Abstract
Among the micronutrients required by humans, zinc has particularly divergent modes of action. cDNA microarray and quantitative PCR technologies were used to investigate the zinc responsiveness of known genes that influence zinc homeostasis and to identify, through global screening, genes that may relate to phenotypic outcomes of altered dietary zinc intake. Human monocytic/macrophage THP-1 cells were either acutely zinc depleted, using a cell-permeable zinc-specific chelator, or were supplemented with zinc to alter intracellular zinc concentrations. Initially, genes associated with zinc homeostasis were evaluated by quantitative PCR to establish ranges for fold changes in transcript abundance that might be expected with global screening. Zinc transporter-1 and zinc transporter-7 expression increased when cellular zinc increased, whereas Zip-2 expression, the most zinc-responsive gene examined, was markedly increased by zinc depletion. Microarrays composed of ≈22,000 elements were used to identify those genes responsive to either zinc depletion, zinc supplementation, or both conditions. Hierarchal clustering and ANOVA revealed that ≈5% or 1,045 genes were zinc responsive. Further sorting based on this pattern of the zinc responsiveness of these genes into seven groups revealed that 104 genes were linearly zinc responsive in a positive mode (i.e., increased expression as cellular zinc increases) and 86 genes that were linearly zinc responsive in a negative mode (i.e., decreased expression as cellular zinc increases). Expression of some genes was responsive to only zinc depletion or supplementation. Categorization by function revealed numerous genes needed for host defense were among those identified as zinc responsive, including cytokine receptors and genes associated with amplification of the Th1 immune response.
Keywords: nutrition, genomics, functional genomics, immunology, microarray
As an essential micronutrient, zinc has defined ranges of intake recommended for health of domestic animals and humans. Considerable attention has been focused on the impaired immune function that accompanies zinc deficiency (1, 2). The therapeutic benefit of zinc in the treatment of infectious diseases and associated morbidities has been demonstrated repeatedly in both developed and developing countries (3–5). Recommendations for adequate dietary zinc intake by humans have been recently revised for the United States and Canada (6). That process has also placed a focus on estimates of tolerable upper intakes that appear not to pose a risk to health. These estimates of normal intake and excess intake are based on metabolic indicators that have considerable variance and are used in lieu of defined biochemical indicators. This situation is in contrast to estimates of iron adequacy and excess, which are supported by functional hematological measures and those related to the translational control of ferritin and transferrin receptor synthesis by iron (6, 7). A stimulus for the research presented here relates to the need for development of a series of markers that define zinc function and metabolism, which are sensitive to the dietary intake of this micronutrient.
Zinc has three documented biological roles: catalytic, structural, and regulatory (8). Activities of zinc metalloenzymes are variable in response and have not proven to be uniformly responsive to changes in the dietary intake of zinc (8, 9). Consequently, it is unlikely the catalytic role of zinc can be expected to yield a functional indicator. Similarly, the tight binding dynamics of zinc occupancy for most proteins is such that the structural role of zinc is not likely to produce an indicator based on currently available information. In contrast, the regulatory role of zinc may provide a responsive indicator. Through an interaction with a metal-responsive transcription factor, (MTF-1), zinc has been shown to regulate a number of genes (10, 11). MTF-1-regulated genes include those that influence zinc trafficking, such as metallothionein (MT) and zinc transporter-1 (ZnT-1), and those associated with a variety of genes not related to zinc in an obvious way (10). The embryonic lethality of MTF-1 null mice points to the critical nature of this mode of gene regulation (11–12). An expanding inventory of rodent genes has been shown responsive to a reduction in the intake of dietary zinc (13–16). Many of these changes may occur through mechanisms that do not involve MTF-1 or other direct modes of gene regulation, but, instead, influence mRNA abundance through other signaling pathways and/or feedback loops (17).
The present experiments were undertaken to identify, under controlled conditions, which genes in human cells are responsive to zinc depletion and to zinc excess. For this global microarray screening, we used THP-1 cells, a human monocytic leukemic cell line that we have previously shown is amenable to depletion of zinc with the cell-permeable chelator, N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN), and to zinc supplementation of the medium (18). A human monocytic cell line was chosen in order for the information derived to be directly applicable to a cell type that can readily be obtained by veni-puncture in clinical or field studies involving human subjects. We have previously demonstrated that THP-1 cells respond to zinc in a comparable fashion as peripheral blood mononuclear cells (PBMCs) from human subjects (18).
Our results show a select number of genes (≈5%) are responsive to zinc deprivation or zinc supplementation of THP-1 human mononuclear cells, based on analysis with 22,216 element microarrays. Both down-regulation and up-regulation of gene expression were demonstrated, suggesting, through appropriate signaling processes, zinc can simultaneously act as a stimulator of expression of some genes and a repressor of others. The data provide targets to evaluate the significance of zinc in human biology and a molecular basis for the nutritional assessment of this micronutrient.
Materials and Methods
Cell Culture and Treatments. THP-1 cells (American Type Culture Collection) were cultured in RPMI medium 1640 (Cellgro, Mediatech, Herndon, VA) containing 10% FBS and other additions as described (18). The zinc concentration of the basal medium was ≈3 μM. Cell viability was assessed by trypan blue exclusion. For experiments, cells were grown in T-25 flasks to a density of 6–9 × 105 cells per ml. Supplemental zinc (Zn+) was added as ZnSO4 in sterile water to adjust the zinc concentration to 40 μM. A normal plasma zinc concentration in humans is 15 μM. Zinc depletion (Zn–) was carried out with 10 μM TPEN (Sigma) dissolved in DMSO. As a control, all other cultures contained DMSO (17 μl/8.5 ml of medium). Four hours after the treatments were initiated, cells were collected by centrifugation (300 × g) and lysed in QIA shredder columns, with RNeasy reagents (Qiagen, Valencia, CA) for isolation of total RNA. RNA integrity was verified by ethidium bromide staining after agarose gel electrophoresis (14, 16).
Real-Time Quantitative PCR (Q-PCR). Primer and probe sets were designed for each of the genes assayed by using PRIMER EXPRESS software, version 2.0 (Applied Biosystems) and cDNA sequences were retrieved from GenBank. Primers and fluorescence resonance energy transfer probes were purchased from BioSource International (Camarillo, CA) and are presented in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. The RT-PCR reagents, including the 18S rRNA assay, and the 5700 sequence detection system were from Applied Biosystems. Relative quantitation was performed by using 5-log10 standard curves with 18S rRNA as the normalizer for each sample, and the average of control samples as the calibrator used as described (14). All assays except 18S rRNA used 900 nM primers and 250 nM probe. Before PCR, each total RNA preparation was processed with DNase (DNA-free; Ambion, Austin, TX) to remove any residual DNA.
Fluorescence-Activated Cell Sorter (FACS) Analysis. THP-1 cells, treated as described above, were collected by centrifugation (300 × g), washed two times, and resuspended in appropriate buffers. To assess various stages of the apoptotic pathway, Rhodamine 123 (Molecular Probes) monitored the mitochondrial membrane potential (ΔΨm), active caspase-3 was a marker for full commitment to the apoptotic cascade, and annexin V was used as a marker for externalized phosphotidylserine (BD Biosciences PharMingen, San Diego). Fluorescence from 104 cells per treatment (n = 3) was measured with a FACSort cell sorter and analyzed with CELLQUANT software, version 3.3 (BD Biosciences, San Jose, CA).
Target RNA Preparation, Hybridization, and Microarray Analysis. Total RNA was reverse transcribed (Superscript II, Invitrogen) by using oligo(dT) linked to a T7 RNA polymerase promoter sequence (Proligo, La Jolla, CA) to prime cDNA synthesis. After second-strand synthesis, biotinylated cRNA was produced by in vitro transcription using biotinylated UTP and CTP (Bioarray high-yield RNA transcript labeling kit, Enzo Diagnostics), and purified with RNeasy mini columns (Qiagen). The biotinylated cRNA was heated in a MgOAc/KOAc buffer to produce 35–200 base fragments. Affymetrix (Santa Clara, CA) GeneChip U133A arrays (n = 3 per treatment) were hybridized for 16 h at 45°C with 15 μg of fragmented cRNA. Arrays were stained with a streptavidin-phycoerythrin conjugate (Molecular Probes) and visualized with a GeneArray scanner (Agilent, Palo Alto, CA).
PROBE PROFILER software, version 1.3.11 (Corimbia, Berkeley, CA), was used to convert hybridization intensity data into quantitative estimates of gene expression.
Bioinformatics Analysis. Gene probes that were not expressed in any of the samples (P = 0.05) were considered absent. Absent probes were removed from the dataset and not included in further analyses. Logarithms of the transformed signal values were analyzed with a one-way ANOVA for three replications from each of three treatments. Probes for which there was not a significant treatment effect (P = 0.0001) were removed from the dataset. A pairwise comparison (P = 0.05) of the treatment means was performed with Tukey's honestly significantly different (HSD) test w-procedure. Finally, signal values were normalized by subtracting the mean of a probe across all arrays, from its value on each array and dividing the difference by the standard deviation of values for that gene, thus generating a population with mean of zero and a standard deviation of one. Hierarchical and k-means clustering were performed on the normalized data by using CLUSTER (19). Dendograms and kmeans groups were visualized with TREEVIEW (19). All transformations, filtering, normalization, and statistical analyses were performed with software developed by the Interdisciplinary Center for Biotechnology Research. The statistical package, R, serves as the backend for ANOVA and Tukey's HSD (20) in this software.
Results
Previous experience with TPEN and Zn in the THP-1 cell model has shown that acute Zn– by TPEN chelation results in a mild degree of apoptosis, but Zn+, under the conditions used, does not (18). The data shown in Table 1, using three markers of early apoptosis, show TPEN produces some indication of early stage of progression to apoptosis. The fact that active caspase-3, the apparent committed step in the apoptotic pathway, is the least influenced of the three markers suggests Zn– conditions in TPEN-treated cells produced mild changes early in the apoptotic cascade. That these changes are mild is supported by a uniform viability across treatments as assessed by 7-amino-actinomycin D (7-AAD) fluorescence (data not shown).
Table 1. Flow-cytometric analysis of apoptotic markers in response to Zn- and Zn+ of THP-1 cells.
| Percent apoptotic cells
|
|||
|---|---|---|---|
| Cells | Annexin V | Active caspase-3 | Loss of ΔΨm |
| Control | 5 ± 2 | 1 ± 1 | 3 ± 1 |
| Zn- | 29 ± 11 | 11 ± 5 | 27 ± 6 |
| Zn+ | 5 ± 2 | 2 ± 1 | 3 ± 1 |
Values are means ± SD (n = 3 separate experiments per treatment). Cells were cultured with 10 μM TPEN or 40 μM Zn. DMSO was added to all cultures.
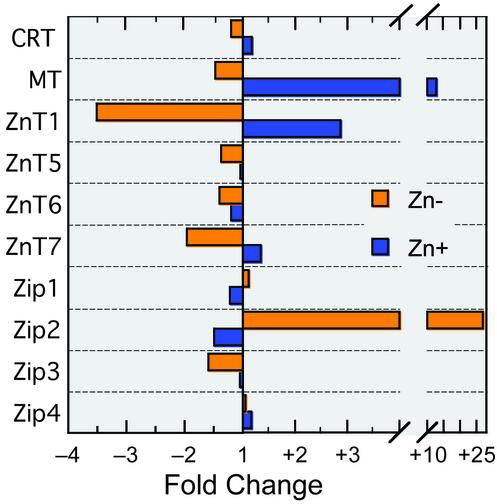
Q-PCR data shown in Fig. 1 present a comprehensive survey of the expression of zinc transporter genes, representing the ZnT and Zip families, and two described zinc-regulated genes (MT and CRT) in response to cellular zinc depletion and supplementation. The purpose of this analysis was to establish, by a very sensitive analytical method, the most likely outer limits expected for zinc regulation of genes. Genes that have been shown to be MTF-1/metal-responsive element (MRE) regulated and those documented as associated with homeostatic control of zinc metabolism were included, as our hypothesis is that these genes will be among those exhibiting the greatest sensitivity to regulation by zinc. In support of that hypothesis, the ZnT-1 and Zip-2 genes were the most sensitive to Zn–, albeit exhibiting decreased and increased relative mRNA quantities, respectively. ZnT-4 expression was below a level detectable by RT-PCR, whereas ZnT-2 and ZnT-3 mRNAs were not measured. These results suggest that Zn– may produce marked changes in expression of genes to at least as much as 27-fold, as in the case of Zip-2. MT-1 and ZnT-1 were the genes most sensitive to Zn+. Similarly, based on MT-1 mRNA levels, a 13-fold increase provided a benchmark for up-regulation.
Fig. 1.
Q-PCR analyses of expression of genes associated with homeostatic control of zinc in Zn– and Zn+ THP-1 cells. Assays were performed in triplicate. Relative quantity calculations used 18S rRNA as the endogenous normalization control. Values shown are means of quantities calibrated to quantities obtained from cells cultured under control conditions.
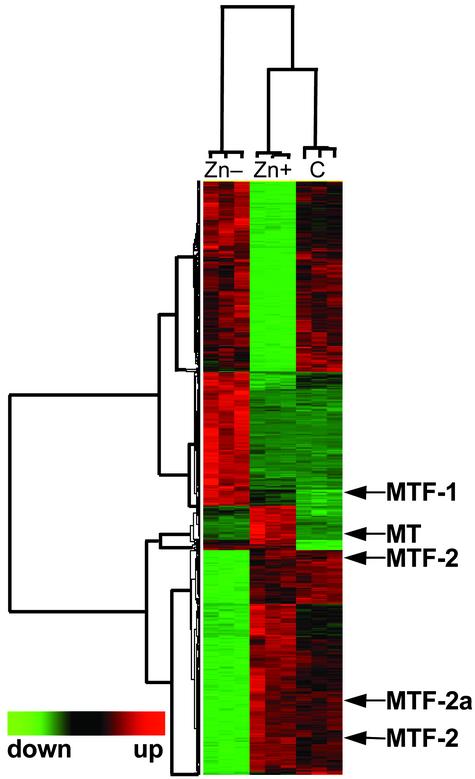
The same conditions of Zn– or Zn+ described for the survey above were used for the microarray analysis. Hybridization signal intensities, derived from >200,000 intensity measurements, were analyzed by ANOVA. Approximately 18,966 genes were considered present in the THP-1 cell RNA. A probability of P < 0.0001 was set to identify those genes most strongly influenced by zinc. These expressed genes were clustered hierarchically, based on the similarity of their normalized expression response. Results from this clustering are presented for each individual microarray chip in Fig. 2. A total of 1,045 genes were identified as either up-regulated or down-regulated by zinc. This total represents somewhat <5% of the genes on these microarrays. Hierarchical clustering of samples suggests that, overall, these genes are more affected by Zn– than by control or Zn+ conditions, as noted in Fig. 2. Of interest is that MT-1 and MTF-1 genes were detected as zinc responsive within this initial analysis. MTF-1 was significantly increased 2.5-fold in Zn– cells. Their mode of expression is predictable within the context of gene regulation by zinc, and such changes in select genes strongly support the methods used. Clustering (Fig. 2) indicated strong but still undefined patterns of gene regulation by zinc.
Fig. 2.
Hierarchal clustering of gene probes based on zinc responsiveness. A total of 1,045 genes expressed in THP-1 cells had transcript levels that were significantly altered at P < 0.0001 based on ANOVA of data from nine individual microarrays. The treatments, Zn–, Zn+, and control (C), are indicated by columns, and probes are indicated by rows. Locations of the MT, MTF-1, and MTF-2 genes are identified by arrows.
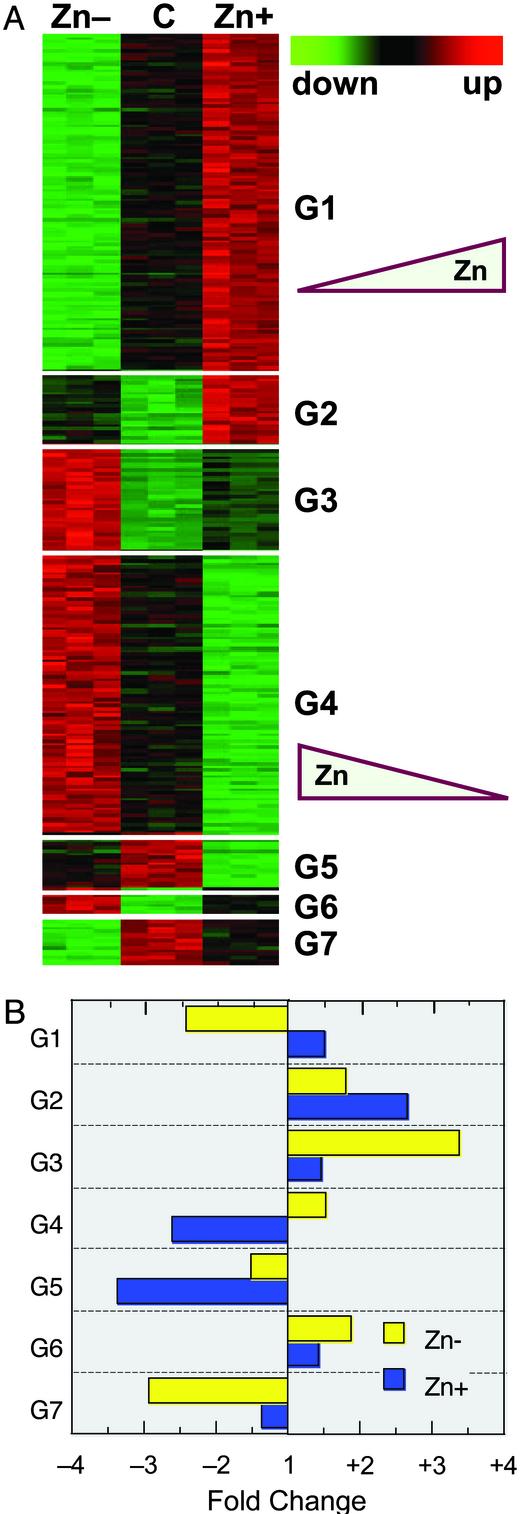
The next level of analytical granularity was achieved through post hoc Tukey's tests. This testing identified 283 genes that were significantly affected by Zn– and Zn+ conditions. These genes were placed into seven groups (k-means clusters; Fig. 3A). The mean fold changes of the transcripts that comprise each of the seven groups are shown in Fig. 3B. The data are presented in comparison to the measured intensities of the control group. The largest groups, G1 and G4, representing 104 and 86 genes, respectively, are of the most immediate interest for an integrative view of zinc functions in biology. Group 1 genes are zinc responsive in a positive mode; i.e., expression is less and more in Zn– and Zn+ conditions, respectively. Conversely, group 4 genes are zinc responsive in a negative mode under the same conditions; i.e., expression is more and less in Zn– and Zn+ conditions, respectively. Names of the individual genes and fold changes in expression for these two groups are presented in Tables 3 and 4, which are published as supporting information on the PNAS web site. These opposing modes of zinc responsiveness have been shown for the ZnT-1 and Zip-2 genes (Fig. 1). For the purposes of understanding functional roles for zinc, genes in other groups are targets for further research. Genes in groups 3, 6, and 7 exhibit marked differences between Zn– and ZnN (control) cells, and in both negative (groups 3 and 6) and positive (group 7) modes of zinc responsiveness. For example, group 3 is up-regulated in Zn– conditions, but not influenced by Zn+ conditions. Genes in groups 2 and 5 exhibit positive and negative zinc responsiveness, respectively, but only in zinc-excess conditions compared with that of control conditions. Those genes may be targets of particular interest for studies of zinc toxicity or to establish recommendations for upper limits of zinc status by using more defined molecular markers than previously available. The magnitude of the fold change of the individual differentially expressed genes that compose groups 1 and 4 is presented in Fig. 5, which is published as supporting information on the PNAS web site. Genes displaying positive zinc responsiveness and negative zinc responsiveness are shown in Fig. 5 A and B, respectively.
Fig. 3.
K-means clusters based on zinc responsiveness. (A) By using a Tukey's post hoc test of data presented in Fig. 2, zinc-responsive genes were identified as having opposite responses in Zn– and Zn+ THP-1 cells. A total of 283 genes were individually clustered into seven groups, each displaying a pattern of responsiveness to zinc. Group 1 exhibits a positive response mode where 104 genes are up-regulated with increasing cellular zinc. Group 4 exhibits a negative response mode with 86 genes down-regulated with increasing cellular zinc. Other groups of genes displayed less linear zinc-responsive characteristics. (B) Mean fold change values among genes in each of the seven groups are shown.
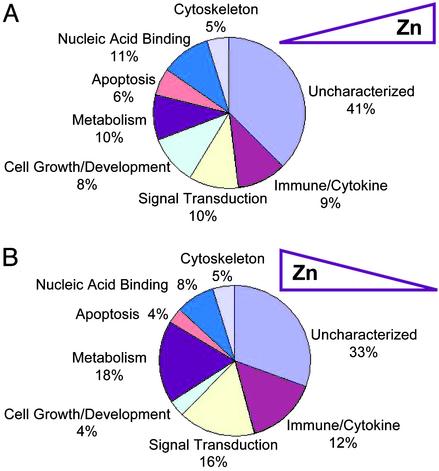
Zinc-responsive genes that compose groups 1 and 4 were sorted into seven functional classes. Of note is that 41% (Fig. 4A) and 33% (Fig. 4B) of the genes in these two respective groups were functionally uncharacterized and compose the eighth class. Relatively few genes fall into the apoptotic class. This finding is consistent with the measures of apoptosis reported in Table 1. Genes involved in nucleic acid binding are a smaller percentage (8–11%) of the total considering the role for zinc in motifs responsible for such functions. Signal transduction has not been viewed as a direct function of zinc; however, 10–16% of the genes in these two zinc-responsive groups point to areas of zinc function yet to be explored. The 10–15% of the genes in the immune/cytokine functional category were of primary interest, considering the monocytic/macrophage lineage of the THP-1 cells used in these profiling studies.
Fig. 4.
Functional grouping of genes that display positive or negative zinc responsiveness (as shown in Fig. 3). The individual genes from groups 1 and 4 were assigned to one of eight functional categories, and the percent within each category is presented. (A) Group 1, genes positively regulated by zinc. (B) Group 4, genes negatively regulated by zinc. Names of the individual genes can be found in Tables 3 and 4.
Discussion
The experiments reported show human monocytic/macrophage THP-1 cells manifest marked changes in gene expression during acute Zn– and Zn+. Many of these genes could influence host defense, as dietary zinc deficiency has been documented to contribute to the incidence of infectious disease (1–5), related in part to diminished macrophage activation (21). Therefore, monocytes as macrophage precursors are likely targets of this nutritional deficiency.
Previous analyses showed that among leukocytes, monocytes are the highest producers of MT. By using competitive RT-PCR, monocytes and PBMCs (85% lymphocytes) obtained from Zn+ (15 mg/d) human subjects displayed comparable changes in MT gene up-regulation (22). Purified monocytes expressed about three times more MT mRNA than PBMCs, however, reflecting their identified greater potential to produce MT. Companion experiments showed THP-1 cells depleted of zinc with TPEN or supplemented with zinc produced comparable down- and upregulation of MT expression, respectively, as PBMCs obtained from human subjects and cultured under the same conditions. Because MT is the prototypical zinc-responsive gene and responds in a fashion expected of a model for human genes regulated by zinc, THP-1 cells were a logical choice for these initial global screening experiments to identify zinc-responsive genes. We also compared expression of the calreticulin (CRT) gene to zinc because it is reported to be MRE regulated (23). Responsiveness of the CRT gene, as expected, was less than that of MT, probably as the result of a lower number of MRE motifs in the regulatory region compared with most MT genes.
The spectrum of zinc responsiveness exhibited by the zinc transporter genes demonstrates both their potential collective importance in coordinating cellular zinc trafficking and influence on zinc homeostasis at the integrative level. ZnT genes have been identified through different experimental approaches employing a variety of cell types (24–31). These data provide a comparison of the influence zinc has on expression of eight transporter genes in one cell type. The up-regulation of the ZnT genes in Zn+ cells shown in these experiments is consistent with a role in cellular efflux and reduced zinc retention. Clearly, the 27-fold up-regulation of Zip-2 expression in zinc-depleted cells is most rare. This profound response supports the convincing experiments demonstrating (in transfected cells) that Zip-2 has a role in cellular zinc uptake (29). Of the transporter genes we evaluated, only Zip-2, and to a limited extent, Zip-1 respond to Zn– in a fashion consistent with a cellular attempt to homeostatically compensate for zinc loss through increased cellular uptake. The extremely low Zip-4 expression in a monocytic cell fits with this protein's apparent role in intestinal zinc absorption and the altered genotype that results in the zinc malabsorption syndrome, acrodermatitis enteropathica (31). The differential expression of specific genes of the ZnT and Zip transporter families in response to zinc will provide opportunities to examine nutritional status of this micronutrient that are based on physiological responses.
The microarrays used in these experiments allow a survey of 22,216 elements. Our ANOVA, which identified 1,045 genes (P < 0.0001) or ≈5% as zinc responsive, was modest in scope. Recently, the association of zinc as a component of the gene expression and protein synthesizing machinery of cells has focused on regulatory roles of zinc participating in signaling processes, perhaps mediated by binding proteins that titer intracellular zinc concentrations (8). Mammalian cells have one recognized zinc sensor that regulates gene expression, MTF-1. Zinc binding by MTF-1 provides a positive signal for transcriptional regulation for genes with MREs in regulatory regions (32). MT serves as the only well characterized MTF-1-regulated gene. Nevertheless, others must exist because, whereas MTF-1 null mice display embryonic lethality (11), MT null mice are viable (33). The exact gene responsible for lethality has not been identified, but a number of candidates have been considered as database searches reveal an ever expanding list of genes with MRE sequences (10). The 104 genes in these microarray studies found responsive to zinc in a mode consistent with MTF-1-regulated genes substantially increase the number of MRE-containing candidate genes. Furthermore, identification of another group of 86 genes that are zinc responsive, but in a mode opposite to that of MTF-1, produces another avenue of inquiry into gene regulation by zinc. We envision, within that context, zinc occupancy by a factor based on cellular zinc status could lead to repression of gene expression through a mechanism that influences transcript abundance. In this context, it is of interest that MTF-1 appears as an up-regulated transcript (2.5-fold) in Zn– cells, while MTF-2, an ortholog of MTF-1, is down-regulated 2.5-fold in the Zn– cells. Expression of these transcription factors by zinc has not been previously observed, to our knowledge. The reciprocal expression of these metal-responsive transcription factors may prove to be important in explaining the two modes of zinc regulation observed in these genome screening experiments.
The cellular pool(s) from which Zn(II) ions are donated to MTF-1 or other regulatory factors, including zinc finger proteins in general, has not been defined. The cytosolic pool of free Zn(II) ions from which occupancy could be derived is considered small. Titration experiments using TPEN suggest the pool of free zinc in Escherichia coli is extremely low (≈10–15 M), suggesting that, in mammalian cells, similar conditions would exist (34). Most likely, under the conditions used in our experiments, where acute Zn– is induced with TPEN (Kd = 6.3 × 10–16 M), these mononuclear cells would be depleted of zinc needed to signal transcription events by MTF-1 or similar factors requiring occupancy of Zn(II) ions.
The conditions of Zn– and Zn+ used in these experiments were mild by design. Previously, we found THP-1 cells display an inverse relationship between loss of cellular zinc concentration and cell viability (18). TPEN can result in apoptosis (35), the sequelae of which usually involves cellular redox perturbation, reduction in ΔΨm, and then caspase-3 activation for commitment to apoptosis. Our data show the effects of TPEN-induced reduction in cellular zinc, but without altered cell viability. This finding is because of our use of lower TPEN concentrations and shorter exposure times than are frequently used. The changes in the various apoptotic markers used here (Table 1) are consistent with previous descriptions of events leading to apoptosis in THP-1 cells (36). Consequently, it was expected that some genes in apoptotic signaling pathways would be identified as zinc responsive. Based on gene ontology, 4–6% of the genes were identified as zinc responsive based on our sorting of genes into functional classes. None of the caspase genes were identified among the group 1 and 4 genes.
Analysis of genes identified as dysregulated in Zn– THP-1 cells fits a pattern that is analogous to defective macrophage activation in intact animals. Once monocytes differentiate into macrophages, and on appropriate stimulation, they display enhanced antimicrobial activity against intracellular pathogens. IFN-γ, a Th1 cytokine, is one of the two signals needed for activation. The other is provided by armed Th1 cells, which secrete tumor necrosis factor (TNF)-α. This augments the activation produced by IFN-γ alone (37). Consequently, our finding that the IFN-γ receptor 2 gene (IFN-γR2; a class II cytokine receptor) is within group 1 genes, and therefore positively regulated by zinc, is of potential importance. Down-regulation of IFN-γR2 leads to proliferation of IFN-γ-stimulated myeloid cells, whereas up-regulation leads to apoptosis (38). Representatives of the class I (IL-4R) and chemokine (c-c motif R2) cytokine receptor families were also identified within genes in group 1 mode of zinc responsiveness. Ligand binding by the latter receptor promotes migration of monocytes to become tissue macrophages. Given the additional 9-fold down-regulation of TNFR-1A, we conclude a representative of each of the cytokine receptor families was found responsive to zinc depletion. A limitation of cytokine receptor expression in a zinc-deficient animal or human would be expected to decrease antimicrobial activity of activated macrophages and prevent amplification of the Th1 immune response to intracellular pathogens. Th1 cytokines have been reported as decreased in human zinc deficiency (2–4).
Of considerable interest is that, of the 104 genes in Group I, the most zinc-responsive gene is tristetraprolin (TTP), exhibiting a 14-fold reduction in expression in Zn–cells and a ≈2-fold increase in Zn+ cells. TTP is a zinc finger protein (CCCH type), also called Zfp36, that destabilizes TNF-α mRNA and acts as a negative feedback regulator of TNF-α production (39). TTP null mice develop chronic inflammation and cachexia as the result of greater than normal TNF-α synthesis. Down-regulation of TTP in Zn– cells should allow increased TNFα mRNA stability and translation, yielding the potential for greater TNF production. The THP-1 cells used in these experiments were not activated with IL-1β or other agents, thus no induction of TNFα gene expression was expected. The marked down-regulation in TTP observed therefore is likely the product of rapid mRNA turnover, because the t1/2 for TTP mRNA is estimated to be on the order of 30 min (39). The identification of TTP among group 1 genes is consistent with its observed induction by zinc noted by others (40). The finding also relates to the greater probability for identification of genes by array profiling that exhibit rapid mRNA turnover (41).
Profiling experiments such as those reported here provide a plethora of genes that will constitute the basis for further inquiry. Among the transcripts listed in Table 3 that are decreased in zinc depletion are IFN regulatory factor 1, DiGeorge syndrome critical region gene 2, TGF-β R II, NFk-β light polypeptide gene enhancer 1, many zinc finger proteins, histocompatibility proteins, and oncogenes. Further experiments are required to relate the functions of these to zinc adequacy. Similarly, among transcripts of particular interest where expression is decreased in Zn+, include amyloid β (A4) precursor, prolactin, iron-responsive element binding protein 2, and several zinc finger proteins. On initial examination, we see many genes responsible for general aspects of transcription or protein synthesis among the zinc-responsive groups. Transcripts for these genes were observed as particularly abundant in PBMCs in a recent evaluation of gene expression in cells from human blood (42).
The genes identified in this global screening may provide targets to evaluate in field and clinical studies as biomarkers of zinc status. An impetus for further investigation related to these goals is the continuing reports citing the contribution of mild degrees of zinc deficiency to underlying causes of morbidity and mortality in various population groups (5). Dried blood spots as the source of total RNA have allowed us to identify changes in MT gene expression in human subjects (22). Appropriate primer design should allow similar PCR-based measures of the expression of any number of the zinc-responsive genes identified in this report in situations where sample size is a limiting factor.
Supplementary Material
Acknowledgments
This paper is dedicated by R.J.C. to the late Professor Hamilton Dean Eaton of the University of Connecticut. We thank Neal A. Benson of the University of Florida Flow Cytometry Core Facility for help and advice with the FACS analyses, and the Shands Cancer Center/University of Florida Interdisciplinary Center for Biotechnology Research Microarray Core Facility for array processing. This work was supported by National Institutes of Health Grant DK 31127, the Boston Family Endowment Funds of the University of Florida, the Florida Agricultural Experiment Station, and approved for publication as journal series R-09465.
Abbreviations: MTF-1, metal-responsive transcription factor 1; ZnT, zinc transporter; PBMC, peripheral blood mononuclear cell; TPEN, N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine; Zn–, zinc depletion; Zn+, zinc supplementation; MRE, metal-responsive element; TNF, tumor necrosis factor.
References
- 1.Fraker, P. J., King, L. E., Laakko, T. & Vollmer, T. L. (2000) J. Nutr. 130, 1399S–1406S. [DOI] [PubMed] [Google Scholar]
- 2.Shankar, A. H. & Prasad, A. S. (1998) Am. J. Clin. Nutr. 68, 447S–463S. [DOI] [PubMed] [Google Scholar]
- 3.Koski, K. G. & Scott, M. E. (2001) Annu. Rev. Nutr. 21, 297–321. [DOI] [PubMed] [Google Scholar]
- 4.Wellinghausen, N., Kirchner, H. & Rink, L. (1997) Immunol. Today 18, 519–521. [DOI] [PubMed] [Google Scholar]
- 5.Black, R. E. (2003) Bull. W. H. O. 81, 79. [PMC free article] [PubMed] [Google Scholar]
- 6.Panel on Micronutrients, Subcommittees on Upper Reference Levels of Nutrients and of Interpretation and Use of Dietary Reference Intakes, and the Standing Committee on the Scientific Evaluation of Dietary Reference Intakes (2002) in Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc (Natl. Acad. Press, Washington, DC), pp. 290–501.
- 7.Wood, R. J. (2002) Nutr. Rev. 60, 144–148. [DOI] [PubMed] [Google Scholar]
- 8.Cousins, R. J. (1996) in Present Knowledge in Nutrition, eds. Filer, L. J. & Ziegler, E. E. (Int. Life Sci. Inst.-Nutr. Foundation, Washington, DC), 7th. Ed., pp. 293–306.
- 9.King, J. D. & Keen, C. L. (1999) in Modern Nutrition in Health and Disease, eds. Shils, M. E., Olson, J. A., Shike, M. & Ross, A. C. (Lippincott Williams & Wilkins, Baltimore), 9th Ed., pp. 223–239.
- 10.Lichtlen, P., Wang, Y., Belser, T., Georgiev, O., Certa, U., Sack, R. & Schaffner, W. (2001) Nucleic Acids Res. 29, 1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Günes, C., Heuchel, R., Georgiev, O., Muller, K. H., Lichtlen, P., Bluthmann, H., Marino, S., Aguzzi, A. & Schaffner, W. (1998) EMBO J. 17, 2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmade, S. J., Ravindra, R., Daniels, P. J. & Andrews, G. K. (2000) J. Biol. Chem. 275, 34803–34809. [DOI] [PubMed] [Google Scholar]
- 13.Blanchard, R. K. & Cousins, R. J. (1996) Proc. Natl. Acad. Sci. USA 93, 6863–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard, R. K., Moore, J. B., Green, C. L. & Cousins, R. J. (2001) Proc. Natl. Acad. Sci. USA 98, 13507–13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, J. B., Blanchard, R. K., McCormack, W. T. & Cousins, R. J. (2001) J. Nutr. 131, 3189–3196. [DOI] [PubMed] [Google Scholar]
- 16.Moore, J. B., Blanchard, R. K. & Cousins, R. J. (2003) Proc. Natl. Acad. Sci. USA 100, 3883–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanchard, R. K. & Cousins, R. J. (2000) J. Nutr. 130, 1393S–1398S. [DOI] [PubMed] [Google Scholar]
- 18.Cao, J., Bobo, J. A., Liuzzi, J. P. & Cousins R. J. (2001) J. Leukocyte Biol. 70, 559–566. [PubMed] [Google Scholar]
- 19.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yandell, B. S., ed. (1997) in Practical Data Analysis for Designed Experiments (Chapman & Hall, New York).
- 21.Wirth, J. J., Fraker, P. J. & Kierszenbaum, F. (1989) Immunology 68, 114–119. [PMC free article] [PubMed] [Google Scholar]
- 22.Cao J. & Cousins R. J. (2000) J. Nutr. 130, 2180–2187. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen, T. Q., Capra, J. D. & Sontheimer, R. D. (1996) Mol. Immunol. 33, 379–386. [DOI] [PubMed] [Google Scholar]
- 24.Palmiter, R. D. & Findley, S. D. (1995) EMBO J. 14, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kambe, T., Narita, H., Yamaguchi-Iwai, Y., Hirose, J., Amano, T., Sugiura, N., Sasaki, R., Mori, K., Iwanaga, T. & Nagao, M. (2002) J. Biol. Chem. 277, 19049–19055. [DOI] [PubMed] [Google Scholar]
- 26.Huang, L., Kirschke, C. P. & Gitschier, J. (2002) J. Biol. Chem. 277, 26389–26395. [DOI] [PubMed] [Google Scholar]
- 27.Kirschke, C. P. & Huang, L. (2003) J. Biol. Chem. 278, 4096–4102. [DOI] [PubMed] [Google Scholar]
- 28.Costello, L. C., Liu, Y., Zou, J. & Franklin, R. B. (1999) J. Biol. Chem. 274, 17499–17504. [DOI] [PubMed] [Google Scholar]
- 29.Gaither, L. A. & Eide, D. J. (2001) J. Biol. Chem. 275, 5560–5564. [DOI] [PubMed] [Google Scholar]
- 30.Gaither, L. A. & Eide, D. J. (2001) Biometals 14, 251–270. [DOI] [PubMed] [Google Scholar]
- 31.Wang, K., Zhou, B., Kuo, Y.-M., Zemansky, J. & Gitschier, J. (2002) Am. J. Hum. Genet. 71, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalton T. P., Bittel, D. & Andrews, G. K. (1997) Mol. Cell. Biol. 17, 2781–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masters, B. A., Kelly, E. J., Quaife, C. J., Brinster, R. L. & Palmiter, R. D. (1994) Proc. Natl. Acad. Sci. USA 91, 584–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Outten, C. E. & O'Halloran, T. V. (2001) Science 292, 2488–2492. [DOI] [PubMed] [Google Scholar]
- 35.Kondoh, M., Tasaki, E., Araragi, S., Takiguchi, M., Higashimoto, M., Watanabe, Y. & Sato, M. (2002) Eur. J. Biochem. 269, 6204–6211. [DOI] [PubMed] [Google Scholar]
- 36.Dinsdale, D., Zhuang, J. & Cohen, G. M. (1999) Am. J. Pathol. 155, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janeway, C. A., Travers, P., Walport, M. & Shlomchik, M., eds. (2001) Immunobiology: The Immune System in Health and Disease (Garland, New York), 5th. Ed.
- 38.Bernabei, P., Coccia, E. M., Rigamonti, L., Bosticardo, M., Forni, G., Pestka, S., Krause, C. D., Battistini, A. & Novelli, F. (2001) J. Leukocyte Biol. 70, 950–960. [PubMed] [Google Scholar]
- 39.Carballo, E., Lai, W. S. & Blackshear, P. J. (1998) Science 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, G. A. & Blackshear, P. J. (1995) J. Cell. Physiol. 162, 378–387. [DOI] [PubMed] [Google Scholar]
- 41.Fan, J., Yang, X., Wang, W., Wood, W. H., III, Becker, K. G. & Gorospe, M. (2002) Proc. Natl. Acad. Sci. USA 99, 10611–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitney, A. R., Diehn, M., Popper, S. J., Alizadeh, A. A., Boldrick, J. C., Relman, D. A. & Brown, P. O. (2003) Proc. Natl. Acad. Sci. USA 100, 1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.