Abstract
A phage λ-based recombination system, Red, can be used for high-efficiency mutagenesis, repair, and engineering of chromosomal or episomal DNA in vivo in Escherichia coli. When long linear double-stranded DNA with short flanking homologies to their targets are used for the recombination, the λ Exo, Beta, and Gam proteins are required. The current model is: (i) Gam inhibits the host RecBCD activity, thereby protecting the DNA substrate for recombination; (ii) Exo degrades from each DNA end in a 5′ → 3′ direction, creating double-stranded DNA with 3′ single-stranded DNA tails; and (iii) Beta binds these 3′ overhangs to protect and anneal them to complementary sequences. We have tested this model for Red recombination by using electroporation to introduce overlapping, complementary oligonucleotides that when annealed in vivo approximate the recombination intermediate that Exo should create. Using this technique we found Exo-independent recombination. Surprisingly, a similarly constructed substrate with 5′ overhangs recombined more efficiently. This 5′ overhang recombination required both Exo and Beta for high levels of recombination and the two oligonucleotides need to overlap by only 6 bp on their 3′ ends. Results indicate that Exo may load Beta onto the 3′ overhang it produces. In addition, multiple overlapping oligonucleotides were successfully used to generate recombinants in vivo, a technique that could prove useful for many genetic engineering procedures.
Homologous recombination is intricately involved in DNA replication and repair of eukaryotic and prokaryotic organisms. Recent developments have provided new insights for the use of homologous recombination in gene therapy and in vivo genetic engineering (1). Recombination mediated by bacteriophage-encoded recombination functions is efficient and requires <50 base homologies to carry out the recombination (2–5). The λ Red phage-mediated recombination system uses PCR products (2, 5–7) or even synthetic single-stranded (ss) oligonucleotides (oligos) (8, 9) as substrates for recombination with homologous targets in the chromosome or episomes of Escherichia coli. Compared with classical in vitro genetic engineering, homologous recombination with PCR products and oligos provides more efficient, precise, and versatile ways to engineer DNA (1). Directly engineering recombinant chromosomes and plasmids in vivo by phage-encoded homologous recombination systems that use short homologies is called recombineering (10).
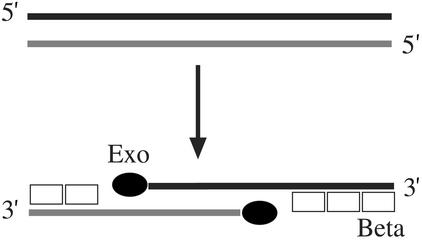
Recombineering between long (≈1 kb) linear duplex DNA [double-stranded DNA (dsDNA)] and the chromosome requires three phage λ Red functions (Gam, Exo, and Beta) and can occur in the absence of the E. coli RecA protein (2, 5). The Gam protein inhibits the nucleases RecBCD and SbcCD (1), thereby protecting linear dsDNA from nuclease degradation. Red Exo, a dsDNA-dependent exonuclease, degrades DNA in a 5′ → 3′ direction, leaving a duplex with 3′ overhangs (11, 12). Red Beta, a ssDNA-binding protein that anneals complementary DNA strands, will bind preferentially to these 3′ overhangs (13, 14). Thus, when dsDNA is used for recombineering, the current model is that the 5′ ends of dsDNA are degraded by λ Exo, leaving ssDNA overhangs at the 3′ ends. Beta binds to these ssDNA ends as an intermediate in the recombination process (Fig. 1). If this model is correct, direct introduction of dsDNA with 3′ DNA extensions will be recombinogenic and will not require λ Exo function.
Fig. 1.
Model for Red-mediated recombination. λ Exo enters a dsDNA end and degrades one DNA strand in a 5′ → 3′ direction. A linear fragment can be degraded from both ends simultaneously. Beta protein, perhaps aided by Exo, loads onto the 3′ ssDNA overhangs created by Exo. This substrate can then be used for strand invasion (26) or ss annealing (26, 28).
Materials and Methods
Bacterial Strains. HME6 is W3110 Δ(argF-lac)U169 [λcI857Δ(cro-bioA)] galKtyr145UAG. Derivatives of HME6 have the following additional markers: HME25 [λ Δgam<>cat], HME27 [λ Δexo<>cat], and HME43, [λ Δ(exo-int)<>cat Δ<>(gam-N)]. As in Yu et al. (2), the <> symbol denotes a replacement generated by recombineering. The Red recombination functions produced by these strains are as indicated in Table 1.
Table 1. λ Red recombination with ssDNA vs dsDNA substrates.
| Prophage
|
Recombinants†
|
||||||
|---|---|---|---|---|---|---|---|
| Strain* | exo | bet | gam | ssDNA | ssDNA + MBN | dsDNA | dsDNA + MBN |
| HME6 | + | + | + | 2 × 105 | 2 × 102 | 9 × 104 | 6 × 104 |
| HME25 | + | + | - | ND | ND | 1 × 104 | 1 × 104 |
| HME27 | - | + | + | ND | ND | 5 × 102 | 2 × 102 |
| HME43 | - | + | - | 9 × 104 | 3 × 101 | 1 × 102 | 1 × 101 |
ND, not determined.
Besides the prophage genotype listed, all strains contain the galKtyr145UAG mutation that renders them Gal-. These strains were induced for λ Red expression (see Materials and Methods) and were electroporated with ≈200 ng of either ssDNA or dsDNA. Samples were either treated with mung bean nuclease (MBN) or not as indicated before electroporation (see Materials and Methods)
Recombinants were normalized to 108 viable cells because this is the approximate number of survivors of electroporation. Viable cells were counted on LB plates and recombinants were selected on M63 plates
DY411 is W3110 [λcI857 Δ(cro-bioA)] galKΔ34bp<>kan. The galK allele was produced by recombineering using oligos described below and in Fig. 2. Derivatives of DY411 had the following additional markers, and the Red functions produced in each strain are indicated in Table 2: DY418 [λ Δgam<>cat], DY419 [λ Δbet<>cat], DY420 [λ Δexo<>cat], and DY433 [λ Δ(exo-int)<>cat Δ<>(gam-N)]. recA mutant versions [Δ(srl-recA)301::Tn10] of these strains were JS451, JS452, JS453, JS454, and JS455, respectively.
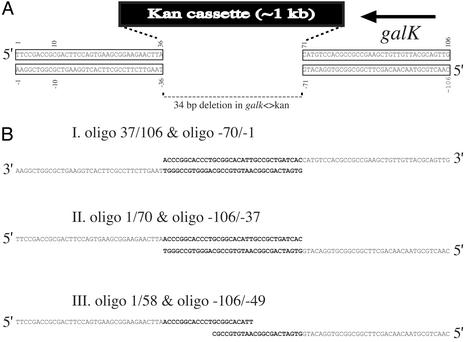
Fig. 2.
galK<>kan construct and oligos used to correct it to Gal+. (A) Diagram of the galK<>kan construct showing the 36-bp flanking homologies. Oligos are numbered from left to right with the top strand + and the bottom strand -. Thus, oligo 1/106 (not shown) contains the top strand in A including the flanking regions and the 34 bases deleted by the galK<>kan mutation. (B) Examples of oligos used in recombineering experiments. Bases in bold denote the bases missing in the 34-bp deletion created in galK<>kan.(I) Oligos used to test 3′ overhangs. (II) Oligos used to test 5′ overhangs. (III) Oligos with 5′ overhangs and a 10-bp overlap.
Table 2. λ Red recombination efficiency with dsDNA with ssDNA overhangs.
| Prophage
|
Recombinants†
|
||||
|---|---|---|---|---|---|
| Strain* | exo | bet | gam | 3′ overhangs‡ | 5′ overhangs‡ |
| DY411 | + | + | + | 3.0 × 102 | 1.4 × 105 |
| DY418 | + | + | - | 4.0 × 101 | 2.0 × 104 |
| DY419 | + | - | + | <1 | <1 |
| DY420 | - | + | + | 2.0 × 102 | 1.2 × 102 |
| DY433 | - | + | - | 3.0 × 101 | 3.8 × 102 |
Besides the prophage genotype listed, all strains contain a 34-bp deletion in galK replaced by Kmr. The cat substitution mutations in exo, bet, and gam were constructed as described (2). The Red-induced cells made from these strains were electroporated with ≈100 ng of each oligo
Recombinant and viable cells were normalized and determined as in Table 1. Values for 3′ and 5′ overhangs in DY411 are the average of 12 and 21 experiments, respectively. Absolute values in experiments vary up to 4-fold from the mean. This variation holds in all other experiments reported. On a given day, however, all experiments yield more internally consistent values than values compared between days
Oligos used for 3′ overhangs were 37/106 and -70/-1. Oligos used for 5′ overhangs were 1/70 and -106/-37 as shown in Fig. 2
Oligos. Oligos used were purchased from Invitrogen as salt free, without a 5′ phosphate, and are described in Fig. 2. Oligos to make galKΔ34bp<>kan were 5′-TTCCGACCGCGACTTCCAGTGAAGCGGAAGAACTTA/TATGGACAGCAAGCGAACCG and 5′-CAACTGCGTAACAACAGCTTCGGCGGCGTGGACATG/TCAGA AGA ACTCGTCA AGA AG. The/denotes the junction between galK homology and the kan cassette. Oligos to correct galKtyr145UAG were as described in Ellis et al. (8).
Oligos containing bases connected via phosphorothioate, “thiol,” groups and/or a 3′ terminal deoxy base were purchased from Oligos Etc. (Wilsonville, OR).
Preparation of Cells for Recombineering. Cells were grown and induced for Red functions for 15 min as in Yu et al. (2). In preparation for electroporation the protocol described by Yu et al. (2) has been modified as follows (Mikhail Bubunenko, personal communication). Cells chilled on ice (15 ml) were centrifuged at 6,500 × g for 7 min. After the supernatant was decanted, the cells were resuspended gently with 1 ml of ice-cold H2O, and an additional 30 ml of ice-cold H2O was added before centrifuging at 6,500 × g for 7 min. Beyond this point, the cell pellets were very loose. The supernatant was carefully discarded, and the pellet was suspended in 1 ml of ice-cold H2O and transferred to a microfuge tube. Cells were pelleted at 14,000 rpm at 4°C for 30 s and resuspended in 200 μl of ice-cold H2O. Electroporations were done with 40–50 μl of these cells and ≈100 ng of each oligo unless noted. After electroporation, using conditions described in Yu et al. (2), 1 ml of LB was immediately added to the electroporation mix, and the cells were grown at 30°C for 2 h before being diluted for plating. With DY411 and its derivatives >20% of the cells survive electroporation. However, there is a 10-fold reduction in cell survival after electroporation with HME6 and its derivatives.
Media. Cells were diluted in TMG buffer (10 mM Tris/10 mM Mg SO4/0.01% gelatin) before plating for Gal+ recombinants on M63 medium containing 0.2% galactose and biotin (0.001%). Plates were incubated at 32°C. Recombinants appeared after 3 days. Viable cell counts were determined on LB plates.
Mung Bean Nuclease. When indicated, oligos were treated with mung bean nuclease (New England Biolabs) according to the manufacturer's recommendations. Oligos were incubated with the nuclease for 30 min at 30°C, chilled on ice, and purified with a Qiagen (Chatsworth, CA) nucleotide removal kit.
Results
Recombineering with Short dsDNA Constructed from Oligos. Previously, it was demonstrated that a short, 70-bp dsDNA made by annealing complementary oligos can be used to transfer a galK point mutation from the dsDNA fragment to the E. coli chromosome (2). Once transferred to the chromsome, this mutation, galKtyr145UAG, is efficiently corrected to Gal+ by using a single-strand oligo (ssDNA) of 70 bases (8). Thus, it is possible that the recombination seen with the original 70-bp dsDNA was actually caused by residual ssDNA left after annealing the two oligos. We annealed two 70-base oligos and treated the mixture with mung bean nuclease, a ss-specific endonuclease, to remove ssDNA. Table 1 shows that the mung bean nuclease treatment reduced recombination generated by ssDNA by 1,000-fold, whereas recombination by dsDNA was largely unaffected. Additionally, the dsDNA required λ Exo (Table 1, lines 1 vs. 3 and 2 vs. 4) as well as Beta (data not shown) functions whereas ssDNA required only Beta (Table 1 and ref. 8). Deletion of gam had only a small effect on the dsDNA recombination. This finding is in contrast to previous experiments (2); however, in those experiments the large (at least 4,000-fold) Gam effect was seen with a long (≈1-kb) dsDNA. The small size of the linear duplex used here may render it more resistant to degradation by cellular nucleases.
As recombination by the dsDNA required Exo, and because mung bean nuclease did not adversely affect it, we conclude that the short dsDNA made from annealing two oligos is itself recombinogenic. However, it was still possible that once in the cell, Exo degrades one entire strand of a linear DNA before another Exo binds to the other end. This process could generate a recombinogenic ssDNA in vivo. To ascertain whether recombination that depends on both DNA strands can occur we set up the following assay.
An Assay for Recombination with Overlapping Oligos. Recombineering was used to create a galK mutation, galKΔ34bp<>kan (hereafter referred to as galK<>kan), with a kanamycin resistance gene cassette substituted for 34 bp of galK (Fig. 2). This strain, DY411, contains a defective prophage that encodes the Red recombination functions (ref. 2; see Materials and Methods), cannot grow on minimal plates with galactose as a sole carbon source (Gal-), and is resistant to kanamycin (KmR).
To test this strain, we generated a 106-bp PCR product containing the 34 bp of galK missing in galK<>kan, plus an additional 36 bp of galK DNA on each side. When electroporated into DY411, this 106-bp duplex repaired the galK mutation to Gal+ as it replaced the kan cassette making the cells KmS. Gal+ recombinants occurred at a frequency of 5 × 105/108 viable cells. Consistent with the 70-bp dsDNA results, this recombination depended on Exo and Beta but was largely Gam-independent (data not shown).
Recombineering Using dsDNA with 3′ Overhangs. To test the model for Red recombination presented in the introduction, we examined dsDNA with ssDNA overhangs at the 3′ ends for recombination activity (Fig. 2). The duplex DNA was made by annealing two 70-base oligos such that the 34 bases at the 5′ ends of the oligos were complementary and corresponded exactly to the 34-bp sequence deleted in galK<>kan. The 36 bases at each 3′ end remained unpaired and were homologous to the DNA flanking the kan substitution (Fig. 2BI). When each oligo was electroporated alone, they yielded no Gal+ recombinants as expected because each has homology to only one side of the deletion/substitution (data not shown). However, when both oligos were simultaneously electroporated into cells expressing the Red functions, 3 × 102 recombinants per 108 viable cells were seen (Table 2). Thus, our assay demonstrated recombination that depended on both DNA strands. We tested various concentrations of oligos and found that under our experimental conditions, 100 ng of each oligo was saturating as maximal recombination frequencies were seen (data not shown). Presumably, at this concentration the oligos finding each other is not rate limiting. Of course, “finding each other” includes getting into the cell, escaping nucleases, locating the complementary oligo and annealing to it, and completing the recombination reaction.
The frequency of Gal+ recombinants was the same whether the oligos were annealed before electroporation or just mixed in water and electroporated together (data not shown). Thus, annealing presumably occurs in vivo and may, in fact, be helped by the presence of Beta protein, which can bind and anneal ssDNAs (13, 15, 16). In all subsequent experiments, the oligos were coelectroporated without prior annealing.
Encouraged by the presence of Gal+ recombinants, we tested whether this recombination requires λ Exo. Consistent with the model, in a strain deleted for the exo gene, recombination remained fully active, showing Exo was not required (Table 2). Recombinant formation, however, fully depended on the Beta function as demonstrated by the lack of Gal+ recombinants in the bet deletion mutant. In fact, of λ functions, only Beta is required as evidenced by the recombination seen in strain, DY433. Thus, recombination with 3′ overhangs requires λ Beta function but is completely independent of λ Exo.
Recombineering with dsDNA with 5′ Overhangs. We next tested whether 5′ overhangs of the same DNA region were recombinogenic, and, if so, what λ Red functions were required. Two 70-base oligos were made such that they were complementary for 34 bases [the bases deleted by galK<>kan) at their 3′ ends; the remaining 36 bases of each oligo were homologous to flanking galK DNA (Fig. 2BII). When each oligo was tested alone, they yielded no recombinants as expected. However, when electroporated together, recombination to Gal+ was surprisingly efficient (Table 2) and depended on the presence of both Beta and Exo. Residual recombination activity in the absence of Exo was nearly identical in frequency to that found for dsDNA with 3′ overhangs. Thus, dsDNA with either 3′ or 5′ overhangs can recombine in the absence of Exo to generate Gal+ recombinants, albeit at low efficiency. dsDNA with 5′ overhangs becomes much more recombinogenic in the presence of Exo.
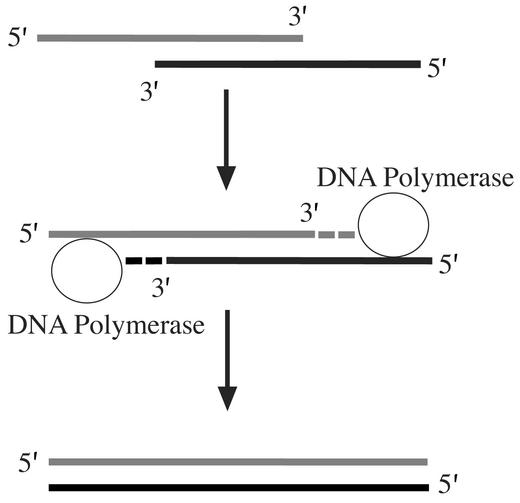
A major difference between dsDNA with 5′ vs. 3′ overhangs is that the former can be used as a substrate for a DNA polymerase to generate full-length duplex DNA (Fig. 3). We believe this occurs in vivo to dsDNA with 5′ overhangs because it would generate full-length dsDNA that then requires Exo for recombination.
Fig. 3.
Model to explain the high recombination frequency of dsDNA with 5′ overhangs. The recessed 3′ ends are accessible to a DNA polymerase, which can use the 5′ overhang as a template generating full-length dsDNA. The dsDNA can then be processed by the Red proteins as shown in Fig. 1.
Recombination Frequencies of dsDNA with 3′ Overhangs Can Be Increased with Additional Oligos. If the critical difference between the recombination efficiencies of 3′ and 5′ overhangs is that the 5′ overhangs are filled in by a DNA polymerase, then addition of oligos complementary to the 3′ overhangs might increase the recombination frequency. Table 3 shows that if we coelectroporate the two oligos needed to create the 3′ overhang substrate, plus an additional 36-base oligo complementary to one of the 3′ overhangs, we observed increased recombination frequency. The third oligo could be complementary to either 3′ overhang; however, oligo -106/-71 was 10-fold more effective than oligo 1/36. Addition of both of the extra oligos (four oligos total) brought the recombination frequency up to that seen with 5′ overhangs or dsDNA. These results are consistent with the model that 5′ overhangs are more recombinogenic than 3′ overhangs because the former are filled in vivo by a DNA polymerase.
Table 3. Additional oligos rescue poor recombination frequency of 3′ overhangs.
*Recombinant and viable cells were normalized and determined as in Table 1. Experiments were done in DY411.
Recombination of dsDNA with 5′ Overhangs Whose 3′ Ends Cannot Be Extended by a DNA Polymerase. If recombination with 5′ overhangs is efficient because a DNA polymerase can chain extend from the recessed 3′ end by using the 5′ overhang as a template to generate dsDNA (Fig. 3), then using modified bases on the 3′ end that cannot be extended should reduce recombination frequencies. Table 4 shows the results of recombination with two 70-base oligos with 34 complementary bases at their 3′ ends and 36-base 5′ overhangs. As noted in Table 4, some oligos had thiol groups between each of the last four bases on the 3′ end to reduce sensitivity to exonucleases. In addition, one set of oligos lacked a 3′ hydroxyl group and thus cannot be extended by a DNA polymerase. In the absence of a 3′ hydroxyl group, recombination was reduced nearly 100-fold compared with oligos containing thiol groups and a 3′ hydroxyl group. Consistent with the model, these results suggest a 3′ hydroxyl is necessary for the high level of recombination seen when using dsDNA with 5′ overhangs.
Table 4. Recombination of overlapping oligos whose 3′ ends are defective for chain extension.
| Recombination functions | Configuration of overlapping oligos | Gal+ recombinants* |
|---|---|---|
| WT | 3′ overhangs | 1.3×102 |
| Exo- | 3′ overhangs | 1.0×102 |
| WT | 5′ overhangs | 1.6×105 |
| Exo- | 5′ overhangs | 2.2×102 |
| WT | 5′ overhangs with thiol groups on 3′ end | 4.2×104 |
| Exo- | 5′ overhangs with thiol groups on 3′ end | 2.4×102 |
| WT | 5′ overhangs with thiol groups on 3′ end and no 3′ -OH group | 6.6×102 |
| Exo- | 5′ overhangs with thiol groups on 3′ end and no 3′ -OH group | 1.4×101 |
The WT strain was DY411, Exo- was DY420. Oligos used for 3′ overhangs were 37/106 and -70/-1. Oligos used for 5′ overhangs were 1/70 and -106/-37 as shown in Fig. 2. Oligos “with thiol groups” contained a phosphorothiate linkage between the last four 3′ bases. Those with “no 3′ -OH group” end in a 3′-deoxy-2′-hydroxyl as suggested by Oligos Etc.
Recombinant and viable cells were normalized and determined as in Table 1
Additionally, recombination of oligos without a 3′ hydroxyl group was reduced another 40-fold in the absence of λ Exo. These results indicate that although the absence of a hydroxyl group greatly reduced recombination, there is still a significant amount of Exo-dependent recombination. It is possible that even with thiol groups, an E. coli 3′ → 5′ exonuclease is degrading the terminal 3′ hydroxyl-free base, exposing a base containing a 3′ hydroxyl, which can be chain-extended by a DNA polymerase. λ Exo can then act on the dsDNA substrate created.
Length of Overlap Required for 5′ Overhang Recombination. As the efficiency of recombination with 5′ overhangs was high, we were able to determine how long the complementary region at the 3′ end (length of overlap of the two oligos) must be to generate recombinants. To do this, we used an oligo that has 36 bp of homology to galK and 22 bp of the 34 bp within the deleted region (oligo 1/58), combined with oligos of various lengths (Fig. 2). For comparison, a 34-bp overlap gave the highest recombination with 1.5 × 105 recombinants per 108 viable cells (Table 5). Reducing the overlap to 9 bp still generated nearly 104 recombinants, whereas further reducing the overlap to 5 bp eliminated all recombination. An overlap of 6 bp appears to be the minimum length necessary for recombination under these conditions. Using pairs of oligos that contain different portions of the deleted region, yet overlap by the same number of base pairs as those shown in Table 5, gave similar results (data not shown). Presumably, these experiments measure the minimal complementary sequence necessary for the two oligos to anneal in vivo. Consistent with our finding that 10-bp overlaps efficiently find each other and produce recombinants, the 12-base cohesive ends of λ rapidly find and anneal to each other after λ infection (17, 18).
Table 5. The effect of complementary base length on 5′ overhang recombination.
Recombineering with 3′ or 5′ Overhangs Is RecA-Independent. Strains producing the various Red recombination functions (Table 2) were deleted for the recA gene and recombination frequencies were determined. Sets of 70-base oligos that were complementary for either their 5′ or 3′ terminal 34 bases were used for recombination as in Table 2.
When all Red functions were present (DY411) and the oligos had 5′ overhangs, we saw ≈1.5 × 105 recombinants per 108 viable cells. Deleting recA (JS451) gave ≈3 × 105 recombinants per 108 viable cells. Likewise, no matter which Red functions were present (see Table 2 and Materials and Methods for recA mutant strains), no significant reductions in recombination frequencies were seen when the recA gene was deleted. The largest effect seen was in cases when gam was mutated. In those cases, mutation of recA gave up to a 10-fold effect in some trials of the experiments. Although the frequency of recombination is lower with 3′ overhangs (see Table 2), the effects of mutating recA were the same as those with 5′ overhangs. Thus, recombination with overlapping oligos with either 5′ or 3′ overhangs is largely recA-independent, but in all conditions tested fully depends on Beta.
Recombineering Can Be Used to Make Constructs That Require Multiple Oligos. Can Red-mediated recombineering be used to create more complex constructs such as full-length genes by using multiple overlapping oligos as has been done in vitro (19)? We used four oligos, any three of which when added to the cells failed to correct the galK<>kan mutation. The oligos each overlapped by 15 bases and there were five base gaps between them. When all four oligos (1/41, 47/80, -106/-66, and -61/-27) were added, recombinants were formed at a frequency of 3.5 × 102/108 viable cells. Although the efficiency of these events was low, it is adequate for strain constructions. In a similar experiment, six overlapping oligos were used to regenerate a complete suppressor tRNA gene, supF, which was recombined into the chromosome (Teresa Baker and D.L.C., unpublished work).
Discussion
Previously we reported that recombineering with short homologous regions flanking long (>1,000 bp) DNA cassettes depends on λ Exo, Beta, and Gam, but is largely independent of the E. coli RecA function (2, 8). Here we have analyzed the recombination of short homologous regions flanking a short (34 bp) DNA cassette. This recombination assay was designed so the flanking homologies (36 bp) could be either dsDNA or ssDNA and the assay could also detect recombinants that depended on both strands of DNA for repair. With the galK<>kan assay strain, two oligos, which were complementary for part of their length and if annealed had overhangs that were homologous to the flanking DNA, were simultaneously electroporated into cells, and Gal+ recombinants were scored.
Recombination of a short dsDNA with ssDNA overhangs was largely Gam-independent, perhaps because these substrates are resistant to degradation by cellular nucleases (Table 2). We found that overlapping complementary ssDNA oligos did not need to be preannealed before electroporation for optimum recombination. We propose that individual complementary oligos entering the cell during electroporation are bound by Beta before annealing to each other and recombining with galK. Beta binding could also provide nuclease resistance, rendering recombination of these substrates Gam-independent. Complementary Beta-bound oligos are expected to anneal to each other efficiently (13, 15, 16). Once annealed, the overlapping oligos may have Beta bound to both the dsDNA region (13, 14, 16) and the free ssDNA overhangs and thus be poised to anneal with homologous sites in the chromosome.
Similar to what has been reported for ssDNA (8), recombineering with overlapping oligos with either 3′ or 5′ ssDNA overhangs can occur with only the λ Beta protein (Table 2). This result seems to disagree with an in vitro study that showed Beta could not bind to 5′ overhangs (14). However, it is likely that Beta binds to the individual ssDNA oligos, anneals them, and remains bound as discussed above. Thus, even the 5′ overhangs would have Beta bound. With Beta alone, recombination was 10-fold more efficient with 5′ ssDNA overhangs than with 3′ overhangs (Table 2). This difference may be a RecBCD effect because when Gam is present, Beta recombined both 5′ and 3′ overhangs with similar efficiencies. It is unclear in this case what Gam is doing. Another unexplained feature of Beta-only recombineering was that recombinant colonies were variable in size. The presence of Exo alleviates this variability for 5′ overhangs, but not for 3′ overhangs. It is possible that the recombination event takes a long time to complete under these conditions and Gal+ recombinants arise sporadically over this time span.
Although Beta alone can use dsDNA with either 3′ or 5′ ssDNA overhangs for recombineering, Exo greatly stimulated recombination of substrates with 5′ overhangs. On the other hand, Exo had no beneficial effect on recombination with 3′ overhangs. To explain this disparity, we propose that when DNA with 5′ overhangs enters the cell, it is a substrate for a DNA polymerase, which can bind to the recessed 3′ end and use the 5′ overhang as a template to generate dsDNA (Fig. 3). This dsDNA can be used as a substrate for Exo- and Beta-mediated recombination (Fig. 1). If the ability to fill in the 5′ overhang by a DNA polymerase is the reason for increased recombination by this substrate, we expected that addition of an oligo complementary to a 3′ overhang might increase the recombination frequency seen with this substrate. We found this to be true. In addition, when the oligos with 5′ overhangs contained a 3′ terminal base lacking a 3′ hydroxyl group, recombination was greatly reduced. Both of these findings support the model that high levels of recombination proceed through a dsDNA intermediate. Thus, the higher frequency of recombination and Exo dependency seen with the 5′ overhangs relative to 3′ overhangs are likely caused by the fact that 3′ overhangs are obliged to go through the mechanism shown in Fig. 1, whereas the 5′ overhangs, once made ds, may go by this mechanism or also may be completely degraded for one strand, leaving ssDNA to be recombined by Beta at high levels. Our results cannot distinguish these possibilities.
Our results indicate that when Exo and Beta work together on dsDNA recombination is more efficient than when Beta alone catalyzes recombination even on the substrate that Exo presumably generates, dsDNA with 3′ overhangs. Because Beta is associated with Exo in vivo (13), Exo may actively load Beta onto the 3′ overhangs it creates similar to how RecBCD (20, 21) or RecORF (22–24) load RecA onto ssDNA in E. coli. We note, however, that this activity doesn't appear to be essential as Beta alone catalyzes efficient recombination with ssDNA (8).
We have used the overhang substrates to test the model that after a linear DNA enters the cell, Exo degrades part of the 5′ chains, producing a dsDNA with 3′ ssDNA overhangs, which Beta can use for recombination (Fig. 1). Our finding of Exo-independent recombination of a dsDNA substrate with 3′ ssDNA overhangs is consistent with this model. These results, however, are in contrast to those of Muyrers et al. (5). They constructed substrates with 3′ overhangs flanking a drug resistance marker. Regardless of the length of 3′ ssDNA overhangs, recombinants fully depended on Exo and Beta. Even with Exo and Beta present, however, recombination efficiency was low in these experiments, perhaps because of how the substrates were constructed (5). 5′ Overhangs were not tested in their experiments. Recombination using substrates with ssDNA overhangs was also tested in Saccharomyces cerevisiae (25). A 20-bp dsDNA “cassette” flanked by 60-base 3′ ssDNA overhangs gave no detectable recombinants. However, a low level of recombination (≈3 × 102 per 108 viable cells) with 60-base 5′ ssDNA overhangs was observed. As their recombination is 500-fold lower than what we observed with 5′ overhangs, perhaps recombination by 3′ overhangs occurs in yeast, but is below detection in the assay used. In addition to these low recombination levels, we note that recombination with oligos in yeast requires at least 100-fold higher DNA concentrations than in E. coli. Regardless, in both yeast and E. coli, dsDNA with 5′ ssDNA overhangs (as opposed to 3′ ssDNA overhangs) was the more recombinogenic substrate, perhaps indicating a similar mechanism.
Two mechanisms for Red recombination have been described, ss annealing and ss invasion (26, 27). In both models, the initial event is Exo-mediated processing of the ds end, creating a dsDNA substrate with a 3′ overhang that is coated with Beta protein (Fig. 1). In the presence of RecA, this end can invade another molecule (26). In the absence of RecA, it has been suggested that Red recombination is catalyzed by Beta-dependent annealing (26, 28) of complementary 3′ ssDNA ends generated by Exo. As our results were largely RecA-independent, this latter mechanism is likely to be relevant. Thus, for recombineering of dsDNA with overhangs, we propose that Beta annealing may occur at a DNA replication fork. Detailed models on how this may be occurring have been published (1).
Recombineering with overlapping oligos can be efficient, especially with 5′ ssDNA overhangs. We have demonstrated these overlaps need to be >5 bp; interestingly, homologies >5bp have been shown to allow dsDNA ends to be joined by Red recombination functions (27). The use of multiple overlapping oligos is efficient enough to make complex constructs or entire genes in vivo without the need for restriction enzymes or DNA ligase. This technique should prove useful for many genetic engineering procedures currently performed in vitro and for new ones designed with it in mind. As demonstrated here, these substrates have also provided insights about the recombination process itself (1).
Acknowledgments
We thank Mikhail Bubuneko, Nina Costantino, and Lynn Thomason for critical reading and discussion of the manuscript.
Abbreviations: ss, single-stranded; dsDNA, double-stranded DNA.
References
- 1.Court, D. L., Sawitzke, J. A. & Thomason, L. C. (2002) Annu. Rev. Genet. 36, 361-388. [DOI] [PubMed] [Google Scholar]
- 2.Yu, D., Ellis, H. M., Lee, E. C., Jenkins, N. A., Copeland, N. G. & Court, D. L. (2000) Proc. Natl. Acad. Sci. USA 97, 5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, Y., Buchholz, F., Muyrers, J. P. & Stewart, A. F. (1998) Nat. Genet. 20, 123-128. [DOI] [PubMed] [Google Scholar]
- 4.Muyrers, J. P., Zhang, Y., Testa, G. & Stewart, A. F. (1999) Nucleic Acids Res. 27, 1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muyrers, J. P., Zhang, Y., Buchholz, F. & Stewart, A. F. (2000) Genes Dev. 14, 1971-1982. [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, E. C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D. A., Court, D. L., Jenkins, N. A. & Copeland, N. G. (2001) Genomics 73, 56-65. [DOI] [PubMed] [Google Scholar]
- 7.Murphy, K. C., Campellone, K. G. & Poteete, A. R. (2000) Gene 246, 321-330. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, H. M., Yu, D., DiTizio, T. & Court, D. L. (2001) Proc. Natl. Acad. Sci. USA 98, 6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swaminathan, S., Ellis, H. M., Waters, L. S., Yu, D., Lee, E.-C., Court, D. L. & Sharan, S. K. (2001) Genesis 29, 14-21. [DOI] [PubMed] [Google Scholar]
- 10.Copeland, N. G., Jenkins, N. A. & Court, D. L. (2001) Nat. Rev. Genet. 2, 769-779. [DOI] [PubMed] [Google Scholar]
- 11.Carter, D. M. & Radding, C. M. (1971) J. Biol. Chem. 246, 2502-2512. [PubMed] [Google Scholar]
- 12.Little, J. W. (1967) J. Biol. Chem. 242, 679-686. [PubMed] [Google Scholar]
- 13.Karakousis, G., Ye, N., Li, Z., Chiu, S. K., Reddy, G. & Radding, C. M. (1998) J. Mol. Biol. 276, 721-731. [DOI] [PubMed] [Google Scholar]
- 14.Li, Z., Karakousis, G., Chiu, S. K., Reddy, G. & Radding, C. M. (1998) J. Mol. Biol. 276, 733-744. [DOI] [PubMed] [Google Scholar]
- 15.Kmiec, E. & Holloman, W. K. (1981) J. Biol. Chem. 256, 12636-12639. [PubMed] [Google Scholar]
- 16.Muniyappa, K. & Radding, C. M. (1986) J. Biol. Chem. 261, 7472-7478. [PubMed] [Google Scholar]
- 17.Kaiser, A. D. & Wu, R. (1968) Cold Spring Harbor Symp. Quant. Biol. 33, 729-734. [DOI] [PubMed] [Google Scholar]
- 18.Wu, R. & Kaiser, A. D. (1968) J. Mol. Biol. 35, 523-537. [DOI] [PubMed] [Google Scholar]
- 19.Stemmer, W. P., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. (1995) Gene 164, 49-53. [DOI] [PubMed] [Google Scholar]
- 20.Anderson, D. G. & Kowalczykowski, S. C. (1997) Cell 90, 77-86. [DOI] [PubMed] [Google Scholar]
- 21.Amundsen, S. K., Taylor, A. F. & Smith, G. R. (2000) Proc. Natl. Acad. Sci. USA 97, 7399-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bork, J. M., Cox, M. M. & Inman, R. B. (2001) EMBO J. 20, 7313-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegde, S. P., Qin, M. H., Li, X. H., Atkinson, M. A., Clark, A. J., Rajagopalan, M. & Madiraju, M. V. (1996) Proc. Natl. Acad. Sci. USA 93, 14468-14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umezu, K., Chi, N.-W. & Kolodner, R. D. (1993) Proc. Natl. Acad. Sci. USA 90, 3875-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storici, F., Lewis, L. K. & Resnick, M. A. (2001) Nat. Biotechnol. 19, 773-776. [DOI] [PubMed] [Google Scholar]
- 26.Stahl, M. M., Thomason, L., Poteete, A. R., Tarkowski, T., Kuzminov, A. & Stahl, F. W. (1997) Genetics 147, 961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, Y., Muyrers, J. P., Testa, G. & Stewart, A. F. (2000) Nat. Biotechnol. 18, 1314-1317. [DOI] [PubMed] [Google Scholar]
- 28.Kuzminov, A. (1999) Microbiol. Mol. Biol. Rev. 63, 751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]