Abstract
The genomes of bacteriophage φ6 and its relatives are packaged through a mechanism that involves the recognition and translocation of the three different plus strand transcripts of the segmented double-stranded RNA genomes into preformed polyhedral structures called procapsids or inner cores. This packaging requires hydrolysis of nucleoside triphosphates and takes place in the order S-M-L. Packaging is dependent on unique sequences of about 200 nucleotides near the 5′ ends of plus strand transcripts of the three genomic segments. Changes in the pac sequences lead to loss of packaging ability but can be suppressed by second-site changes in RNA or amino acid changes in protein P1, the major structural protein of the procapsid. It appears that P1 is the determinant of the RNA binding sites, and it is suggested that the binding sites overlap or are conformational changes of the same domains.
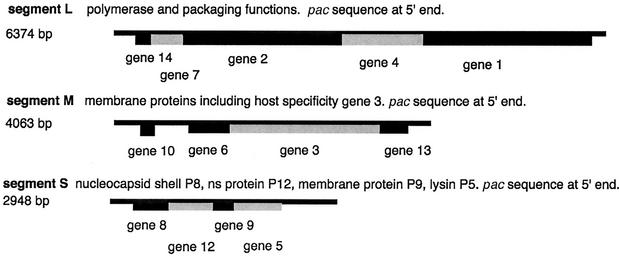
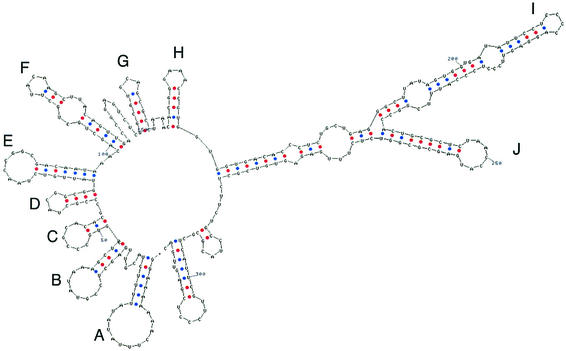
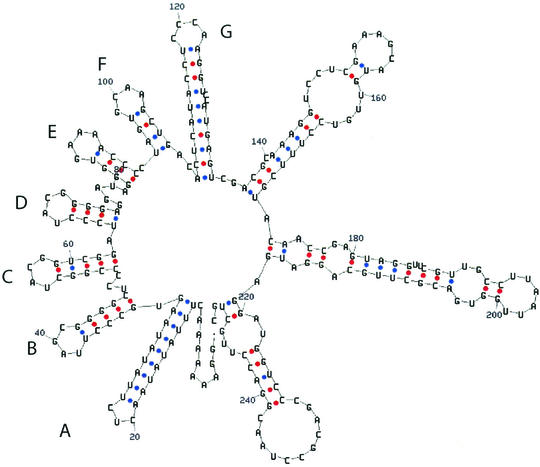
The genomes of bacteriophage φ6 and its relatives are packaged through a mechanism that involves the recognition and translocation of the three different plus strand transcripts of the segmented double-stranded RNA (dsRNA) genomes into preformed polyhedral structures called procapsids or inner cores. This packaging requires hydrolysis of nucleoside triphosphates and takes place in the order S-M-L (15) (Fig. 1). Minus strand synthesis begins after the completion of plus strand packaging (5). The packaging and replication reactions can be studied in vitro with purified components. Packaging of plus strands is dependent on a region called the pac sequence, which begins about 50 nucleotides away from the 5′ 18-base consensus sequence on each plus strand. This region contains approximately 200 nucleotides and has extensive secondary structure (7, 22) (Fig. 2 and 3). There is little or no similarity among the pac sequences of the three genomic segments.
FIG. 1.
Physical map of the genomic segments of bacteriophage φ6.
FIG. 2.
Secondary structure of the 5′ end of the transcript of genomic segment M according to Zuker et al. (13).
FIG. 3.
Secondary structure of the 5′ end of the transcript of genomic segment S according to Zuker et al. (13).
In this study, we prepared plus strands with directed changes in the pac sequences to determine the consequences of these changes on the frequencies of acquisition of the mutated plus strands and to examine the resulting live phages for compensating mutations in RNA or in the composition of proteins of the viral procapsid. These experiments are facilitated by the ability of the virus to acquire plasmid transcripts as replacements for defective genomic segments by simply propagating deletion mutants of the virus on host strains harboring plasmids with complete cDNA copies of the corresponding genomic segment (19).
We had previously found mutants of φ6 that had lost genomic segment S. Procapsids prepared from proteins directed by cDNA copies of segment L did not require the packaging of the plus strand of S in order to package the plus strands of segments M and L. A mutation was found in the gene for protein P1, the major structural protein of the procapsid, that was sufficient to cause this behavior (21).
Purified procapsids are competent to perform in vitro genomic packaging and minus strand synthesis. This facilitates analysis of the consequences of pac sequence alterations (23) because it is possible to study genomic packaging in vitro with procapsids containing proteins with suppressor mutations.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Pseudomonas syringae HB10Y was the host for φ6 (28). Escherichia coli strain JM109 [recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 λ− Δ(lac-proAB)] [F′ traD36 proAB lacIq ΔM15] (29) was used for the propagation of all plasmids and for the production of procapsids. Epicurian Super Competent cells of E. coli (Stratagene) were used for transformation after directed mutagenesis. Plasmids pLM659, pLM656, and pLM682 contain cDNA copies of genomic segments S, M, and L, respectively, in the pT7T319U vector (18). Plasmid pLM1157 contains a cDNA copy of segment L with a deletion of the XhoI fragment from position 545 to position 5366. Its transcript is about 1.5 kb long and is packaged more efficiently than the normal L segment in vitro. Plasmid pKT230 is a shuttle vector that is propagated by E. coli and pseudomonads (1).
Directed mutagenesis.
Oligonucleotides of about 30 bases, listed in Table 1, were prepared by MWG Biotech and used in the Quick Change procedure (Stratagene), which involves PCR on a plasmid template and subsequent treatment with DpnI to eliminate the template DNA.
TABLE 1.
Directed changes in pac sequences of segments S and M
| Oligonucleotide(s) used to change pac | Changes in RNA pac | Segmenta | Acquisition frequency of ch/wt by wild-type phageb | Acquisition frequency of ch/wt by suppressor phagec | RNA changes upon acquisitiond |
|---|---|---|---|---|---|
| olm454/olm455 | G142A | MG | <10−6 wt | 2R, A144G | |
| olm424/olm425 | AC111GU | MF | 0.001 wt | 0 | |
| olm428/olm429 | AC249GU | MJ | 0.01 wt | 0 | |
| olm426/olm427 | GC142AU | MG | <10−6 wt | 0.005 wt | Rcb, R, C143U |
| olm456/olm457 | AC249UG | MJ | 0.1 wt | 0 | |
| olm453 | AC111GA | MF | 0.3 wt | 0 | |
| olm483/olm484 | AA160GG | MH | 10−2 wt | 0 | |
| olm488/olm489 | CCC212UUU | MI | 10−2 wt | 0 | |
| olm490/olm491 | CC120UU | SG | Wt | 0 | |
| olm492/olm493 | GC99AU | SF | <10−6 wt | 0.2 wt | 1/7 U100C |
| olm494/olm495 | AA87GG | SE | Wt | 0 |
Acquisition frequency of changed (ch) pac RNA relative to that of wild-type (wt) RNA by wild-type phage.
Acquisition frequency of changed (ch) pac RNA relative to that of wild-type (wt) RNA by suppressor mutant phage.
Rcb, recombinants; R, revertants; O, acquisition without change.
Preparation of procapsids.
Procapsids were isolated from E. coli JM109. They were purified by sucrose gradient centrifugation from French press lysates produced at 7,000 lb/in2 as described by Gottlieb et al. (8). Purified procapsids were divided into aliquots and frozen at −70°C. Aliquots were thawed immediately prior to use.
In vitro synthesis of radioactive plus sense transcripts by T7 polymerase.
Plasmids derived from pLM659, pLM2502, and pLM1299 produced transcripts identical to the plus strands of wild-type segment S, segment S with GC142AU, and segment S with a four-base deletion at position 62, respectively. They were cut with restriction endonuclease XbaI. The resulting 5′ overhang was removed with mung bean nuclease before transcription with T7 RNA polymerase (18). This procedure generated 5′ and 3′ ends of the transcripts identical to those of the φ6 virus-produced mRNA. The polymerase reaction mixture contained 2 mM each UTP, ATP, GTP, and CTP and 400 μCi of α-32P-labeled UTP per ml. The RNA was purified by filtering through Sephadex G-50 spin columns (Boehringer Mannheim).
RNA polymerase reaction conditions.
Conditions for minus strand synthesis were similar to those reported previously (9), except that the pH of the reaction buffer was 8.9 (5). An equimolar amount of each segment was used, and the total RNA concentration was about 120 μg/ml in a 12.5-μl reaction volume. The reaction was stopped by addition of 3× sample buffer (27) and EDTA, each to a final concentration of 10 mM. The reaction products were analyzed with 1 μg of carrier RNA on 1% agarose gels containing 0.1% sodium dodecyl sulfate in 0.5× Tris-borate-EDTA buffer that were subsequently dried, and the 32P-labeled RNA was visualized after autoradiography with a Cronex enhancing screen.
Packaging reaction conditions.
Frozen purified procapsid preparations were thawed and incubated for 60 min at 28°C in a 12.5-μl packaging reaction mixture consisting of 50 mM Tris-Cl (pH 8.9), 3 or 4 mM MgCl2, 100 mM ammonium acetate, 20 mM NaCl, 5 mM KCl, 5 mM dithiothreitol, 0.1 mM Na2 EDTA, 1 mM ATP, 100 ng of Macaloid, 5% polyethylene glycol 4000, and about 150 ng of [32P]UTP-labeled single-stranded φ6 RNA for each segment. Approximately 1 μg of procapsid was used per reaction mixture. The samples were then treated with 10 U of RNase I (RNase One; Promega) (14) and incubated for 30 min at 28°C. Ten microliters of stop solution (3× sample buffer [27]), 1 μg of carrier RNA, and 25 mM EDTA were added, and the samples were heated at 85°C for 5 min. The samples were then electrophoresed in 1.5 or 2% agarose gels as described above.
RESULTS
Strategy.
Our goal was to prepare single- and double-base changes in the pac sequences of segments S and M that would sabotage normal packaging and to isolate viral mutants that have changed their RNA binding sites so as to be able to accommodate the sequence alterations. A virus that carries a deletion of gene 3 in genomic segment M can be propagated on a host strain carrying a plasmid with the cDNA copy of normal segment M. The deletion virus can be propagated by complementation; however, the virus can also acquire the plasmid transcript as a replacement for the deletion genomic segment (16). This acquisition works at high efficiency even though the 5′ and 3′ ends of the transcript must be trimmed in vivo in order to obtain the correct ends. We have introduced base changes into the pac region of the plasmid and determined the frequency of acquisition of these transcripts by plating the resulting virus on host strains with no plasmid. In some cases, the frequency of acquisition was several orders of magnitude lower than that for a plus strand with the normal pac sequence but the segment was maintained without a suppressor mutation. This was probably due to the lower competitiveness of the altered segment versus that of the wild type. But once acquired, the altered pac gene could be maintained normally. In some cases, the frequency of acquisition of the mutated plasmid transcript was so low that pickup occurred only with deletion phage that had been previously mutagenized with nitrosoguanidine (26). In Table 1, we list the frequency of acquisition relative to that for a segment with the wild-type pac sequence. Reverse transcription-PCR was then performed to clone the 5′ end of segment M so as to determine whether the base sequence remained that of the alteration or whether reversions or suppressor mutations had taken place. In the cases in which the alterations persisted, we then determined whether the virus had actually mutated so as to accommodate the base change. This was done by crossing the virus with a virus that had a deletion in genomic segment L and a segment M that carried the host attachment genes of bacteriophage φ13 (17, 24). Products of the cross were plated on strain LM2509, which does not support the attachment of bacteriophage φ6. These phages were then used to test the reacquisition of the original mutated segment M. If the relative frequency of acquisition was higher than in the first test, the virus was presumed to have a mutation and genomic segment L was subjected to reverse transcription-PCR and the product was sequenced and used to construct procapsids for the testing of in vitro packaging. Plasmids with complete cDNA copies of segment L with the particular suppressor mutations were also used to produce live phages, and these phages were tested for acquisition of the altered pac sequences so as to confirm that the suppressor mutations were sufficient to enable acquisition.
Mutation of the pac sequence of genomic segment M.
Plasmid pLM1053, which contains a cDNA copy of segment M with a deletion of gene 3, was mutagenized with oligonucleotides olm426 and olm427 (Table 1) by the Stratagene Quick-Change protocol. After approximately 12 cycles of PCR, restriction enzyme DpnI was added to destroy the original template. E. coli was transformed, colonies were isolated, and plasmids were prepared. This resulted in a change of GC142AU in the pac sequence of segment M. The mutated DNA was inserted into a plasmid having a cDNA copy of segment M and designated pLM2371, which was then ligated to shuttle vector pKT230 to produce plasmid pLM2379.
Nitrosoguanidine-mutagenized φ2007 (20, 26), which has a deletion of gene 3 in genomic segment M, was used to pick up the transcript of pLM2379. No plaques were obtained when unmutagenized phage was used for the acquisition. After mutagenesis, single plaques were picked and purified and these phages were then propagated on LM2632, which carries a plasmid with normal M pac but the attachment protein genes of φ13. Phages that had acquired the new host range were isolated on LM2509, which lacks the type IV pilus to which φ6 attaches. These phages were then tested for the ability to acquire the transcript of pLM2379.
Mutations in gene 1 were found at A4348G (E133G), C5100A (Q384K), A5101G (Q384R), and A4252G (E101G). It is noteworthy that all of the changes result in a loss of negative charge or an increase in positive charge. Other changes within the pac region of M had less dramatic effects (Table 1). In many cases, the frequency of acquisition was less than that found for wild-type RNA but the altered RNA was acquired without the need for a suppressor mutation in either the RNA or the protein. A two-base change at nucleotides 111 and 112 (olm424) could be packaged by phage that had not mutated. The same was found for changes in J (olm428). A one-base change in G (olm454) gave same-site revertants in two of the pickups and an A-to-G change two bases away in another. This indicates that even one base can cause a significant effect in G. Even with the mutagenized phage, we have found that some of the acquisitions involve changes in the RNA. Of four phages from two plasmids, we found that two lost the A-to-G change but kept the U-to-C change; one lost both changes, and one kept both changes. In the cases in which the changes remained, suppressor mutations were found in gene 1.
Mutation of the pac sequence of genomic segment S.
Plasmid pLM2428, which is a cDNA copy of segment S with a deletion of the 3′ end, was mutagenized with oligonucleotides to change bases in segment S from nucleotide 87 to nucleotide 120 (Table 1). The mutant with the poorest frequency of acquisition was produced by oligonucleotides olm492 and olm493, which caused the GC99AU change. The mutagenized DNA was cut with ScaI and ligated to plasmid pLM2469, which had been cut with ScaI. Plasmid pLM2469 is a derivative of pLM1743, which is a chimera of segments S and M (20) and contains lacα in the BamHI site at N62 to facilitate the screening of ligation products. The resulting plasmids were then ligated to pKT230, a shuttle vector for pseudomonads. Acquisition of the altered transcripts was performed by plating φ2606 on strains carrying the mutant plasmid, pLM2507. φ2606 is a mutant of φ6 that has a chimeric segment of S and M with a deletion in gene 3. The phage was able to acquire transcripts of the plasmid as a replacement of its own chimeric SM segment. The resulting phage was tested by replacing the chimeric segment with a normal S segment and an M segment that carries the pac sequence of M and the attachment genes of φ13. This phage was then used to pick up segment M of φ2007, and the resulting phage was then used to pick up the original mutated chimeric SM segment of plasmid pLM2507. φ2665 is a mutant phage that showed high-frequency pickup of the RNA with the altered pac sequence.
cDNA copies of segment L with suppressor mutations were used to prepare plasmid pLM2564 in LM3027 and plasmid pLM2541 in LM3000. Plasmid pLM2541 was a cDNA copy of segment L with the sequence of gene 1 of φ2665. Plasmid pLM2564 contained gene 4, as well as gene 1 of φ2665. Procapsids prepared from these strains were purified and used to study the packaging behavior for normal and altered plus strands.
The packaging alteration was localized to gene 1, and sequencing of pLM2541 showed that A5119C, which results in E390A, was the only new change in P1. This change was found in an additional number of phage mutants that were able to use the pac GC99AU change. One mutant, cloned in pLM2553, had the A4375G suppressor mutation, which resulted in E142G in P1. Again, we see that the suppressor mutations in P1 result in a loss of negative charge.
In vitro packaging and minus strand synthesis.
Procapsids were prepared from French press lysates of JM109 carrying plasmids with cDNA copies of genomic segment L.
Although the suppressor mutants of the segment M pac changes were able to acquire the altered plus strands about 1,000 times better than wild-type phage could, they still preferred wild-type pac sequences about 1,000 times more. In in vitro packaging or minus strand synthesis, the mutant procapsids showed little more activity than the wild type. It should be noted that although the frequency of acquisition of many of the altered pac plus strands was much lower than that for the wild type in phages that were able to propagate with the altered RNA, the acquisition involves competition between the altered RNA and the resident wild-type RNA produced by the phage used for pickup. Once the RNA is acquired, it no longer has to compete and phage production is virtually normal. This is true for the altered RNAs that are acquired by wild-type phage and for the altered RNAs that are acquired by phage with suppressor mutations in gene 1.
However, the suppressor mutants with the segment S pac changes were much more active in both in vitro packaging and minus strand synthesis (Fig. 4 and 5). The packaging of the plus strand of segment S with the GC99AU change was almost as efficient as that of wild-type S. The minus strand synthesis of an SM chimeric plus strand with the altered pac site was again similar to that seen for wild-type RNA. A striking change in behavior was the competition shown by M versus S in the mutant procapsids. The suppressor mutations in P1 for changes in M pac did not suppress changes in S, and vice versa.
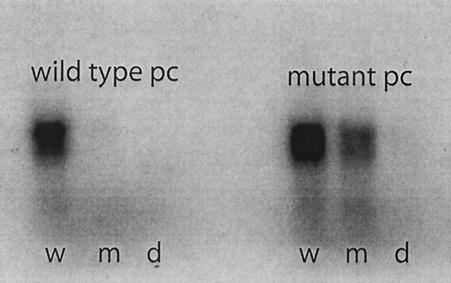
FIG. 4.
Packaging of radioactive plus strands of genomic segment S. The wild-type pac sequence (w) is packaged by both. Altered pac sequence GC99AU (m) is only packaged by mutant procapsid (pc). A pac sequence, pLM1299 (d), that is missing four nucleotides at position 62 is not packaged by either.
FIG. 5.
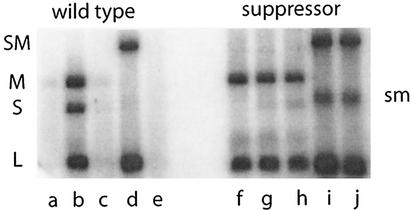
Minus strand synthesis to demonstrate packaging. Wild-type procapsids do not make minus strands when incubated with only M and L (a). With S, M, and L, they do make minus strands (b). With M, L, and an altered S pac sequence, there is no minus strand synthesis (c). There is minus strand synthesis with L and a chimeric segment that includes normal S and M (SM) (d). If the chimera has an altered S pac gene, there is no minus strand synthesis (e). A mutant procapsid that suppresses the poor packaging of the altered S pac site is able to package the altered chimera (j) and to synthesize minus strands as well as it does with the wild-type sequence (i). However; the mutant procapsid is also able to package segment M in the absence of segment S (f). The mutant procapsid shows competition between the plus strand of M and that of S. In lane g, synthesis of the minus strand of S is greatly reduced; however, the response is similar with the plus strand carrying the GC99AU alteration in the pac sequence (h). The band labeled sm is the plus strand transcript of chimeric segment SM.
DISCUSSION
Bacteriophage φ6 contains three dsRNA genomic segments (25). Packaging of the genome involves serial dependent packaging of plus strand transcripts of the segments in the order S-M-L (15). The specificity of packaging is determined on the outside of the procapsid (15). In this study, we attempted to determine which proteins of the procapsid are involved in the determination of packaging specificity. Packaging specificity is determined by sequences of about 200 nucleotides near the 5′ ends of the plus strands (7). These pac sequences show striking secondary structure (15, 22) (Fig. 2 and 3). We made directed changes in the pac sequences of segments S and M and isolated mutants of the phage that are able to accommodate these changes. We found that all of the suppressor mutations are in gene 1 of segment L. Protein P1 is the major structural protein of the procapsid. The procapsid contains 120 molecules of P1, and the arrangement of these molecules is believed to be similar to that found in the members of the families Reoviridae and Totiviridae, two families of dsRNA viruses in eukaryotes (2, 4). Protein P4 is located on the outside of the procapsid but does not seem to be involved in the specificity of binding, although it is the motor for the translocation of the RNA into the procapsid (4, 10). The location of P7 has not been determined, but it does not seem to be involved in the specificity of binding on the basis of the findings in this study and the previous finding that packaging is reduced but specific in procapsids that lack P7 (11, 12). Protein P2, which is the RNA polymerase of φ6, is located inside the particle and was shown previously to have no role in the determination of packaging specificity (11). We had also found previously that mutant forms of φ6 that lack genomic segment S can be isolated and that procapsids prepared from cDNA copies of segment L package plus strands of M and L without the requirement of S and even exclude plus strands of S when those of M and L are present. The mutation responsible for this behavior was found in gene 1 at position C3990G, and it results in the amino acid change R14G (21). This mutant, however, does not package either S or M plus strands with altered pac sequences. Neither in the case of the mutant that excludes S nor in those of the mutants that can package abnormal S and M pac sequences do we know whether the changes are in the binding sites of P1. The changes may modulate the conformation of the protein so as to present a modified binding region.
There is no competition between plus strands of the various genomic segments during normal packaging in φ6. This suggests that the binding sites are unique for each segment. This condition could be attained by having binding sites that are spatially distinct or by having the same site change with the progression of the packaging program. It is of interest that the suppressor mutation shown in Fig. 5 causes competition between the packaging of the plus strand of M with the packaging of the plus strand of S. This suggests that the modification that allows the packaging of S with an altered pac sequence has resulted in a change in the binding site for M, as well as that for S, and that the two sites may overlap.
In the tailed bacteriophages, all of which package their DNA genomes into preformed particles, packaging specificity is determined by a complex called terminase (3). This complex is not usually found to be a structural component of the mature virion. Single-stranded RNA viruses assemble through an interaction of the genomic RNA with a site on the capsid protein that ends up in the interior of the capsid. In those cases, the capsid forms around the RNA (6). The case for φ6 is unique in that the exterior surface of the major structural protein of the procapsid has the binding specificity for the three plus strand precursors of the genomic segments. And it does this through a program of changes in binding specificity that is dependent on orderly filling of the procapsid with plus strand RNA (15).
Acknowledgments
This work was supported by grant GM34352 from the National Institutes of Health.
REFERENCES
- 1.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 2.Day, L. A., and L. Mindich. 1980. The molecular weight of bacteriophage φ6 and its nucleocapsid. Virology 103:376-385. [DOI] [PubMed] [Google Scholar]
- 3.de Beer, T., J. Fang, M. Ortega, Q. Yang, L. Maies, C. Duffy, N. Berton, J. Sippy, M. Overduin, M. Feiss, and C. E. Catalano. 2002. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol. Cell 9:981-991. [DOI] [PubMed] [Google Scholar]
- 4.de Haas, F., A. O. Paatero, L. Mindich, D. H. Bamford, and S. D. Fuller. 1999. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage φ6. J. Mol. Biol. 294:357-372. [DOI] [PubMed] [Google Scholar]
- 5.Frilander, M., P. Gottlieb, J. Strassman, D. H. Bamford, and L. Mindich. 1992. Dependence of minus strand synthesis upon complete genomic packaging in the dsRNA bacteriophage φ6. J. Virol. 66:5013-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golmohammadi, R., K. Fridborg, M. Bundule, K. Valegard, and L. Liljas. 1996. The crystal structure of bacteriophage Q beta at 3.5 A resolution. Structure 4:543-554. [DOI] [PubMed]
- 7.Gottlieb, P., X. Qiao, J. Strassman, M. Frilander, and L. Mindich. 1994. Identification of the packaging regions within the genomic RNA segments of bacteriophage φ6. Virology 200:42-47. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb, P., J. Strassman, D. H. Bamford, and L. Mindich. 1988. Production of a polyhedral particle in Escherichia coli from a cDNA copy of the large genomic segment of bacteriophage φ6. J. Virol. 62:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb, P., J. Strassman, A. Frucht, X. Qiao, and L. Mindich. 1991. In vitro packaging of the bacteriophage φ6 ssRNA genomic precursors. Virology 181:589-594. [DOI] [PubMed] [Google Scholar]
- 10.Gottlieb, P., J. Strassman, and L. Mindich. 1992. Protein P4 of the bacteriophage φ6 procapsid has a nucleoside triphosphate-binding site with associated nucleoside triphosphate phosphohydrolase activity. J. Virol. 66:6220-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb, P., J. Strassman, X. Qiao, A. Frucht, and L. Mindich. 1990. In vitro replication, packaging, and transcription of the segmented double-stranded RNA genome of bacteriophage φ6: studies with procapsids assembled from plasmid-encoded proteins. J. Bacteriol. 172:5774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juuti, J. T., and D. H. Bamford. 1997. Protein P7 of phage φ6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J. Mol. Biol. 266:891-900. [DOI] [PubMed] [Google Scholar]
- 13.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 14.Meador, J., B. Cannon, V. J. Cannistraro, and D. Kennell. 1990. Purification and characterization of Escherichia coli RNase I: comparisons with RNase M. Eur. J. Biochem. 187:549-553. [DOI] [PubMed] [Google Scholar]
- 15.Mindich, L. 1999. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage φ6. Microbiol. Mol. Biol. Rev. 63:149-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mindich, L. 1999. Reverse genetics of the dsRNA bacteriophage φ6. Adv. Virus Res. 53:341-353. [DOI] [PubMed] [Google Scholar]
- 17.Mindich, L., X. Qiao, J. Qiao, S. Onodera, M. Romantschuk, and D. Hoogstraten. 1999. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J. Bacteriol. 181:4505-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olkkonen, V. M., P. Gottlieb, J. Strassman, X. Qiao, D. H. Bamford, and L. Mindich. 1990. In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 87:9173-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onodera, S., X. Qiao, J. Qiao, and L. Mindich. 1995. Acquisition of a fourth genomic segment in bacteriophage φ6: a bacteriophage with a genome of three segments of dsRNA. Virology 212:204-212. [DOI] [PubMed] [Google Scholar]
- 20.Onodera, S., X. Qiao, J. Qiao, and L. Mindich. 1998. Directed changes in the number of dsRNA genomic segments in bacteriophage φ6. Proc. Nat. Acad. Sci. USA 95:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onodera, S., X. Qiao, J. Qiao, and L. Mindich. 1998. Isolation of a mutant that changes genomic packaging specificity in φ6. Virology 252:438-442. [DOI] [PubMed] [Google Scholar]
- 22.Pirttimaa, M. J., and D. H. Bamford. 2000. RNA secondary structures of the bacteriophage φ6 packaging regions. RNA 6:880-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiao, X., G. Casini, J. Qiao, and L. Mindich. 1995. In vitro packaging of individual genomic segments of bacteriophage φ6 RNA: serial dependence relationships. J. Virol. 69:2926-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao, X., J. Qiao, S. Onodera, and L. Mindich. 2000. Characterization of φ13, a bacteriophage related to φ6 and containing three dsRNA genomic segments. Virology 275:218-224. [DOI] [PubMed] [Google Scholar]
- 25.Semancik, J. S., A. K. Vidaver, and J. L. Van Etten. 1973. Characterization of a segmented double-helical RNA from bacteriophage φ6. J. Mol. Biol. 78:617-625. [DOI] [PubMed] [Google Scholar]
- 26.Sinclair, J. F., J. Cohen, and L. Mindich. 1976. The isolation of suppressible nonsense mutants of bacteriophage φ6. Virology 75:198-208. [PubMed] [Google Scholar]
- 27.Studier, F. W. 1973. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J. Mol. Biol. 79:237-248. [DOI] [PubMed] [Google Scholar]
- 28.Vidaver, A. K., R. K. Koski, and J. L. Van Etten. 1973. Bacteriophage φ6: a lipid-containing virus of Pseudomonas phaseolicola. J. Virol. 11:799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]