) and GMP. The glutamic acid is then added to the lactyl carboxyl group of F420-0 (Fig. 1) to generate the F420-1, which is followed by the repeated addition of single glutamates to generate the polyglutamate derivatives.
As part of our work to establish the genes and pathway involved in F420 biosynthesis in Methanococcus jannaschii (13-15) we examined the F420 species present in this euryarchaeon. These analyses established that the F420s present in these cells are unique and consist of a series of γ-linked F420s capped with a single terminal α-linked l-glutamate.
A cell extract of M. jannaschii was prepared by sonication of 4.67 g of frozen cells suspended in 10 ml of TES extraction buffer {50 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH-10 mM MgCl2 [pH 7.5]} under Ar for 5 min at 3°C. The resulting mixture was centrifuged under Ar (10 min at 27,000 × g) and stored frozen at −20°C until used. The protein concentration of the M. jannaschii extract was 38 mg/ml. Protein concentrations were measured using the BCA total protein assay (Pierce, Rockford, Ill.) with bovine serum albumin as a standard. Similar procedures were used to prepare cell extracts of Methanosarcina thermophila, Methanosarcina barkeri, Methanobacterium thermoautotrophicum ΔH, and Archaeoglobus fulgidus, having protein concentrations of 38, 6.8, 37, and 20 mg/ml, respectively. Mycobacterium smegmatis cell pellet (104 mg) was extracted by heating for 10 min at 100°C with 0.5 ml of water. After the addition of 0.6 ml of methanol and after centrifugation (14,000 × g for 5 min) to remove insoluble material, the resulting clear yellow extract was separated, evaporated, and dissolved in water prior to high-performance liquid chromatography (HPLC) analysis.
F420 present in the cell extracts produced by sonication was isolated by precipitation of the proteins from the cell extracts (50 μl) with the addition of 80 μl of methanol, followed by centrifugation (14,000 × g for 5 min). After separation from the pellet, the clear liquid was diluted to 1 ml with water and 20-μl portions were analyzed by HPLC. Analyses were performed on a Shimadzu SCL-6B HPLC using a C-18 reversed phase column (AXXI-Chrom octyldecyl silane column; 5-μm particle size; 4.6 mm [internal diameter] by 25 cm) eluted isocratically with 15% methanol in 25 mM sodium acetate (pH 6.5) buffer at a flow rate of 0.5 ml per min. The eluent was monitored by fluorescence (excitation wavelength, 420 nm; emission wavelength, 480 nm) and by absorbance at 280 nm. By use of this HPLC method, the different F420 derivatives showed the following retention times: γ-F420-7, 4.88 min; γ-F420-6, 5.04 min; γ-F420-5, 5.84 min; γ-F420-4, 5.99 min; α-F420-5, 6.05 min; α-F420-4, 6.51 min; γ-F420-3, 6.95 min; α-F420-3, 7.41 min; γ-F420-2, 9.07 min; F420-1, 13.0 min; F420-0, 26.5 min; FO-P, 27.8 min; and FO, 30.4 min. The γ-F420s were identified by coinjection with previously characterized γ-F420 samples from Methanosarcina barkeri and Mycobacterium smegmatis (1). Those peaks not corresponding with these known samples were the α-F420s and were identified with regard to the number of glutamates by their elution positions relative to the known samples and by the observed difference between the elution times of F420s that differed by one glutamate.
HPLC analysis of the F420s from M. jannaschii showed that they consisted of four major fluorescent peaks (percent of total fluorescence): γ-F420-2 (2.9%), α-F420-3 (85.9%), α-F420-4 (10.1%), and α-F420-5 (1.5%) (Table 1). The measured total amount of F420 in this organism (>2.0 μmol/g [dry weight]) was among the highest reported for any organism (16). Except for the γ-F420-2 peak, each peak had chromatographic retention times different from that of the γ-linked F420 present in any of the other archaea tested, which included Methanosarcina thermophila, Methanosarcina barkeri, Methanobacterium thermoautotrophicum, Archaeoglobus fulgidus, and Mycobacterium smegmatis. These organisms were found to have the distributions of γ-F420 (percent of total fluorescence) shown in Table 1. No F420-1 was detected in any sample.
TABLE 1.
Distribution and types of F420 in the assayed cells
| F420 | Distribution of F420 type (% of total F420s) in:
|
||||
|---|---|---|---|---|---|
| Methano- coccus jannaschii | Methano- sarcina thermophila | Methano- sarcina barkeri | Archaeo- globus fulgidus | Myco- bacterium smegmatis | |
| γ-F420-2 | 2.9 | 2.4 | 31 | 0.3 | Trace |
| γ-F420-3 | 19 | 6.8 | 0.5 | ||
| α-F420-3 | 85.9 | ||||
| γ-F420-4 | 62 | 32 | 89a | ||
| α-F420-4 | 10.1 | ||||
| γ-F420-5 | 16 | 25 | 67 | ||
| α-F420-5 | 1.5 | ||||
| γ-F420-6 | 1.0 | 4.0 | 10b | 31 | |
| γ-F420-7 | Trace | 0.7 | 7.2 | ||
The sum of γ-F420-4 and -5 was 89%.
The sum of γ-F420-6 and -7 was 10%.
To establish the nature of the linkages in the samples of F420, they were incubated with peptidases of known specificity and the products were measured by HPLC. The enzymes used were carboxypeptidase Y (a peptidyl-l-amino acid hydrolase that specifically removes carboxyl-terminal α-amino acids) and glutamyltranspeptidase and carboxypeptidase G (both γ-glutamyl hydrolases that specifically cleave terminal γ-linked glutamyl peptide bonds). Thus, treatment of 2 μl of M. jannaschii cell extract with 10 μl of a solution of carboxypeptidase Y from baker's yeast (Sigma) (4.35 U in 50 mM TES-Na+-10 mM MgCl2 [pH 7.5]) for 1 h at room temperature converted the sample to a mixture of γ-F420-2, γ-F420-3, and γ-F420-4 based on their HPLC retention times. The abundances of F420s in the resulting mixture matched the ratios of the starting F420s. Treatment of the extracts containing the F420 analogs from the other organisms in the same manner had no effect on their HPLC profiles. Treatment of all of the cell extracts with glutamyltranspeptidase type IV from porcine kidney (Sigma) as previously described (15) had no effect on the M. jannaschii α-F420s but degraded all of the other γ-F420s, including the γ-F420-2 present in M. jannaschii, to F420-1. Treatment of 2 μl of M. jannaschii cell extract with 10 μl of carboxypeptidase G from Pseudomonas spp. (Sigma) (50 U per ml of 50 mM TES-Na+-10 mM MgCl2 [pH 7.5]) for 2 h at room temperature had no effect on the major M. jannaschii α-F420s but did hydrolyze the γ-F420-2 present in M. jannaschii to F420-1. A similar treatment of the F420 samples from the other organisms degraded all of them to F420-0.
For the purification of the F420 analogs from M. jannaschii, cell extracts were separated on a MonoQ HR 5/5 column on a BioLogic HR chromatographic system (Bio-Rad) using a linear sodium chloride gradient. Buffer A was 25 mM Tris-HCl [pH 7.5] buffer, and buffer B was the same but containing 1 M NaCl. The total flow was 0.5 ml per min for 40 min. In this system, the F420 eluted at 0.6 M NaCl and was identified by its fluorescence intensity (excitation, 420 nm; emission, 480 nm). The F420-containing fractions were combined and concentrated and applied to a C-18 column (0.5 by 10 cm; 55-105 μm; Waters) equilibrated with water. The F420 was eluted with 10% methanol in water. The F420-containing fractions were combined and concentrated, formic acid was added to a concentration of 1%, and the sample was applied to a C-18 column (0.5 by 10 cm; 55-105 μm; Waters) equilibrated with 1% formic acid. The F420, which was tightly bound to the column, was then eluted with a step gradient of 1% formic acid-methanol. The F420 was eluted around 50% methanol. The F420, which has no color at this pH, was observed via its fluorescence under a UV light. The final material was then purified by preparative thin-layer chromatography using the solvent system acetonitrile-water-formic acid (80:20:10 [vol/vol/vol]) with a Rf of 0.22. This material was subjected to acid hydrolysis (6 M HCl; 110°C; 12 h), and the resulting amino acids were converted into their N-trifluoroacetyl methyl ester derivatives for gas chromatography-mass spectrometry (GC-MS) analysis. GC-MS analysis as previously described (25) showed only the presence of the glutamate derivative. GC-MS analysis of the sample using an Alpha Dex 120 fused chiral silica capillary column (30 m by 0.25 mm by 0.25-μm film thickness; Supelco, Bellefonte, Pa.) showed that it was composed of only l-glutamic acid.
In total, these data are consistent with the conclusion that the F420s in M. jannaschii, consisting of γ-F420-2, γ-F420-3, and γ-F420-4, each have a single α-linked terminal glutamate, as shown in Fig. 1. These structures will be referred to as α-F420. The cells also contain a small amount of γ-F420-2, which could serve as the biosynthetic precursor to the other structures.
Although no example of α-linked glutamates in F420 has been described in any organism, the occurrence of α-linked glutamates in folates in E. coli (8) and an α-glutamylmethanopterin, sarcinapterin, in Methanosarcina and some Methanococcus species has been described (24). In the case of the folates, it has been established that a bifunctional dihydrofolate synthetase-folylpolyglutamate synthetase (FolC) adds the first glutamate to pteroic acid and then up to three additional γ-linked glutamates. A second enzyme then extends the folylpolyglutamate chain via the addition of α-linked glutamates (9). The enzyme adding these α-linked glutamates was purified and shown to differ from FolC, but the gene for its generation was not identified. It is very likely that the same sequence of events is occurring in M. jannaschii. In M. jannaschii, there could be one enzyme, which would add the glutamate to F420-0 to generate the series of γ-linked glutamate F420 analogs. This process would be analogous to that with the bifunctional FolC. The resulting γ-linked glutamate F420 analogs would then serve as substrates for the addition of the α-linked glutamate. In each case, the addition of the glutamates would likely proceed via acylphosphate intermediates, generated from a nucleotide triphosphate, as is known to occur with FolC (2). The major difference however, between the biosynthesis of the α-linked glutamates in folates and the M. jannaschii F420s is that only one α-linked glutamate would be added.
Since M. jannaschii contains α-linked glutamate F420 analogs, many enzymes in these cells which are known to use the γ-linked glutamate F420 analogs in other bacteria and archaea must have adapted in M. jannaschii to use the α-linked analogs. This list of enzymes for M. jannaschii in which F420 is essential would include hydrogenase, formate dehydrogenase, methylene-tetrahydromethanopterin dehydrogenase, alcohol dehydrogenase, methylene-tetrahydromethanopterin reductase, and F420-dependent NADP+ oxidoreductase. Questions still to be answered are how and why this specific change in coenzyme structure specifically occurred in M. jannaschii and what enzymes are involved.
Acknowledgments
We thank Kim Harich for help with the GC-MS analysis and D. E. Graham for help in editing the manuscript.
National Science Foundation grant MCB 9985712 supported this work.
REFERENCES
- 1.Bair, T. B., D. W. Isabelle, and L. Daniels. 2001. Structures of coenzyme F420 in Mycobacterium species. Arch. Microbiol. 176:37-43. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, R. V., B. Shane, J. J. McGuire, and J. K. Coward. 1988. Dihydrofolate synthetase and folylpolyglutamate synthetase: direct evidence for intervention of acyl phosphate intermediates. Biochemistry 27:9062-9070. [DOI] [PubMed] [Google Scholar]
- 3.Daniels, L. 1993. Biochemistry of methanogenesis, p. 41-112. In M. Kates, D. J. Kushner, and A. T. Matheson (ed.), The biochemistry of the archaea (Archaebacteria). Elsevier, Amsterdam, The Netherlands.
- 4.DiMarco, A. A., T. A. Bobik, and R. S. Wolfe. 1990. Unusual coenzymes of methanogenesis. Annu. Rev. Biochem. 59:355-394. [DOI] [PubMed] [Google Scholar]
- 5.Ebert, S., P. G. Rieger, and H. J. Knackmuss. 1999. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J. Bacteriol. 181:2669-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eirich, L. D., G. D. Vogels, and R. S. Wolfe. 1978. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry 17:4583-4593. [DOI] [PubMed] [Google Scholar]
- 7.Eker, A. P., P. Kooiman, J. K. Hessels, and A. Yasui. 1990. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J. Biol. Chem. 265:8009-8015. [PubMed] [Google Scholar]
- 8.Ferone, R., M. H. Hanlon, S. C. Singer, and D. F. Hunt. 1986. α-Carboxyl-linked glutamates in the folylpolyglutamates of Escherichia coli. J. Biol. Chem. 261:16356-16362. [PubMed] [Google Scholar]
- 9.Ferone, R., S. C. Singer, and D. F. Hunt. 1986. In vitro synthesis of α-carboxyl-linked folylpolyglutamates by an enzyme preparation from Escherichia coli. J. Biol. Chem. 261:16363-16371. [PubMed] [Google Scholar]
- 10.Gorris, L., and C. van der Drift. 1988. Separation and quantification of cofactors from methanogenic bacteria by high-performance liquid chromatography: optimum and routine analysis. J. Microbiol. Methods 8:175-190. [Google Scholar]
- 11.Gorris, L. G., and C. van der Drift. 1994. Cofactor contents of methanogenic bacteria reviewed. Biofactors 4:139-145. [PubMed] [Google Scholar]
- 12.Gorris, L. G., A. C. Voet, and C. van der Drift. 1991. Structural characteristics of methanogenic cofactors in the non-methanogenic archaebacterium Archaeoglobus fulgidus. Biofactors 3:29-35. [PubMed] [Google Scholar]
- 13.Graham, D. E., H. Xu, and R. H. White. 2002. A member of a new class of GTP cyclohydrolases produces formylaminopyrimidine nucleotide monophosphates. Biochemistry 41:15074-15084. [DOI] [PubMed] [Google Scholar]
- 14.Graupner, M., and R. H. White. 2001. Biosynthesis of the phosphodiester bond in coenzyme F420 in the methanoarchaea. Biochemistry 40:10859-10872. [DOI] [PubMed] [Google Scholar]
- 15.Graupner, M., H. Xu, and R. H. White. 2002. Characterization of the 2-phospho-l-lactate transferase enzyme involved in coenzyme F420 biosynthesis in Methanococcus jannaschii. Biochemistry 41:3754-3761. [DOI] [PubMed] [Google Scholar]
- 16.Isabelle, D., D. R. Simpson, and L. Daniels. 2002. Large-scale production of coenzyme F420-5,6 by using Mycobacterium smegmatis. Appl. Environ. Microbiol. 68:5750-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, X. L., and R. H. White. 1986. Occurrence of coenzyme F420 and its γ-monoglutamyl derivative in nonmethanogenic archaebacteria. J. Bacteriol. 168:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick, J. R. D., and G. O. Morton. 1982. Identity of cosynthetic factor I of Streptomyces aureofaciens and fragment FO from coenzyme F420 of Methanobacterium species. J. Am. Chem. Soc. 104:4014-4015. [Google Scholar]
- 19.Peck, M. W. 1989. Changes in concentrations of coenzyme F420 analogs during batch growth of Methanosarcina barkeri and Methanosarcina mazei. Appl. Environ. Microbiol. 55:940-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peschke, U., H. Schmidt, H. Z. Zhang, and W. Piepersberg. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137-1156. [DOI] [PubMed] [Google Scholar]
- 21.Purwantini, E., and L. Daniels. 1996. Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J. Bacteriol. 178:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purwantini, E., T. P. Gillis, and L. Daniels. 1997. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence from Streptomyces and Corynebacterium species and methanogenic archaea. FEMS Microbiol. Lett. 146:129-134. [DOI] [PubMed] [Google Scholar]
- 23.Stover, C. K., P. Warrener, D. R. VanDevanter, D. R. Sherman, T. M. Arain, M. H. Langhorne, S. W. Anderson, J. A. Towell, Y. Yuan, D. N. McMurray, B. N. Kreiswirth, C. E. Barry, and W. R. Baker. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962-966. [DOI] [PubMed] [Google Scholar]
- 24.van Beelen, P., J. F. Labro, J. T. Keltjens, W. J. Geerts, G. D. Vogels, W. H. Laarhoven, W. Guijt, and C. A. Haasnoot. 1984. Derivatives of methanopterin, a coenzyme involved in methanogenesis. Eur. J. Biochem. 139:359-365. [DOI] [PubMed] [Google Scholar]
- 25.Zheng, L., R. H. White, V. L. Cash, and D. R. Dean. 1994. Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33:4714-4720. [DOI] [PubMed] [Google Scholar]