Abstract
The transfer of T-DNA from Agrobacterium to plant cells is mediated by a system which involves the virB operon of the Ti plasmid. We report that VirE2 and VirD2, two T-DNA-associated proteins, as well as VirF, a protein known to be secreted into plant cells, are present in the periplasm and supernatant fractions of growing cells of Agrobacterium as are VirJ and ChvE, two known periplasmic proteins. Two cytoplasmic proteins, Ros and chloramphenicol acetyl transferase, and a VirE2∷green fluorescent protein construct were not detected in the above fraction. Export of VirE2 into the culture supernatant did not require any Ti plasmid genes, except for VirE1, a specific chaperone for VirE2. The levels of the VirE2 and VirD2 proteins in the supernatant increased significantly when cells were grown at 19°C as compared with 28°C. When Agrobacterium expressed the oncogenic suppressive activity protein (Osa), VirE2 and VirF proteins could not be detected in the supernatant or the periplasm and the level of VirD2 was greatly reduced. However, oncogenic suppressive activity protein did not block the accumulation of VirJ and ChvE in the periplasm. Our data suggest that VirD2, VirE2, and VirF are transported across the cytoplasmic membrane by a specific pathway, independent of virB. Thus, transfer of the T-complex of Agrobacterium may take place in two steps, the first mediated by an unidentified pathway and the second by the virB system.
Keywords: virulence protein export, secretion system
Agrobacterium tumefaciens induces tumors in plants by transferring a piece of single-stranded DNA, T-DNA, from its tumor-inducing (Ti) plasmid into plant cells (1). The subsequent integration and expression of T-DNA in the plant genome leads to the formation of crown gall tumors. Because any DNA region on the T-DNA, regardless of its sequence, will be transferred to the host, A. tumefaciens has been widely adapted as a vehicle to transfer foreign genes into plants. However, fundamental questions as to how the T-DNA is exported from bacteria and integrated into the plant genome remain to be answered.
T-DNA is transported into plant cells as part of a nucleoprotein complex, the T-complex, consisting of single-stranded DNA, VirD2, and perhaps VirE2. The T-DNA is delineated by two imperfect repeat sequences, the T-borders (2, 3). VirD2 cleaves the bottom strand of the T-DNA at the T-borders (2–4) and remains covalently bound to the 5′ end (5, 6). VirE2 is a nonsequence specific single-stranded DNA-binding protein which presumably coats the T-DNA and protects it from host nucleases (7). VirE2 functions primarily in plants because plants expressing VirE2 can be successfully infected by A. tumefaciens lacking VirE2 (8). Both VirD2 and VirE2 possess functional nuclear localization signals that guide the T-DNA to the plant cell nucleus (9, 10).
Transfer of the T-complex in A. tumefaciens relies on the T-pilus, a structure encoded by the virB operon (11–13). Mutations in any of the 11 genes in this operon abolish both the pilus and tumor formation (13–15). Although the VirB proteins possess homology to components of other conjugal transfer systems (11, 14, 15), no direct evidence exists that the T-pilus is the transport channel for the T-complex. Protein homologues of the virB system can be found in many animal pathogens, including Bartonella henselae, Bordetella pertussis, Brucella abortus, Brucella suis, Helicobacter pylori, Legionella pneumophila, and Rickettsia prowazekii (14, 16). In mammalian pathogens, these systems are required for the export of toxins and other unidentified virulence factors as well as for intracellular survival (14, 17). For example, the transfer of the pertussis toxin of Bordetella and the CagA protein of Helicobacter depends on these systems (17, 18). Two virB-like secretion systems have been reported in L. pneumophila (19). The icm/dot system contributes to transfer of pRSF1010 and intracellular survival, whereas the lvh system facilitates only pRSF1010 transfer.
Current models for T-complex export propose a one-step transfer process from the bacterial cytoplasm to the plant cell via the T-pilus (11, 15). In part, this is based on the fact that neither VirD2 nor VirE2 have typical signal peptides for sec-dependent transport into the periplasm. In B. pertussis, however, pertussis toxin appears to be exported by a two-step process because its subunits contain typical signal peptides (17, 20). The subunits are transported independently into the periplasm where the toxin is assembled, and then secreted by a virB-like system (20). In Agrobacterium, it is unclear whether VirE2 and the T-strand with VirD2 covalently linked to T-DNA enter plants as a preassembled complex or as separate entities. Although a preassembled T-complex has been detected inside A. tumefaciens (21), extracellular complementation experiments suggest that the T-strand and VirE2 can originate in separate bacteria (22).
We report that VirE2, VirD2, and VirF, the three exported virulence proteins, are present in the supernatant of growing cells of A. tumefaciens. In Agrobacterium, the secretion of these proteins does not depend on the T-pilus, suggesting that a novel pathway or additional components may participate in the export of virulence proteins in A. tumefaciens.
Materials and Methods
Strains, Plasmids, and Molecular Techniques.
DNA fragments of virE2 (23), virE1 + E2 (23), oncogenic suppressive activity (osa) (24), phoA (25), chloramphenicol acetyltransferase (CAT), and gfpmut2 (26) were amplified with sequence-specific primers by using Vent DNA polymerase (New England Biolabs). For all genes, a unique NdeI site was introduced into the start codon in the 5′ primers and a different unique restriction site after the stop codon in the 3′ primers. These restriction sites were used to fuse these genes into the ptac expression vector, pPR1068 (New England Biolabs). Extracellular complementation experiments were performed according to Otten et al. (22). Standard molecular biology techniques were performed as described in Ausubel et al. (27).
Growth of Bacteria and Cell Fractionation.
Bacterial strains were grown in mannitol glutamate/Luria (MG/L), AB, or induction media as indicated (25). Cultures were incubated at 28°C unless stated otherwise. All strains were first grown overnight in MG/L to saturation and then diluted 1:50 into fresh media. After an additional 6–8 h growth (OD600 of about 1.2 for MG/L, 0.17 for induction and AB medium), the cells were centrifuged at 16,300 × g for 30 min. This pellet was designated the total cell fraction. The supernatant was centrifuged again at 27,300 × g for an additional 30–60 min to remove any residual bacteria. Trichloroacetic acid was then added to a final concentration of 10%. The mixture was incubated at 4°C for 16 h and then centrifuged to pellet the precipitated proteins. This pellet was designated the supernatant fraction. The pellets recovered after trichloroacetic acid precipitation were washed twice with 80% acetone before analysis. Periplasmic proteins were isolated from the total cell fraction as described by Lu and Lory (28). The pellet remaining after the isolation of the periplasmic proteins was resuspended in buffer containing Tris (pH 7.0)/10% glycerol/0.1 μM NaCl/0.1 μM PMSF, and was solubilized by two 10-s sonication treatments at 0°C. This preparation was then centrifuged and separated into a soluble fraction, designated the cytoplasmic-soluble fraction, and an insoluble fraction, designated the cytoplasmic-insoluble fraction.
Western Blot Analysis.
Proteins were resuspended in SDS/PAGE buffer, electrophoresed in either N-[tris(hydroxymethyl)]glycine or glycine gels (29), and blotted onto poly(vinylidene difluoride) membranes (Millipore) by using a semidry electrophoresis transfer apparatus (Bio-Rad). Unless specified otherwise, each lane was loaded with the equivalent of 50 ml of culture supernatant derived from cultures grown to similar densities. In most cases, the membranes were cut at the region corresponding to 32–42 kDa. The high molecular weight portion was developed with VirE2 (30), VirD2 (Alpha Diagnostic International, San Antonio, TX), PhoA (5′ to 3′), or GFP (Boehringer Mannheim) antibodies. The low molecular weight portion was developed with Ros (13), VirJ (31), VirF (32), or CAT (5′ to 3′) antibodies. Western blotting was performed according to Harlow and Lanes (33) with enhanced chemiluminescence or enhanced chemiluminescence/plus (Amersham International).
Results
VirE2 and VirD2 Are Secreted into the Culture Supernatant.
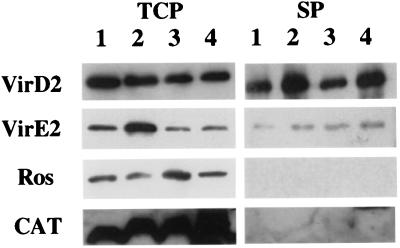
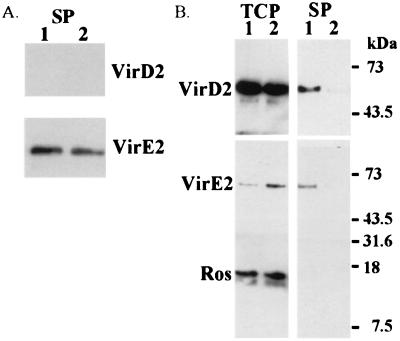
Western blot analyses of the supernatant and the total cell fractions from induced cells of A. tumefaciens A348 were performed by using VirE2 and VirD2 antibodies. The analysis of VirE2 shows a protein band of ≈66 kDa in both the total cell protein and the supernatant fractions (Fig. 1). This band corresponds to the predicted mobility of VirE2 and is not seen in virE2 mutant controls (At12516; KE1; ref. 34). A similar analysis of the VirD2 Western also shows a protein band at 63 kDa in the total cell and supernatant fractions, but not in a virD2 mutant (At10002). After compensating for the volumes analyzed, we conservatively estimate that about 1% of the total VirE2 and VirD2 proteins are in the supernatant fraction.
Figure 1.
Western blots of the supernatant and total cell fractions from different constructs in A. tumefaciens strain A348. Cells were grown at 28°C under vir gene-inducing conditions, separated into supernatant and total cell fractions (noted on the top). The proteins were detected by using specific antibodies (noted on the left). Lane 1, A348; lane 2, A348 (pUFR047); lane 3, A348 (ptac-ros/pUFR047); and lane 4, A348 (ptac-CAT/pUFR047). Total cell proteins, TCP; and supernatant proteins, SP.
Because VirE2 and VirD2 in the supernatant fraction could result from either cell lysis or trace amounts of residual bacteria, two cytoplasmic proteins, Ros and CAT, which served as cytoplasmic controls in other studies (13, 35, 36), were used to detect cell lysis. Neither Ros (18 kDa) nor CAT (26 kDa) was detected in the supernatant fraction. To further increase the sensitivity, we expressed Ros and CAT from the strong promoter, ptac. Even at the elevated levels of expression, we did not detect either control protein in the supernatant fraction. We also passed the supernatant fraction through a 0.22 μM low protein-binding filter before trichloroacetic acid precipitation. VirE2 was still detected in these fractions, suggesting that residual bacteria did not contribute VirE2 to the supernatant fraction. These data all suggest that the VirE2 and VirD2 are exported into the supernatant of Agrobacterium.
VirE2∷PhoA, But Not VirE2∷GFPmut2, Is Released into the Supernatant Fraction.
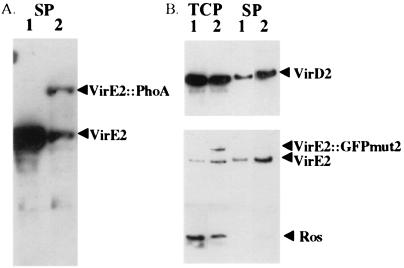
Studies of two fusion forms of VirE2 also support the above conclusion. The bacterial phoA gene lacking a leader sequence was fused to the C-terminal domain of VirE2. This construct (ptac-virE1 + E2∷phoA) was transformed into a phoA− strain of A. tumefaciens, A6007 (25). Although a wild-type copy of the virE operon exists in A6007, native virE2 is not expressed in MG/L. Colonies carrying ptac-virE1 + E2∷phoA were light blue on a MG/L plate containing 5-bromo-4-chloro-3-indolyl phosphate (data not shown), indicating that the VirE2∷PhoA protein was exported from the cytoplasm. Western blot analysis with VirE2 antibodies confirmed that VirE2∷phoA was present in the supernatant fraction of cells grown in MG/L (Fig. 2A). Two bands are present. A 115-kDa band corresponds to the predicted size of the VirE2∷PhoA protein; the second 66-kDa bands corresponds to VirE2. PhoA antibodies detected only one band at the same position as VirE2∷PhoA, verifying that the fusion protein was present in the supernatant fraction (data not shown). VirE2∷PhoA appears to be unstable because several bands, presumably representing degradation products, were detected in the total cell fraction (data not shown). However, multiple bands were not detected in the supernatant fraction. In this case, either the degradation products were not exported, were readily degraded in the supernatant, or beyond the level of antibody detection.
Figure 2.
Western blots of A. tumefaciens containing VirE2 fusions with PhoA and GFPmut2. (A) Supernatant fraction from PhoA constructs detected with VirE2 antibodies. A348 (lane 1) was grown under inducing conditions, and A136 (phoA−) (ptac-virE1 + E2∷phoA/pUFR047) (lane 2) was grown under noninducing conditions. (B) Cells were grown at 28°C under inducing conditions, separated into the supernatant and total cell fractions (noted on the top), and detected with protein-specific antibodies (noted on the right). Lane 1, A348 (pUFR047); and lane 2, A348 (ptac-virE1 + E2∷gfpmut2).
A second fusion construct, ptac-virE1 + E2∷gfpmut2, was transformed into A348 and a virE deletion mutant, KE1. These strains exhibited a green fluorescence (data not shown), indicating that the fusion protein was expressed. This protein could be detected in the total cell fraction by using both VirE2 and GFP antibodies. The VirE2∷GFPmut2 was not degraded. Unlike VirE2∷phoA, the VirE2∷GFPmut2 was not detected in the supernatant fraction of either A348 or KE1, suggesting that VirE2∷GFPmut2 is not exported. This would explain why ptac-virE1 + E2∷gfpmut2 could not complement KE1 in tumor formation. Although VirE2 could still be detected in the supernatant fraction in A348 (ptac-virE1 + E2∷gfpmut2) (Fig. 2B), the virulence of A348 (ptac-virE1 + E2∷gfpmut2) was attenuated (data not shown).
Secretion of VirE2 and VirD2 Is Independent of the virB Pilus.
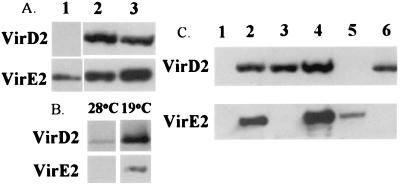
In A348 (ptac-virE1 + E2∷phoA), VirE2∷PhoA was present in the supernatant fraction even when cells were grown in MG/L at pH 7 (Fig. 2A). Under these conditions, no vir genes other than ptac-driven virE1 and virE2∷phoA should be expressed (23). These results suggest that the VirB proteins are not required for the secretion of VirE2. To confirm this suggestion, two strains cured of their Ti plasmid, A136 and AD802 (37), were transformed with ptac-virE1 + E2. VirE2 was detected in the supernatant fractions of both strains (Fig. 3A, lane 1). Similar results were obtained by using KE1 (ptac-virE1 + E2) grown under noninducing conditions in MG/L or AB media (data not shown). To further demonstrate that neither the VirB proteins nor VirD4 is required, we analyzed the supernatant fractions of two virB mutants [At11067 (a polar virB1 mutant; Fig. 3A, lane 3) and A348ΔB2 (a virB2 deletion; data not shown)] and a virD4 mutant [At12506 (a virD4 polar mutant; Fig. 3A, lane 2)]. Both VirE2 and VirD2 were detected in the supernatant of the above mutants.
Figure 3.
VirE2 and VirD2 secretion under various conditions and genetic backgrounds. (A) Western blots of supernatants from different virB secretion system mutant constructs detected with antibodies (noted on the left). Lane 1, A136 (ptac-virE1 + E2/pUFR047); lane 2, virD4 mutant (At12506); and lane 3, virB1 polar mutant (At10067). (B) Western blots of supernatants of A348 cells grown under inducing conditions at two different temperatures (19 and 28°C) detected with antibodies (noted on the left). Each lane was loaded with the equivalent of 10 ml of culture supernatant (the equivalent of 50 ml was loaded in other assays) from cells with a similar OD600. (C) Western blots of supernatants from different A. tumefaciens strains detected with antibodies (noted on the left). Lane 1, A136; lane 2, A348; lane 3, virE mutant (KE1); lane 4, KE1 (ptac-virE1 + E2/pUFR047); lane 5, virD2 mutant (At10002); and lane 6, virE2 mutant (At12516).
Low Temperature Enhances the Secretion of VirE2 and VirD2.
Fullner and Nester (38) provided the first evidence that temperature affects the virB secretory machinery of A. tumefaciens when they reported that pili could be readily observed on the surface of cells grown at 19 but not 28°C (12). We investigated whether temperature affects the levels of secreted VirE2 and VirD2. Both proteins were detected in the supernatant and in the total cell fractions at 28 and 19°C (Fig. 3B). Because the volume of supernatant used in this experiment was only 1/5 of the normal amount used, the VirE2 and VirD2 bands at 28°C are much weaker than in other sets of experiments. No significant difference in the level of either protein can be seen in the total cell fractions at 28 and 19°C, indicating that the production of both proteins is unaffected by temperature. However, after normalizing to the same OD, significantly more VirE2 and VirD2 was observed in the supernatant fraction of cells grown at 19°C. We estimate that at least fivefold more VirE2 and VirD2 proteins are present in the supernatant fraction of cells grown at 19°C compared with 28°C.
VirE1 Is Required for VirE2 Secretion.
We next determined if VirE1, a molecular chaperone for VirE2 (23, 39), is required for secretion of VirE2. A6007 (ptac-virE2∷phoA) was assayed for phoA activity under noninducing conditions. In contrast to what was observed with A6007 (ptac-virE1 + E2∷phoA), A6007 (ptac-virE2∷phoA) did not turn blue on 5-bromo-4-chloro-3-indolyl phosphate plates and the VirE2∷phoA protein was not detected in the supernatant fraction (data not shown). To confirm these results, Western blotting was used to determine whether VirE2 was secreted in KE1 carrying ptac-virE2 or ptac-virE1 + E2, when cells were grown under noninduced conditions. In all cases, VirE2 was only detected in the supernatant when VirE1 was coexpressed (Fig. 3C). In addition, even though VirE2 was not secreted, VirD2 was detected in the supernatant, indicating that VirD2 can be exported independently of VirE2.
VirF Is Also Secreted.
VirF is a Ti plasmid-encoded protein that is found in some, but not all, strains of Agrobacterium (32). It functions to extend the host range of the strain on certain plants, but its mechanism of action is not understood. VirF is transferred into plant cells like VirE2 and VirD2. Western blot analysis determined that VirF was present in the supernatant fraction of cultures grown under inducing conditions (Fig. 4B).
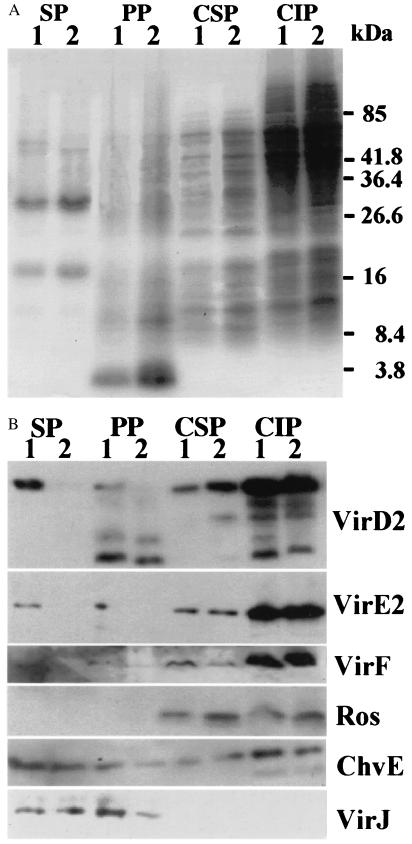
Figure 4.
Coomassie Brilliant Blue staining and Western blotting of protein gels from different fractions of A. tumefaciens cells containing ptac-osa. (A) Cells were grown under inducing conditions and then collected, and separated into supernatant, periplasmic, cytoplasmic-soluble, and cytoplasmic-insoluble fractions (noted on the top). The fractions were run on SDS/PAGE gels, and stained with Coomassie Brilliant Blue. Lane 1, A348 (pUFR047); and lane 2, A348 (ptac-osa/pUFR047). (B) Western blots of the proteins from different fractions (noted on the top) detected with antibodies (noted on the right). Lane 1, A348 (pUFR047); and lane 2, A348 (ptac-osa/pUFR047). Periplasmic proteins, PP; cytoplasmic-soluble proteins, CSP; and cytoplasmic-insoluble proteins, CIP.
Avirulent Chromosomal Mutants Secret VirE2.
Our results indicate that the factor(s) involved in the secretion of VirE2 is not encoded in the Ti plasmid. The supernatant fractions from previously described avirulent chromosomal mutants were analyzed to determine if these genes played a role in VirE2 secretion. Mutants included chvA (40), chvB (40), exoC (41), chvE (42), and acvB/virJ (43) mutants. ChvA is an inner membrane protein that is involved in the transport of β1,2-glucan synthesized by ChvB and ExoC. β1,2-glucan is required for attachment of Agrobacterium to plant cells. ChvE is a periplasmic sugar-binding protein that is required for optimal vir gene induction. AcvB and presumably its functional homolog VirJ are periplasmic proteins that have been reported to bind to the T-strand. VirE2 was detected in the supernatant fractions from all mutants at levels comparable to wild type (data not shown). An acvB mutant strain A208, which lacks virJ in its Ti plasmid, secreted VirE2 when the virE construct under ptac control was introduced. We conclude that none of the avirulent chromosomal mutants tested affect VirE2 secretion.
Vir Proteins Are Exported Across the Inner Membrane.
To gain some insight as to whether the exported molecules were first being secreted into the periplasm and were then exiting through the outer membrane, we determined whether proteins known to be in the periplasm could also be observed in the culture supernatant. Cells were grown under inducing conditions, and after isolation of the supernatant fraction, the total cell fraction (pellet) was further subdivided into periplasmic, cytoplasmic-soluble, and cytoplasmic-insoluble fractions (see Materials and Methods). SDS/PAGE analysis of all four fractions stained with Coomassie brilliant blue suggests that the proteins in the supernatant and periplasmic fractions are distinctly different from those in the cytoplasmic-soluble and -insoluble fractions. A putative flagellin band (44), at ≈32 kDa, is observed only in the supernatant fraction (Fig. 4A). In Agrobacterium, two periplasmic proteins, VirJ and ChvE, contain leader peptides and are presumably exported in a sec-dependent manner into the periplasm. Both proteins were found in the periplasmic fraction and also in the supernatant fraction (Fig. 4B). In addition, ChvE was found in the cytoplasmic-soluble and -insoluble fractions. VirD2, VirE2, and VirF proteins were found in all four fractions. However, Ros was located only in the cytoplasmic-soluble and -insoluble fractions. We conclude that the supernatant fraction contained only proteins that were exported across the inner membrane, i.e., VirE2, VirD2, and VirF, as well as the two known periplasmic proteins ChvE and VirJ.
Osa, but Not pRSF1010, Inhibits the Secretion of VirE2, VirD2, and VirF.
Although our data show that three proteins known to enter plant cells are found in the supernatant, the biological significance of these observations is not clear. In the absence of mutants in which transport is blocked, we looked at the effect of macromolecules that have been shown to block transfer of VirD2 and VirE2. We determined whether VirE2 is secreted into the supernatant of A. tumefaciens carrying either a derivative of the RSF1010 plasmid, pML122ΔGm (45), or an Osa expression construct ptac-osa in pUFR047 (46). RSF1010 can be transferred into other bacteria and plant cells in a virB-dependent manner. RSF1010 competitively inhibits the transfer of the T-complex, hereby inhibiting tumor formation (47). Osa, a protein encoded by the pSa plasmid of Shigella flexneri, reportedly inhibits the export of VirE2 leading to avirulence (48, 49).
Western blot analysis shows that the level of VirE2 was not reduced in the supernatant fraction of cells carrying pML122ΔGm (Fig. 5A). We did not analyze VirD2 levels because these assays were performed in a strain cured of its Ti plasmid [A136 (pML122ΔGm and ptac-virE1 + E2)].
Figure 5.
Western blots of A. tumefaciens cells containing pRSF1010 (pML122ΔGm) and Osa (ptac-osa/pUFR047). (A) Supernatant fractions were harvested from cells grown under noninducing conditions, and proteins were detected with VirD2 and VirE2 antibodies. Lane 1, A136 (ptac-virE1 + E2/pUFR047 + pML122ΔGm); and lane 2, A136 (ptac-virE1 + E2/pUFR047). (B) Western blot analysis of supernatant and total cell fractions of A. tumefaciens strains carrying Osa. Fractions (noted on the top) were collected from cells grown under inducing conditions, and proteins were detected with specific antibodies (noted on the left). Lane 1, A348 (pUFR047); and lane 2, A348 (ptac-osa/pUFR047).
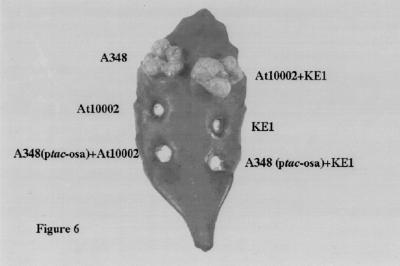
However, in A348 (ptac-osa), neither VirE2 nor VirF was detected in the supernatant or the periplasmic fractions. Both proteins were expressed normally in the total cell fraction (Fig. 5B). The levels of VirD2 in the supernatant and the periplasmic fractions were significantly reduced, but were still detectable. In contrast, Osa did not affect the levels of VirJ and ChvE in the periplasm (Fig. 4B). These results indicate that Osa selectively blocks the passage of VirE2, VirD2, and VirF proteins across the inner membrane. We tested our ptac-osa construct in extracellular complementation assays on leaves of kalanchöe. As anticipated, A. tumefaciens carrying ptac-osa did not serve as either a VirD2 or VirE2 donor (Fig. 6). These results are consistent with our previous data that Osa blocks the exit of both VirD2 and VirE2 from cells of Agrobacterium.
Figure 6.
Extracellular complementation of A. tumefaciens strains on kalanchöe. A total of eight leaves on eight different plants were inoculated, and the photos were taken 21 days later. The strains inoculated are noted beside the infection sites.
Discussion
VirD2 and VirE2 proteins and the T-DNA are presumably transferred directly from the bacterial cytoplasm into plant cells in a virB-dependent manner (11, 15). The data presented in this report indicate that these proteins, and VirF, were present in the periplasm and the supernatant. The presence of VirD2, VirE2, and VirF in the above two fractions did not result from cell lysis, because two cytoplasmic proteins (CAT and Ros) were absent in the same two fractions. Because VirJ and ChvE were also present in the supernatant, it is not clear whether VirE2, VirD2, and VirF entered the supernatant via a dedicated transport pathway or by passive leakage through the outer membrane. However, there is no doubt that VirE2, VirD2, and VirF entered the periplasm. Because VirD2 and VirE2 do not contain typical leader peptides and Osa selectively blocked the export of VirD2, VirE2, and VirF but not VirJ and ChvE, it is unlikely that a sec-dependent pathway participated in the process. In addition, the T-pilus was also not required for the export process.
A number of features of the secretion of the virulence proteins are consistent with other observations of T-complex transfer in A. tumefaciens. First, we observed that virulence proteins could be secreted independently of one another, a conclusion previously reached in the transfer of VirD2 and VirE2 into host cells. Second, several studies have shown that VirE1, the molecular chaperone for VirE2, is required for the secretion of VirE2 (23, 39, 50). We observed in the present study that VirE2 could be detected in the supernatant only when VirE1 was also synthesized.
The levels of virulence proteins we observed in the supernatant were only about 1% of the total virulence proteins synthesized at 28°C. The reason for this low level of secretion is not clear, but it is likely that the conditions for secretion we used were not optimal. For example, contact with host plants might enhance secretion similar to what has been observed in the case of type III secretion systems (51, 52). We also observed that both the levels of VirD2 and VirE2 in the supernatant increased at least fivefold when cells were grown at a lower temperature. This was also observed in the type III-mediated secretion of proteins into the supernatant (53). Although the low temperature is correlated with an increased formation of the T-pilus (12), this cannot be the explanation because secretion was independent of the T-pilus. Thus, temperature must impact other factors. Why temperature should be an important virulence factor and what cell component(s) was affected by temperature remain intriguing but unanswered questions.
Because proteins were secreted by cells expressing the virulence proteins at high levels in the absence of host plants, the VirD2 and VirE2 secretion might not be related to the actual transfer of the T-complex. The RSF1010 plasmid was reported to compete with the transport substrates, especially VirE2, for the transport pore (47), yet we have not observed differences in the levels of VirE2 and VirD2 in the supernatant whether or not pRSF1010 was present. It is possible that pRSF1010 might have influenced a later step in the transport process or caused a more general inhibition not restricted to any specific step. In addition, the question of whether T-DNA is required for secretion of VirD2 and VirE2 has not been addressed in this report.
One piece of evidence supporting the notion that the transfer of VirD2 and VirE2 into the supernatant plays a role in virulence is the observation that Agrobacterium expressing Osa was both avirulent and deficient in the export of the three virulence proteins. This suggests that the entry of VirE2 and VirD2 into the periplasm, and perhaps the supernatant, is required for virulence. Osa is located in the inner membrane of Agrobacterium (54), and the cellular location of Osa is consistent with its inhibitory effect on the export of the virulence proteins. Osa inhibition is selective because it did not alter the export of VirJ and ChvE whose export depends on a sec-dependent pathway. Additionally, it has been reported that many of the VirB proteins are produced and targeted properly in A. tumefaciens cells expressing Osa (54), implying that Osa does not interfere with the assembly of the T-pilus. This is consistent with our observation that the secretion of VirE2 and VirD2 into the periplasm did not require the virB system.
This report raises a number of unanswered questions. The most important one is does the secretion of these virulence proteins into the periplasm play a role in virulence? Although VirE2 and VirD2 were exported across the inner membrane in a virB-independent manner, critical genetic evidence linking this pathway to tumor formation is lacking. We have assayed some previously described avirulent chromosomal mutants; however, all were still able to transfer at least VirE2 (data not shown). We are currently attempting to isolate mutants that are defective in the transport of these proteins into the periplasm. Having such a mutant would test the current model of T-complex transfer in Agrobacterium (11, 15). Is it a one- or two-step process? Does the T-pilus play a role only in the second step? Pertussis toxin export in B. pertussis, mediated by a virB-like system, occurs in two stages (17, 20). First, the toxin subunits are exported into the periplasm by a sec-dependent pathway. This is followed by a second step that transfers the toxin into the host cell via the virB-like secretion system. It is possible that all virB-like secretion systems share a mechanistic similarity in addition to their protein homology. However, one difference between the transported proteins of Agrobacterium and Bordetella is that VirD2, VirE2, and VirF do not have signal sequences that would allow them to cross the inner membrane in a sec-dependent manner (11, 15). Our data suggest a model similar to the B. pertussis toxin transport, in which an additional step independent of VirB is involved that can be specifically blocked by Osa.
Acknowledgments
We thank Drs. Erh-min Lai and Clarence Kado for the gift of anti-Ros antibodies, Dr. Yasunori Machida for anti-ChvE antibodies, Dr. Paul J. J. Hooykaas for the anti-VirF antibodies, Dr. Stephen Farrand for pSa, and Dr. Anath Das for the strain AD802. Also thanked are Dr. Derek Wood for numerous discussions, and helpful insights into the observations. Ms. Yu Ching Chen helped to isolate the periplasmic proteins, and assisted in many other experiments. Dr. Walt Ream provided unpublished data and helpful discussions; Dr. Drucilla Burns shared her insight of the virB-like system in B. pertussis. Drs. Derek Wood, Wanyin Deng, and Beth Traxler critically reviewed the manuscript. We would also like to thank members of the Nester lab for many helpful discussions. This work was supported by National Institutes of Health Grant GM32618 and National Science Foundation Grant MCB-9723735.
Abbreviations
- CAT
chloramphenicolacetyl transferase
- Osa
oncogenic suppressive activity
- T-DNA
transferred DNA
- Ti
tumor-inducing
- MG/L
mannitol glutamate/Luria
- GFP
green fluorescent protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120156997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120156997
References
- 1.Kado C I. In: Genetic Engineering. Setlow J K, editor. Vol. 20. New York: Plenum; 1998. pp. 1–24. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Herrera-Estrella L, van Montagu M, Zambryski P. Cell. 1984;38:455–462. doi: 10.1016/0092-8674(84)90500-2. [DOI] [PubMed] [Google Scholar]
- 3.Albright L M, Yanofsky M F, Leroux B, Ma D, Nester E W. J Bacteriol. 1987;169:1046–1055. doi: 10.1128/jb.169.3.1046-1055.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yanofsky M F, Porter S G, Young C, Albright L M, Gordon M P, Nester E W. Cell. 1986;47:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- 5.Ward E R, Barnes W M. Science. 1988;242:927–930. [Google Scholar]
- 6.Young C, Nester E W. J Bacteriol. 1988;170:3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gietl C, Koukolikova-Nicola Z, Hohn B. Proc Natl Acad Sci USA. 1987;84:9006–9010. doi: 10.1073/pnas.84.24.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citovsky V, Vos G D, Zambryski P. Science. 1988;240:501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- 9.Tinland B, Koulkolikova-Nicola Z, Hall M N, Hohn B. Proc Natl Acad Sci USA. 1992;89:7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 11.Christie P J. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fullner K J, Lara J C, Nester E W. Science. 1996;23:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 13.Lai E-M, Kado C I. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 15.Winans S C, Burns D L, Christie P J. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 17.Burn D L. Curr Opin Microbiol. 1999;2:25–29. doi: 10.1016/s1369-5274(99)80004-6. [DOI] [PubMed] [Google Scholar]
- 18.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Hass R. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 19.Segal G, Russo J J, Shuman H A. Mol Microbiol. 1999;34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 20.Covacci A, Rappuoli R. Mol Microbiol. 1993;8:429–434. doi: 10.1111/j.1365-2958.1993.tb01587.x. [DOI] [PubMed] [Google Scholar]
- 21.Christie P J, Ward J E, Winans S C, Nester E W. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. Mol Gen Genet. 1984;195:159–163. [Google Scholar]
- 23.Deng W, Chen L, Peng W-T, Liang X, Sekiguchi S, Gordon M P, Comai L, Nester E W. Mol Microbiol. 1999;31:1795–1807. doi: 10.1046/j.1365-2958.1999.01316.x. [DOI] [PubMed] [Google Scholar]
- 24.Farrand S K, Kado C I, Ireland C R. Mol Gen Genet. 1981;181:44–51. [Google Scholar]
- 25.Cangelosi G A, Best E A, Martinetti G, Nester E W. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 26.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 27.Ausubel E M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 28.Lu H, Lory S. EMBO J. 1996;15:429–436. [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Deng W, Chen L, Wood D W, Metcalfe T, Liang X, Gordon M T, Comai L, Nester E W. Proc Natl Acad Sci USA. 1998;95:7040–7045. doi: 10.1073/pnas.95.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan S Q, Jin S, Boulton M I, Hawes M, Gordon M P, Nester E W. Mol Microbiol. 1995;17:259–269. doi: 10.1111/j.1365-2958.1995.mmi_17020259.x. [DOI] [PubMed] [Google Scholar]
- 32.Schrammeijer B, Hemelaar J, Hooykaas P J. Mol Plant–Microbe Interact. 1998;11:429–433. doi: 10.1094/MPMI.1998.11.5.429. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E, Lanes D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 34.McBride K E, Knauf V C. J Bacteriol. 1988;170:1430–1437. doi: 10.1128/jb.170.4.1430-1437.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossier O, Wengelnik K, Hahn K, Bonas U. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chilcott G S, Hughes K T. Mol Microbiol. 1998;30:1029–1040. doi: 10.1046/j.1365-2958.1998.01131.x. [DOI] [PubMed] [Google Scholar]
- 37.Das A, Xie Y-H. Mol Microbiol. 1998;27:405–414. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- 38.Fullner K J, Nester E W. J Bacteriol. 1996;178:1498–1503. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundberg C D, Ream W. J Bacteriol. 1999;181:6850–6855. doi: 10.1128/jb.181.21.6850-6855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cangelosi G A, Martinetti G, Nester E W. J Bacteriol. 1990;172:2172–2174. doi: 10.1128/jb.172.4.2172-2174.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cangelosi G A, Hung L, Puvanesarajah V, Stacey G, Ozga D A, Leigh J A, Nester E W. J Bacteriol. 1987;169:2086–2091. doi: 10.1128/jb.169.5.2086-2091.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang M L, Cangelosi G A, Halperin W, Nester E W. J Bacteriol. 1990;172:1814–1822. doi: 10.1128/jb.172.4.1814-1822.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wirawan I G P, Kang H W, Kojima M. J Bacteriol. 1993;175:3208–3212. doi: 10.1128/jb.175.10.3208-3212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 45.Buchanan-Wollaston V, Passiatore J, Cannon F. Nature (London) 1987;328:172–175. [Google Scholar]
- 46.DeFeyter R D, Yang Y, Gabrial D W. Mol Plant–Microbe Interact. 1993;6:225–237. doi: 10.1094/mpmi-6-225. [DOI] [PubMed] [Google Scholar]
- 47.Stahl L E, Jacobs A, Binns A N. J Bacteriol. 1998;180:3933–3939. doi: 10.1128/jb.180.15.3933-3939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, Y, Kado C I. J Bacteriol. 1994;176:5697–5703. doi: 10.1128/jb.176.18.5697-5703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee L Y, Gelvin S B, Kado C I. J Bacteriol. 1999;181:186–196. doi: 10.1128/jb.181.1.186-196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundberg C, Meek L, Carroll K, Das A, Ream W. J Bacteriol. 1996;178:1207–1211. doi: 10.1128/jb.178.4.1207-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 52.Collmer A. Curr Opin Plant Biol. 1998;1:329–335. doi: 10.1016/1369-5266(88)80055-4. [DOI] [PubMed] [Google Scholar]
- 53.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C Y, Kado C I. FEMS Microbiol Lett. 1996;135:85–92. doi: 10.1111/j.1574-6968.1996.tb07970.x. [DOI] [PubMed] [Google Scholar]