Abstract
Bacteria growing in biofilms often develop multicellular, three-dimensional structures known as microcolonies. Complex differentiation within biofilms of Pseudomonas aeruginosa occurs, leading to the creation of voids inside microcolonies and to the dispersal of cells from within these voids. However, key developmental processes regulating these events are poorly understood. A normal component of multicellular development is cell death. Here we report that a repeatable pattern of cell death and lysis occurs in biofilms of P. aeruginosa during the normal course of development. Cell death occurred with temporal and spatial organization within biofilms, inside microcolonies, when the biofilms were allowed to develop in continuous-culture flow cells. A subpopulation of viable cells was always observed in these regions. During the onset of biofilm killing and during biofilm development thereafter, a bacteriophage capable of superinfecting and lysing the P. aeruginosa parent strain was detected in the fluid effluent from the biofilm. The bacteriophage implicated in biofilm killing was closely related to the filamentous phage Pf1 and existed as a prophage within the genome of P. aeruginosa. We propose that prophage-mediated cell death is an important mechanism of differentiation inside microcolonies that facilitates dispersal of a subpopulation of surviving cells.
Bacteria often switch from a free-living lifestyle to a surface-adapted, multicellular lifestyle known as a biofilm. Bacteria in biofilms become highly differentiated from free-living bacteria (51, 59) and often exhibit a developmental sequence, forming complex, matrix-encased, multicellular structures (microcolonies) which become surrounded by a network of water channels.
The differentiated microcolony phenotype has been found in most bacterial biofilms studied to date, including those of Escherichia coli (10), Pseudomonas aeruginosa (14, 34), and Vibrio cholerae (65). In the cystic fibrosis lung, P. aeruginosa forms prominent microcolony structures (55) and is associated with antibiotic-resistant, often fatal infections (9, 25, 55). However, much remains to be learned about the ecological and physiological roles of microcolonies in biofilms. Cell-cell signaling is involved in microcolony development in at least three organisms, P. aeruginosa (13), Burkholderia cepacia (28), and Aeromonas hydrophila (38). A recent study demonstrated that water channels around P. aeruginosa microcolonies are actively maintained by the quorum-sensing-controlled production of rhamnolipid surfactants (12). Moreover, dispersal of free-living cells from voids inside P. aeruginosa microcolonies has recently been demonstrated (51). Taken together, these data suggest that microcolony formation is a coordinated, adaptive response that facilitates continued biofilm development and dispersal. However, the key processes regulating multicellular differentiation and dispersal inside microcolonies are poorly understood.
It is hypothesized that multicellular interactions among prokaryotes have evolved in surface-associated biofilms (59). Thus, it may be relevant to examine these biofilm bacterial populations for the origins of key multicellular traits that occur in other, more complex organisms. In multicellular organisms, cell death is a normal component of development (18, 43). Indeed, programmed cell death is generally believed to be an adaptation that evolved to meet the specialized needs of multicellular life (48, 57, 63). Increasing evidence suggests that programmed cell death also occurs in bacterial development (19, 35, 56). For example, development of multicellular fruiting bodies in Myxococcus xanthus requires autolysis of a subpopulation of M. xanthus cells (50, 67). Autolysis, which appears to be undesirable for a single-cell organism, may be advantageous for a bacterial population at the multicellular level. Autolysis has not yet been examined within the context of multicellular bacterial biofilm development.
Autolysis has previously been observed in P. aeruginosa isolates (5, 6, 26). In early descriptions of this phenomenon the workers reported bacteriophage plaque-like zones of lysis in P. aeruginosa cultures on agar. More recently, P. aeruginosa mutants that overproduce the antibiotic and quorum-sensing signal molecule 2-heptyl-3-hydroxy-4-quinolone showed pronounced lysis on agar plates (11). However, the molecular mechanism and physiological role of autolysis in P. aeruginosa remain to be elucidated.
In this study, we explored the hypothesis that cell death in P. aeruginosa may function in multicellular biofilm development. We observed that killing and lysis occur in localized regions in wild-type P. aeruginosa biofilms, inside microcolonies, by a mechanism that involves a genomic prophage of P. aeruginosa. We propose that prophage-mediated cell death benefits a subpopulation of surviving cells and has an important role in subsequent biofilm differentiation and dispersal.
MATERIALS AND METHODS
Bacterial strains and culture media.
P. aeruginosa strain PAO1, obtained from B. Iglewski, was used unless indicated otherwise. Additional PAO1 isolates used were ATCC 15692 and strains obtained from the culture collection of the Center for Biofilm Engineering at Montana State University and the culture collection at the University of New South Wales, Sydney, Australia. P. aeruginosa cystic fibrosis isolates were obtained from the Prince of Wales Hospital, Sydney. Batch cultures of P. aeruginosa strains were grown at 37°C with shaking in Luria-Bertani (LB) medium. For cultivation of biofilms, M9 medium containing 48 mM Na2HPO4, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 2 mM MgSO4, 100 μM CaCl2, and 5 mM glucose was used. Twitching and swimming motility assays were carried out in LB medium supplemented with 1 and 0.3% agar, respec-tively, as described elsewhere (49). P. aeruginosa PAO1 rpoN::Gmr (61), pilA::TelrfliM::Gmr, and double pilA::TelrfliM:Gmr (30) mutants were maintained on LB medium containing 30 μg of gentamicin ml−1 and/or 50 μg of sodium tellurite ml−1. P. aeruginosa rhl cell-cell signaling mutants were obtained from H. Schweizer, S. Beatson (4), and D. Ohman (7). A P. aeruginosa lasI signaling mutant (JP1) was obtained from B. Iglewski (46).
Biofilm experiments.
Biofilms were grown in continuous-culture flow cells (channel dimensions, 1 by 4 by 40 mm; flow rate, 150 μl min−1) at room temperature as previously described (45). Channels were inoculated with overnight cultures and incubated without flow for 1 h at room temperature. Bacterial viability was determined by using a BacLight LIVE/DEAD bacterial viability staining kit (Molecular Probes Inc., Eugene, Oreg.). Two stock solutions of stain (SYTO 9 and propidium iodide) were each diluted to a concentration of 3 μl ml−1 in biofilm medium and injected into the flow channels. Live SYTO 9-stained cells and dead propidium iodide-stained cells were visualized with a scanning confocal laser microscope (Olympus) by using fluorescein isothiocyanate and tetramethyl rhodamine isothiocyanate optical filters, respectively.
Free intracellular radicals within biofilms were detected by using dihydrorhodamine 123 (DHR) (Sigma). A solution containing 5 μg of DHR ml−1 in M9 medium was prepared from a stock solution containing 2.5 mg of DHR ml−1 in ethanol. The DHR solution was injected into the flow channels (approximately 0.5 ml per channel), and the flow cells were incubated in the dark for 2 h without flow. Biofilms were viewed without further processing by using an epifluorescence microscope (Leica model DMR) and a rhodamine optical filter.
Bacteriophage experiments.
Bacteriophage capable of superinfecting and killing host P. aeruginosa cells were detected in the fluid effluent from biofilms by using a top-layer agar method (17). Droplets (20 μl) of supernatant were placed onto an LB medium top layer containing 0.8% agar and seeded with P. aeruginosa cells from an overnight culture (1:10, vol/vol; approximately 1 × 108 cells ml−1). The plates were incubated at 37°C overnight. Similarly, to determine phage titers from the biofilm effluent (or other phage preparations), serial dilutions of preparations were made in SM buffer (39) and dropped onto top-layer agar seeded with P. aeruginosa cells.
For preparation of phage stocks, 10-ml overnight P. aeruginosa cultures were each infected with a single plaque, and the infected cultures were seeded into top-layer agar (1:10, vol/vol; final concentration, approximately 1 × 108 cells ml−1). The plates were incubated at 37°C overnight, after which confluent lysis was observed. The top-layer agar was then scraped off, 2 ml of SM buffer was added to each plate, and the mixture was vortexed vigorously for 1 min and incubated at 4°C for 2 h. The phage mixture was then centrifuged at 8,000 × g for 10 min at 4°C, the supernatant (representing the phage stock) was collected and filtered through a 0.2-μm-pore-size syringe filter (Pall Corporation), and the phage titer was determined. The phage stock was stored at 4°C.
For large-scale preparation and purification of phage, LB medium cultures (1 liter) were inoculated with 5 ml of an overnight P. aeruginosa culture and allowed to grow to the mid-log phase (∼1 × 106 CFU ml−1) at 37°C. The cultures were then infected with phage stock at a multiplicity of infection of 10 PFU cell−1 and grown overnight at 37°C. The cultures were then centrifuged twice at 13 000 × g for 20 min, and each supernatant was passed through a 0.45-μm-pore-size filter. DNase I and RNase I (Sigma) were each added to the filtrate to a concentration of 1 μg ml−1 and incubated at room temperature for 2 h. An equal volume of a solution containing 4% (wt/vol) polyethylene glycol 8000 (Sigma) and 2 M NaCl was added to the filtrate with stirring, and the mixture was left to precipitate overnight at 4°C. The supernatant was centrifuged at 11,000 × g for 20 min, and the pellet was resuspended in 8 ml of SM buffer. The phage suspension was then ultracentrifuged at 40,000 rpm by using an SW41Ti rotor (Beckman-Coulter), and the supernatant was discarded. The pellet was resuspended in 8 ml of SM buffer and centrifuged at 7,000 × g for 20 min to remove debris. The supernatant was then subjected to CsCl gradient purification as previously described (70). The phage band was extracted and dialyzed against two changes of buffer containing 50 mM Tris (pH 7.5) and 7 mM MgSO4, and the titer of phage was determined.
To identify the infecting phage, we first carried out a transmission electron microscopic examination of CsCl-purified phage preparations. Carbon Formvar-coated 300-mesh copper grids were dipped into appropriate dilutions of phage preparations for 1 min, negatively stained in 1% uranyl acetate for 30 s, air dried, and then analyzed by using a Hitachi-7000 transmission electron microscope. We then manufactured a digoxigenin-labeled Pf1-specific DNA probe (Roche Diagnostics GmbH) using primers 437F (5′-ACCCGGCGAAAGAGAACTGC-3′) and 437R (5′-CGAGGTTGATGATTTCCGCCG-3′), corresponding to bp 8314 to 8333 and 9188 to 9208 of GenBank accession no. AE004507, respectively. These primers amplify a region of Pf1 open reading frame 437 (24). Top-layer LB agar plates containing 30 to 50 plaques were used for plaque hybridization. Cell lysis, DNA transfer to nylon membranes, and hybridization of the Pf1-specific DNA probe were carried out according to the manufacturer's instructions (Roche Diagnostics GmbH). Hybridization of the probe to individual plaques was carried out against a background of P. aeruginosa genomic DNA. We also used primers 437F and 437R to investigate whether the Pf1-like prophage was present in other laboratory P. aeruginosa PAO1 strains and in P. aeruginosa cystic fibrosis isolates. PCRs were carried out by using genomic DNA extracted from P. aeruginosa isolates as previously described (3).
In order to add back the CsCl-purified phage to P. aeruginosa biofilms, we first grew biofilms for 1 day. After this time, approximately 1 × 107 PFU ml−1 was added aseptically to the reservoir of sterile biofilm medium, and growth of biofilms was allowed to continue. After 3 days of biofilm growth, the biofilms were stained by using a BacLight LIVE/DEAD kit and examined by confocal laser microscopy.
To determine whether DNA damage or reactive oxygen species (ROS) could induce infective bacteriophage from P. aeruginosa cells, different concentrations of mitomycin C (1 to 50 μg ml−1) and hydrogen peroxide (1 to 100 mM) were added to 3-ml aliquots of a mid-log-phase (approximately 1 × 107 CFU ml−1) P. aeruginosa culture. After 2 h of incubation at 37°C, 1-ml aliquots of the cultures were centrifuged at 12,000 × g for 5 min, and the supernatants were passed through a 0.2-μm-pore-size Acro-disk filter (Pall Corporation) to remove the cells. The bacteriophage titer in each filtrate was then determined.
Construction of a P. aeruginosa Pf1::Gmr mutant.
Preparation of chromosomal and plasmid DNA, restriction endonuclease digestion and ligation, and PCRs were carried out by using standard protocols (3). A Pf1::Gmr cassette was constructed by (i) PCR amplifying Pf1 genes from genomic DNA of P. aeruginosa PAO1 with primers Pf1 3 (5′-GACTGAAGAAGAAGCTCGC-3′) and Pf1 4 (5′-TCTGTTCGGTTAGAAGAATTCG), corresponding to bp 5300 to 5318 and 7352 to 7331 of GenBank accession no. AE004507 and amplifying a 2,031-bp region of the Pf1 genome, (ii) cloning the blunted and NcoI-digested 1,946-bp PCR fragment into EcoRI-SmaI-digested pUC18Not (23), resulting in pUC18NotPf1, and (iii) inserting an 800-bp SmaI-digested gentamicin resistance gene into a blunted BstEII site of pUC18NotPf1 41 bp from the start of the gene V helix destabilizing protein of Pf1 (PA0720) to obtain pUC18NotPf1Gm.
The knockout plasmid pCK318Pf1Gm was constructed by cloning the Pf1::Gmr-containing NotI fragment into the NotI site of gene replacement vector pCK318, which carries RP4mob, oriR6K, and the sacB gene for sucrose counterselection in homologous recombination.
The P. aeruginosa Pf1 mutant was constructed by allelic displacement by using triparental mating as described previously (1). Transconjugants with knockout constructs inserted were selected on plates supplemented with gentamicin (60 μg/ml). Subsequent sucrose-based screening for double-crossover mutants was performed as described by Schweizer (52). In order to confirm that the Pf1::Gmr cassette had been inserted into the genome and not into the replicative form (RF) of the phage, we carried out a PCR using primers that recognize both the insertion cassette and the adjacent chromosomal DNA. The primers used were Genta1 (5′-GCGTAACATCGTTGCTGCTG-3′) and PA0716 (5′-CGCAAACCCTTAGTGACTTCC-3′; recognizing the hypothetical P. aeruginosa protein PA0716 and corresponding to bp 4253 to 4273 of GenBank accession no. AE004507).
RESULTS AND DISCUSSION
Cell death and lysis occur within P. aeruginosa microcolonies.
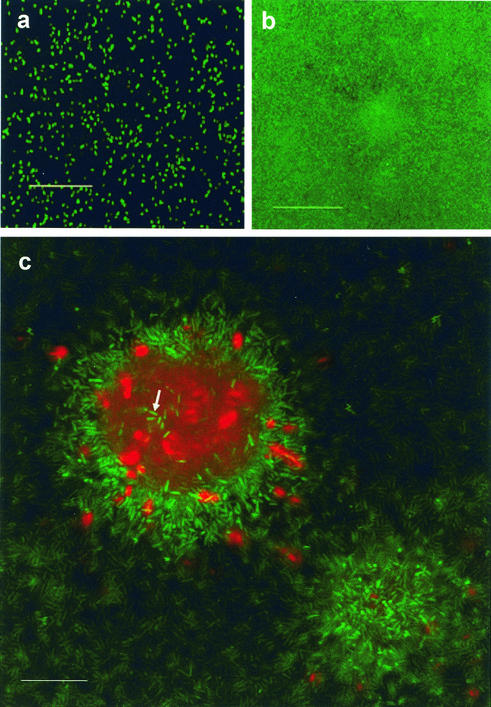
Killing and lysis within P. aeruginosa PAO1 biofilms were observed when they were allowed to develop in glass flow cell reactors for at least 1 week (Fig. 1). By using the BacLight LIVE/DEAD viability probe (Molecular Probes), we observed that cell death occurred with temporal and spatial organization in the biofilm, inside microcolonies. After 12 days of biofilm development (the maximum length of time for which experiments were conducted), up to 50% of the microcolonies in biofilms showed killing and lysis in their centers. We observed dead cells and partially lysed cells, as well as amorphous red propidium iodide-stained material which may have been DNA-containing debris from lysed cells. Subpopulations of cells in these regions that did not die were always observed (Fig. 1c). Recent studies have revealed similar losses of viability inside microcolonies formed by oral bacteria (2, 27), and workers in our laboratory are currently examining cell death that occurs inside microcolonies formed by El Tor and classical strains of V. cholerae and the marine biofilm-forming bacterium Pseudoalteromonas tunicata. Thus, cell death inside microcolonies may be widespread among biofilm-forming bacteria.
FIG. 1.
Cell death occurs in localized regions in microcolonies of mature P. aeruginosa biofilms: confocal micrographs of P. aeruginosa biofilm development in glass flow cells visualized by using the BacLight LIVE/DEAD viability stain. Green fluorescent cells are viable, whereas red fluorescent cells have a compromised cell membrane and are dead. Images were obtained 3 h (a), 3 days (b), and 7 days (c) after the flow cells were inoculated. (a and b) Images taken at the biofilm-substratum interface. Scale bars = 50 μm. (c) x-y plane view through the center of a microcolony that was 50 μm high. A subpopulation of cells inside the microcolony was not killed (arrow). Scale bar = 25 μm. Similar results were obtained in five other experiments.
Genes involved in P. aeruginosa cell death.
Our observations of cell death in P. aeruginosa microcolonies are strikingly similar to the autolysis that is required for the development of fruiting bodies in the social bacterium M. xanthus (50, 68). We hypothesized that genes that control fruiting body development and cell death in M. xanthus may similarly influence P. aeruginosa development. In M. xanthus, development of fruiting bodies is controlled in part by RpoN (21, 22) and by cell-cell signaling (54). In P. aeruginosa, microcolony development is also influenced by RpoN (61) and cell-cell signaling (13). Therefore, to investigate the role of cell death in biofilm development, we examined P. aeruginosa mutants that were defective in rpoN and cell-cell signaling.
Cell death did not occur in biofilms of a P. aeruginosa rpoN mutant strain (Fig. 2b) and could be restored by complementing the rpoN gene in trans (Fig. 2c). The P. aeruginosa cell-signaling systems include the las circuit, which has previously been reported to be involved in biofilm development (13), and the rhl circuit. These signaling systems act through intercellular acylated homoserine lactone signal molecules. We found that a las mutant and two different rhl mutants showed wild-type cell death in biofilms (Fig. 2d and e). Cell death in P. aeruginosa biofilms therefore requires a functional rpoN gene but can occur independent of the P. aeruginosa acylated homoserine lactone cell-signaling circuits.
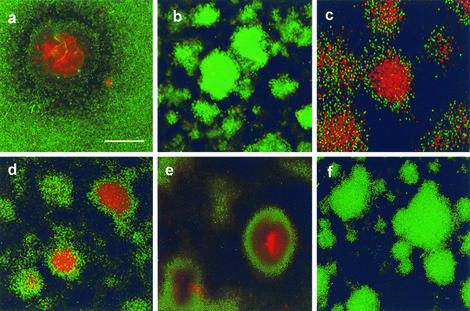
FIG. 2.
Confocal photomicrographs showing viability within microcolonies in 10-day-old biofilms containing the P. aeruginosa wild type (a), rpoN mutant (b), rpoN mutant with rpoN complemented in trans (c), lasI mutant (d), rhlI mutant (S. Beatson) (e), and an fliM pilA double mutant (f). Scale bar = 50 μm. The results are representative of the results of three experiments.
One feature of the rpoN mutant is that it is defective in two types of cell surface structures, type IV pili (T4P) and flagella (62). While screening P. aeruginosa cell-signaling mutants, we found that one rhl mutant (PDO100) was defective in twitching motility, a type of bacterial motility that requires T4P, and swimming motility, which requires flagella. These defects are likely due to secondary mutations (4), but interestingly, this mutant did not exhibit cell death and lysis in biofilms. Thus, all the mutants resistant to biofilm killing were defective in T4P and flagella. We therefore examined biofilm development in P. aeruginosa mutants that specifically lacked T4P (pilA) and flagella (fliM). Both P. aeruginosa pilA and fliM mutants exhibited cell death inside microcolonies that was similar to that of the wild-type strain (data not shown). However, a double pilA fliM knockout mutant did not exhibit cell death inside microcolonies (Fig. 2f). Thus, cell death in P. aeruginosa microcolonies may require either T4P or flagella.
Analysis of the fluid effluent from mature biofilms showed bacteriophage activity.
Flagella and T4P are both common receptors for bacteriophages. We hypothesized that cell death and lysis inside microcolonies may be due to T4P- and flagellum-mediated bacteriophage infection and lysis. We therefore examined the effluent from flow cell biofilms for phage particles capable of superinfecting and killing the P. aeruginosa parent strain. During the onset of cell death and during biofilm development thereafter, effluent from biofilms caused lysis of P. aeruginosa on agar plates (Fig. 3a). However, there was no evidence of lytic phage in the effluent during the first week of biofilm growth. Similarly, infective phage particles were not present in growing or stationary-phase planktonic cultures (data not shown). We diluted the effluent from mature (10-day) biofilms and observed individual plaques with titers of 106 to 108 PFU ml−1. All plaques consisted of a central clear zone of lysis (diameter, <0.5 mm) surrounded by a turbid zone that was 1 to 2 mm in diameter. Only one type of plaque was observed.
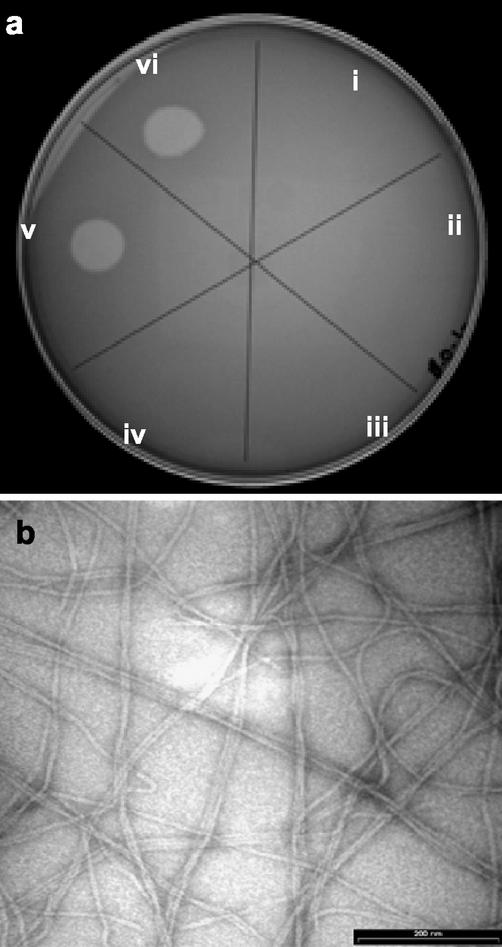
FIG. 3.
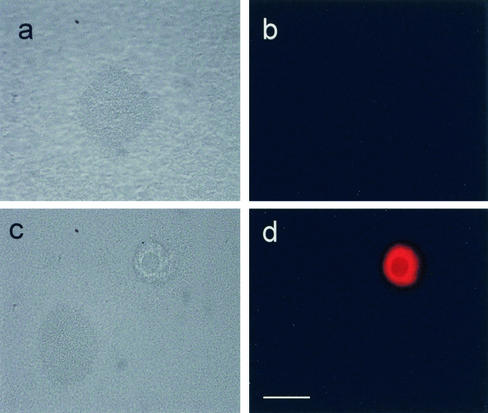
Analysis of bacteriophage activity in the fluid effluents from wild-type P. aeruginosa biofilms. (a) Droplets of biofilm effluent fluids were placed on a lawn of target cells of the P. aeruginosa strain. Fluids from 1-day (sector i), 2-day (sector ii), 3-day (sector iii), 5-day (sector iv), 7-day (sector v), and 9-day (sector vi) biofilms were used. Lysis of P. aeruginosa cells occurred after 1 week, which correlated with the onset of biofilm killing inside microcolonies. (b) Transmission electron microscopy examination of the purified phage revealed filamentous phage particles. Bar = 200 nm.
Because P. aeruginosa rpoN and double pilA fliM mutants did not show cell death in the biofilms, we tested the ability of the phage to infect rpoN, T4P, and flagellar mutants. The phage caused plaque formation on single pilA and fliM mutants in top-layer agar plates, whereas rpoN and double pilA fliM mutants were resistant to the phage in plaque assays (data not shown). Interestingly, we found that biofilms of rpoN and double pilA fliM mutants could also produce phage that formed plaques with the wild-type strain. These plaques had the same morphology as those produced by phage from the wild-type strain. Because these mutants could produce the phage but were resistant to subsequent infection and cell death, our data suggest that the mechanism of cell death in P. aeruginosa biofilms involves infection by the phage and not phage induction.
In order to demonstrate that the phage could kill biofilm cells, we first purified the phage in a preformed CsCl gradient by using ultracentrifugation. The phage formed a single blue band at a density of approximately 1.33 g cm−3 that typically had a titer of 1 × 1010 PFU ml−1. We then added 1 × 107 PFU of the purified phage ml−1 to the biofilm medium. Addition of the phage caused early cell death inside microcolonies and also killed other cells within the biofilm (Fig. 4a and b). Because we had already established that biofilm killing required T4P or flagella, we tested mutants defective in these cell surface structures for sensitivity to the phage in biofilms. The results of these experiments mirrored the results of previous experiments with P. aeruginosa mutants in that single fliM and pilA mutants were sensitive to the phage like the wild-type strain but rpoN and double fliM pilA mutants, which did not show biofilm killing, were resistant to phage added to the biofilm medium (Fig. 4c to f).
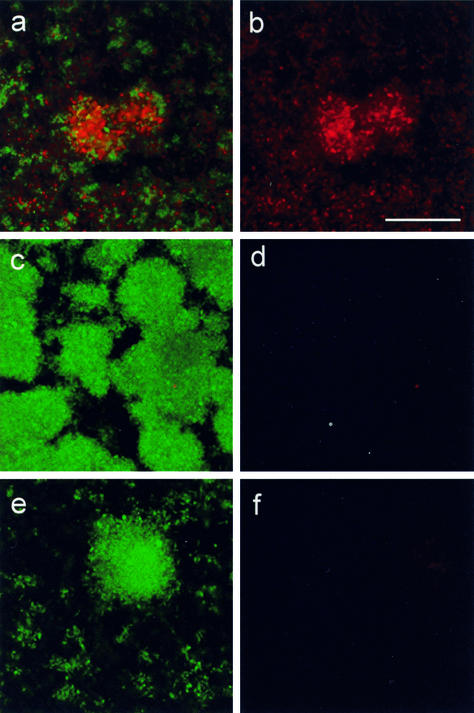
FIG. 4.
Add-back of purified phage to young (3-day) biofilms of P. aeruginosa strains before the onset of normal cell death. (a and b) Wild-type strain; (c and d) rpoN mutant; (e and f) pilA fliM double mutant. Biofilms were visualized by using the BacLight LIVE/DEAD stain after 2 days of growth in the presence of phage. (a, c, and e) Combined live (green) and dead (red) channels; (b, d, and f) corresponding dead channel only. Bar = 50 μm.
A Pf1-like phage is involved in P. aeruginosa biofilm killing.
Electron microscopic examination of the CsCl-purified phage revealed filamentous phage particles that were approximately 1.5 μm long (Fig. 3b). The genome of P. aeruginosa contains a filamentous prophage that is closely related to phage Pf1, and Pf1 genes are known to be upregulated in P. aeruginosa biofilms (66). Moreover, it is known that Pf1 can infect a cell by using T4P (24). Flagella have also been reported to be receptors for filamentous phage (44), and our data suggest that the P. aeruginosa Pf1-like phage may additionally infect a cell through the flagellum. We also carried out PCR with Pf1-specific primers 437F and 437R using DNA extracted from the CsCl-purified phage band. The 894-bp PCR product was sequenced, and the sequence showed 100% identity with the sequence of the Pf1-like prophage from P. aeruginosa. We also hybridized a PCR-labeled, Pf1-specific DNA probe with individual plaques generated from the biofilm effluent, linking the P. aeruginosa Pf1-like phage with host killing.
Many filamentous phages establish a symbiotic state in which continuous replication and release of phage from the cell occur (40, 41). Similarly, with the Pf1-like phage of P. aeruginosa there is continual release of phage particles from cells (66). Our finding that the Pf1-like prophage is involved in biofilm cell death was therefore surprising because (i) filamentous phages are generally thought not to harm host cells and (ii) bacteria harboring prophages (lysogens) are normally immune to superinfection and plaque formation by the phages. Indeed, P. aeruginosa cells harboring the Pf1-like prophage are normally resistant to superinfection by the phage. Intriguingly, however, filamentous phages may exhibit a high frequency of mutation and reversible switching between nonlytic and superinfective, lytic phenotypes (32, 33, 53). Moreover, the first known mechanism of immunity of a filamentous phage has recently been described (8). In this study, immunity-defective phage mutants that could form plaques on lysogenized cells arose spontaneously during normal culture. We propose that similar genetic variations in the Pf1-like filamentous phage result in killing of P. aeruginosa cells. The frequency with which such genetic variations occur in biofilms is probably important in the regulation of cell death and lysis in P. aeruginosa microcolonies. The processes which may increase the frequency of variation in biofilms include adaptive mutation. In E. coli, adaptive mutation is induced by the SOS response (42), which is likely to occur in slowly growing, nutrient-limited populations of cells inside microcolonies (58, 60).
Our observation of phage infection-mediated killing in P. aeruginosa biofilms raises the question of the organization of cell death. I.e., why are most cells in the wall of a microcolony not killed? It is possible that expression of receptors for the phage is developmentally regulated, as is the case for type IV pili in M. xanthus (69). Indeed, RpoN controls morphogenesis and development in M. xanthus and also regulates T4P and flagella in P. aeruginosa (21, 22, 62). T4P have also been reported to be downregulated during P. aeruginosa biofilm development (66). In addition, more subtle regulation of cell death may occur. For example, filamentous phage infection requires the outer membrane protein TolA (29). TolA is both differentially expressed in biofilms (66) and controlled by cell-cell signaling (64). In order to identify genes that are involved in the organization of killing by the Pf1-like phage of P. aeruginosa, workers in our laboratory are currently performing a proteomic analysis of a subpopulation of cells that emerges from P. aeruginosa biofilms and is resistant to the phage.
Additionally, we noted that phage-mediated cell death may have clinical significance. By using the Pf1-specific PCR primers 437F and 437R, which recognize Pf1 open reading frame 437, we detected and confirmed by sequencing the presence of the Pf1-like prophage in each of three P. aeruginosa PAO1 strains commonly used by different laboratories worldwide and in 8 of 11 P. aeruginosa isolates from different cystic fibrosis patients.
In order to examine in more detail the role of the Pf1-like prophage in biofilm development, a P. aeruginosa mutant strain with an insertion in the Pf1-like prophage was generated. DNA replication in all filamentous phages involves the generation of double-stranded circular DNA molecules known as RF molecules, which are stably maintained between generations of bacterial cells (40, 41). We therefore used primers Genta1 and PA0716 (which amplify a region overlapping both the Pf1 prophage and the adjacent chromosomal DNA) and generated a 2,235-bp PCR product, confirming that the gentamicin gene had been inserted into the Pf1 prophage and not the RF. However, the Pf1::Gmr mutant still showed cell death in microcolonies and generated superinfective filamentous phage particles in biofilms with plaque morphology identical to that observed with the wild-type strain. This result is most likely explained by the persistence of wild-type RF molecules within the cell. We found evidence of wild-type RF molecules in the Pf1::Gmr strain by using PCR primers Pf1 3 and Pf1 4, which amplify a region in the phage genome encoding the Gmr insertion. These primers generated two products whose sizes correspond to the size of this region in the mutant prophage (encoding Gmr) and to the size of the same region in the wild-type RF (data not shown). Compared with what is known about the classical lysogenic bacteriophages, such as λ, relatively little is known about the regulation of integration and excision of filamentous prophages. It is possible that dynamic excision of the prophage and insertion of the RF into the chromosome occur, which may hinder inactivation of the prophage. Future studies should involve targeted mutagenesis of the Pf1 insertion site and should be directed towards curing P. aeruginosa of the RF phage.
ROS accumulate inside microcolonies and cause release of phage.
The data presented above suggest a model in which cell death in P. aeruginosa biofilms is due to mutation of the Pf1-like phage, which results in superinfection and killing of P. aeruginosa cells. In order to obtain further details concerning the mechanism of cell death and lysis in biofilms, we hypothesized that conditions which typically induce such a mutation (e.g., bacterial SOS responses and increased recombinase activity) can occur in localized regions inside microcolonies. Physiological conditions that induce the SOS response include DNA damage and the accumulation of ROS, which can damage cellular lipids, proteins, and DNA (15, 20, 31). We therefore examined whether (i) exposure of P. aeruginosa cells to DNA damage and ROS can induce the phage and (ii) ROS can accumulate inside microcolonies and thus provide one possible mechanism of phage release.
Addition of mitomycin C and hydrogen peroxide to planktonic cultures of P. aeruginosa caused the release of phage with plaque morphology identical to that of the phage obtained from biofilm effluent. The titers of PFU liberated from cultures treated with a range of concentrations of mitomycin C were as follows: control (no treatment), no PFU; 1 μg of mitomycin C ml−1, no PFU; 10 μg of mitomycin C ml−1, 15 PFU ml−1; and 50 μg of mitomycin C ml−1, 2 × 103 PFU ml−1. The titers of phage liberated after hydrogen peroxide treatment were as follows: control, no PFU; 0.5 mM hydrogen peroxide, no PFU; 1 mM hydrogen peroxide, 1 × 102 PFU ml−1; and 10 mM hydrogen peroxide, 3 × 103 PFU ml−1. We also demonstrated that ROS can accumulate inside microcolonies. P. aeruginosa biofilms stained with DHR showed localized, bright fluorescence inside microcolonies in mature biofilms but not in young biofilms (Fig. 5). Moreover, bright fluorescence was detected only in microcolonies that had developed internal voids. Microcolonies at this stage of development, after the onset of phage-mediated lysis, increasingly were hollow colonies with less fluorescence in the void spaces (presumably due to the release of ROS and other material from the voids). Microcolonies that had not undergone this differentiation generally did not exhibit fluorescence. These data strongly suggest that prophage-mediated cell death and lysis are correlated with the accumulation of ROS inside microcolonies.
FIG. 5.
Microcolonies in mature P. aeruginosa biofilms can accumulate ROS: phase-contrast images of DHR-stained biofilms (a and c) and the corresponding rhodamine 123 fluorescence displays (b and d). Young (3-day) biofilms (a and b) and mature (10-day) biofilms (c and d) were used. Bar = 50 μm.
Evolutionary and ecological implications.
The importance of cell death in a number of bacterial adaptive responses is well established. In Myxococcus spp. and Bacillus subtilis, autocide-induced killing events occur as part of an adaptive program culminating in sporulation (16, 50, 67, 68). This study demonstrated that cell death also occurs during P. aeruginosa biofilm differentiation and involves a genomic prophage. Parallels between the regulation of prophages and cellular differentiation and developmental processes have been discussed previously (47). More specifically, an evolutionary link between a prophage and differentiation in B. subtilis has been described in detail. SinR, a repressor protein which regulates the master controller of sporulation, Spo0A, in B. subtilis is structurally identical to the repressor protein of a Bacillus bacteriophage in the DNA binding domain (36). Moreover, a number of B. subtilis cell wall endolysins (enzymes that mediate autolysis during sporulation) are derived from bacteriophage lytic enzymes (37). Thus, bacteriophages appear to have played an important role in the evolution of cellular differentiation processes in B. subtilis.
We propose that cell death inside microcolonies is an important physiological event that plays a role in subsequent differentiation and dispersal of a subpopulation of surviving biofilm cells. Recently, a proteomic comparison showed that dispersing cells of P. aeruginosa are more similar to planktonic cells than to mature biofilm cells (51). This suggests that dispersing biofilm cells revert to the planktonic mode of growth. However, the mechanism(s) by which voids are created within microcolonies and by which cells inside disperse is unclear. The data obtained in this study suggest a model in which prophage-mediated cell lysis and cell dispersal may both contribute to void formation inside microcolonies and in which cell death benefits a subpopulation of surviving cells which undergo continued differentiation and dispersal. Our observations may also have medical importance. P. aeruginosa biofilms are linked with chronic infection and mortality in cystic fibrosis patients (9, 25). In the cystic fibrosis lung, regulation of mechanisms that control the susceptibility of cells to prophage-mediated cell death is a possible target for therapeutic control of P. aeruginosa biofilm infections.
Acknowledgments
We thank our colleagues at the University of New South Wales and the Technical University of Denmark for their support and P. Steinberg for comments on the manuscript.
This research was supported by grants from the Leverhulme Trust, United Kingdom, and the Australian Research Council to J.S.W., by a grant from The Villum Kann Rasmussen Foundation to M.G., and by the Centre for Marine Biofouling and Bio-innovation.
REFERENCES
- 1.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auschill, T. M., N. B. Arweiler, L. Netuschil, M. Brecx, E. Reich, A. Sculean, and N. B. Artweiler. 2001. Spatial distribution of vital and dead microorganisms in dental biofilms. Arch. Oral Biol. 46:471-476. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology. Greene Publishing and John Wiley & Sons, New York, N.Y.
- 4.Beatson, S. A., C. B. Whitchurch, A. B. Semmler, and J. S. Mattick. 2002. Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 184:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berk, R. S. 1965. Effect of antibacterial agents on the autoplaque phenomenon of Pseudomonas aeruginosa. Can. J. Microbiol. 11:213-219. [DOI] [PubMed] [Google Scholar]
- 6.Berk, R. S. 1963. Nutritional studies on the “auto-plaque” phenomenon in Pseudomonas aeruginosa. J. Bacteriol. 86:728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, C. M., H. J. Wang, H. J. Bau, and T. T. Kuo. 1999. The primary immunity determinant in modulating the lysogenic immunity of the filamentous bacteriophage cf. J. Mol. Biol. 287:867-876. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 10.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 14.Debeer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structures on oxygen distribution and mass-transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 15.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 16.Doi, R. H. 1989. Sporulation and germination, p. 167-215. In C. R. Harwood (ed.), Bacillus. Plenum Press, London, United Kingdom.
- 17.Eisenstark, A. 1967. Bacteriophage techniques, p. 449-524. In K. Maramorosch and H. Koprowski (ed.), Methods in virology, vol. 1. Academic Press, New York, N.Y.
- 18.Ellis, R. E., J. Y. Yuan, and H. R. Horvitz. 1991. Mechanisms and functions of cell death. Annu. Rev. Cell Biol. 7:663-698. [DOI] [PubMed] [Google Scholar]
- 19.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 20.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorski, L., T. Gronewold, and D. Kaiser. 2000. A σ54 activator protein necessary for spore differentiation within the fruiting body of Myxococcus xanthus. J. Bacteriol. 182:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorski, L., and D. Kaiser. 1998. Targeted mutagenesis of σ54 activator proteins in Myxococcus xanthus. J. Bacteriol. 180:5896-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, D. F., N. J. Short, R. N. Perham, and G. B. Petersen. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349-364. [DOI] [PubMed] [Google Scholar]
- 25.Hoiby, N., H. Krogh Johansen, C. Moser, Z. Song, O. Ciofu, and A. Kharazmi. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3:23-35. [DOI] [PubMed] [Google Scholar]
- 26.Holloway, B. W. 1969. Genetics of Pseudomonas. Bacteriol. Rev. 33:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hope, C. K., D. Clements, and M. Wilson. 2002. Determining the spatial distribution of viable and nonviable bacteria in hydrated microcosm dental plaques by viability profiling. J. Appl. Microbiol. 93:448-455. [DOI] [PubMed] [Google Scholar]
- 28.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson, F., C. A. Borrebaeck, N. Nilsson, and A. C. Malmborg-Hager. 2003. The mechanism of bacterial infection by filamentous phages involves molecular interactions between TolA and phage protein 3 domains. J. Bacteriol. 185:2628-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wildtype, flagella, and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed]
- 31.Konola, J. T., K. E. Sargent, and J. B. Gow. 2000. Efficient repair of hydrogen peroxide-induced DNA damage by Escherichia coli requires SOS induction of RecA and RuvA proteins. Mutat. Res. 459:187-194. [DOI] [PubMed] [Google Scholar]
- 32.Kuo, M. Y., M. K. Yang, W. P. Chen, and T. T. Kuo. 2000. High-frequency interconversion of turbid and clear plaque strains of bacteriophage f1 and associated host cell death. Can. J. Microbiol. 46:841-847. [PubMed] [Google Scholar]
- 33.Kuo, T. T., C. C. Chiang, S. Y. Chen, J. H. Lin, and J. L. Kuo. 1994. A long lytic cycle in filamentous phage Cf1tv infecting Xanthomonas campestris pv. citri. Arch. Virol. 135:253-264. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, R. J., J. A. Brannigan, W. A. Offen, I. Smith, and A. J. Wilkinson. 1998. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J. Mol. Biol. 283:907-912. [DOI] [PubMed] [Google Scholar]
- 37.Loessner, M. J., S. K. Maier, H. Daubek-Puza, G. Wendlinger, and S. Scherer. 1997. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J. Bacteriol. 179:2845-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 39.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Marvin, D. A. 1998. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8:150-158. [DOI] [PubMed] [Google Scholar]
- 41.Marvin, D. A., and B. Hohn. 1969. Filamentous bacterial viruses. Bacteriol. Rev. 33:172-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenzie, G. J., R. S. Harris, P. L. Lee, and S. M. Rosenberg. 2000. The SOS response regulates adaptive mutation. Proc. Natl. Acad. Sci. USA 97:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meier, P., A. Finch, and G. Evan. 2000. Apoptosis in development. Nature 407:796-801. [DOI] [PubMed] [Google Scholar]
- 44.Merino, S., S. Camprubi, and J. M. Tomas. 1990. Isolation and characterization of bacteriophage PM3 from Aeromonas hydrophila, the bacterial receptor for which is the monopolar flagellum. FEMS Microbiol. Lett. 57:277-282. [DOI] [PubMed] [Google Scholar]
- 45.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ptashne, M. 1987. A genetic switch. Blackwell Scientific, Cambridge, United Kingdom.
- 48.Raff, M. C. 1992. Social controls on cell survival and cell death. Nature 356:397-400. [DOI] [PubMed] [Google Scholar]
- 49.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbluh, A., and E. Rosenberg. 1990. Role of autocide AMI in development of Myxococcus xanthus. J. Bacteriol. 172:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 53.Shieh, G. J., Y. C. Charng, B. C. Yang, T. Jenn, H. J. Bau, and T. T. Kuo. 1991. Identification and nucleotide sequence analysis of an open reading frame involved in high-frequency conversion of turbid to clear plaque mutants of filamentous phage Cf1t. Virology 185:316-322. [DOI] [PubMed] [Google Scholar]
- 54.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 55.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 56.Steinmoen, H., E. Knutsen, and L. S. Havarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steller, H. 1995. Mechanisms and genes of cellular suicide. Science 267:1445-1449. [DOI] [PubMed] [Google Scholar]
- 58.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 60.Taddei, F., I. Matic, and M. Radman. 1995. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc. Natl. Acad. Sci. USA 92:11736-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, L. S., J. S. Webb, S. A. Rice, and S. Kjelleberg. 2003. The alternative sigma factor RpoN regulates rhlI expression in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220:187-195. [DOI] [PubMed] [Google Scholar]
- 62.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaux, D. L., G. Haecker, and A. Strasser. 1994. An evolutionary perspective on apoptosis. Cell 76:777-779. [DOI] [PubMed] [Google Scholar]
- 64.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 67.Wireman, J. W., and M. Dworkin. 1977. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 129:798-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wireman, J. W., and M. Dworkin. 1975. Morphogenesis and developmental interactions in myxobacteria. Science 189:516-523. [DOI] [PubMed] [Google Scholar]
- 69.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 179:7748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamamoto, K. R., B. M. Alberts, R. Benzinger, L. Lawhorne, and G. Treiber. 1970. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40:734-744. [DOI] [PubMed] [Google Scholar]