Abstract
DNA polymerases of the A and B families, and reverse transcriptases, share a common mechanism for preventing incorporation of ribonucleotides: a highly conserved active site residue obstructing the position that would be occupied by a 2′ hydroxyl group on the incoming nucleotide. In the family Y (lesion bypass) polymerases, the enzyme active site is more open, with fewer contacts to the DNA and nucleotide substrates. Nevertheless, ribonucleotide discrimination by the DinB homolog (Dbh) DNA polymerase of Sulfolobus solfataricus is as stringent as in other polymerases. A highly conserved aromatic residue (Phe12 in Dbh) occupies a position analogous to the residues responsible for excluding ribonucleotides in other DNA polymerases. The F12A mutant of Dbh incorporates ribonucleoside triphosphates almost as efficiently as deoxyribonucleoside triphosphates, and, unlike analogous mutants in other polymerase families, shows no barrier to adding multiple ribonucleotides, suggesting that Dbh can readily accommodate a DNA–RNA duplex product. Like other members of the DinB group of bypass polymerases, Dbh makes single-base deletion errors at high frequency in particular sequence contexts. When making a deletion error, ribonucleotide discrimination by wild-type and F12A Dbh is the same as in normal DNA synthesis, indicating that the geometry of nucleotide binding is similar in both circumstances.
INTRODUCTION
The recently discovered lesion bypass, or family Y, DNA polymerases are characterized both by their ability to bypass DNA lesions and by their error-prone behavior during copying of undamaged DNA (1). The cellular role of this polymerase family appears to be to facilitate replication past sites of DNA lesions where a high-fidelity ‘classical’ replicative polymerase has stalled. Within the family of lesion bypass polymerases, four subgroups have been identified, based on amino acid sequence homology; these are DinB, UmuC, Rev1 and Rad30 (2). We have chosen the DinB homolog (Dbh) bypass polymerase from the thermophilic archaebacterium Sulfolobus solfataricus (3) as our model system to investigate structural and mechanistic aspects of error-prone DNA synthesis. An advantage of Dbh as a model system is that several relevant X-ray crystal structures have been solved: two of the uncomplexed Dbh, and two polymerase–DNA– deoxyribonucleoside triphosphate (dNTP) ternary complexes of a close homolog, Dpo4, from another S.solfataricus strain (4–6). In one Dpo4 complex (Dpo4.1), the active site contains an apparently normal pairing of an incoming nucleotide with the complementary base in the templating position. The other Dpo4 complex (Dpo4.2) represents a putative frameshift intermediate, with the incoming nucleotide paired opposite a template base that is the 5′ neighbor of the first unpaired template base. The frameshift structure may be significant in the light of observations, by us and others, that Dbh and other DinB polymerases have a remarkable propensity for generating a particular type of deletion error, in which the polymerase ‘skips over’ a template pyrimidine situated 3′ to a G residue (7–9).
The overall structures of lesion bypass polymerases show a global resemblance to those of the classical polymerases, but with some significant differences. The architecture of the polymerase domain conforms to the familiar ‘right hand’ structure, with fingers, thumb and palm subdomains (4–6,10). The palm subdomain contains the conserved ‘polymerase fold’ motif which provides the scaffold for important active site residues (11,12). Despite such global structural similarities, a comparison of ternary complex structures suggests that bypass polymerases make fewer contacts than classical polymerases with their DNA and nucleotide substrates (4,13–16). Moreover, unlike other polymerases, the bypass polymerases appear not to undergo a conformational transition, corresponding to closing of the fingers subdomain, on forming the polymerase–DNA–dNTP ternary complex (4,6, 17). Even in the uncomplexed state, the polymerase domain of the bypass polymerases resembles the closed conformation of the classical polymerases, implying that there is no need for a fingers-closing transition on the bypass polymerase reaction pathway. The structural comparisons, together with the low fidelity of DNA synthesis by family Y polymerases, suggest that the active site of this polymerase family might be arranged differently from that of the classical DNA polymerases, and might function differently with respect to substrate selection. We have chosen to address this question by investigating whether the Dbh polymerase selects the correct sugar structure of the incoming nucleotide by the same mechanism as in other DNA polymerases.
Although DNA polymerases from different families carry out diverse functions to maintain the integrity of the genome, they must all have a mechanism to select deoxyribonucleotides over ribonucleotides, given that the intracellular concentration of ribonucleotides is ∼10-fold greater (18). Structural and biochemical data from DNA polymerase families A and B, and from reverse transcriptases, support a conserved mechanism for ribonucleoside triphosphate (rNTP) discrimination, in which an amino acid side chain within the nucleotide binding pocket makes an unfavorable steric interaction with the 2′-OH of an incoming rNTP (19–25). The identity of the so-called ‘steric gate’ residue differs among polymerase families, but the side chain is strictly conserved, often invariant, within a family. Replacement of the steric gate side chain with a smaller side chain reduces the selectivity against single rNTP incorporation by several thousand fold; however, the resulting mutant proteins are not true RNA polymerases because they are extremely limited in their ability to synthesize long RNA products, indicating that there exist additional constraints to extension of an RNA primer by a DNA polymerase (19,20,22–25). From the Dbh and Dpo4 crystal structures, the steric gate residue of family Y polymerases is predicted to be a conserved Phe or Tyr (Phe12 in Dbh, Tyr12 in Dpo4) whose location, with respect to an incoming nucleotide, is analogous to the positions of steric gate residues in other polymerase structures (4,6,13–16).
In this study, we have tested the hypothesis that Phe12 of Dbh is the steric gate residue by making the F12A mutation, and we have used pre-steady-state kinetics to investigate sugar specificity in nucleotide incorporation by a family Y polymerase.
MATERIALS AND METHODS
Materials
DNA oligonucleotides were synthesized by the Keck Biotechnology Resource Laboratory at Yale Medical School. Oligonucleotides for PCR mutagenesis were desalted using a Sephadex G-25 spin column (Roche) and those used for kinetic measurements were gel purified as described previously (26). DNA duplex substrates for kinetic experiments were prepared by annealing a 5′-labeled 32P primer strand to its complementary template. Ultrapure dNTPs and rNTPs were purchased from Amersham Biosciences. Purification of the 3′-5′ exonuclease-deficient (D424A) E710A Klenow fragment derivative followed our standard procedure (27).
F12A Dbh mutagenesis
The plasmid expressing full-length wild-type Dbh (pDBH) was kindly provided by Bo-Lu Zhou. PCR mutagenesis of pDBH, to produce a plasmid (pF12A) expressing the F12A mutant derivative of Dbh was carried out using the QuikChange Site-Directed Mutagenesis kit, according to the manufacturer’s instructions (Stratagene). The Dbh coding region of pF12A was sequenced to ensure that F12A was the only mutation present.
Expression and purification of wild-type and F12A Dbh
Wild-type and F12A Dbh were overexpressed and purified from a 1 l cell culture as previously described (6). Briefly, pelleted cells were resuspended, lysed and heat treated (20 min at 70°C). After centrifugation, the supernatant was purified on a HiTrap SP–Sepharose column (Amersham Biosciences) and the eluted fractions containing pure Dbh were pooled, dialyzed into 20 mM HEPES–NaOH (pH 7.5), 100 mM NaCl, 5 mM 2-mercaptoethanol, 0.5 mM EDTA and 50% (v/v) glycerol, concentrated using a Centriprep-10 spin concentrator (Amicon) and stored at –20°C. The Ni-NTA column in the published purification was omitted because the proteins used for these studies were not His-tagged.
Kinetic measurements
Reactions were carried out at 22, 37 and 50°C under single-turnover conditions as previously described (7). To measure the kinetic parameters for incorporation of a single dNTP or rNTP, the relevant nucleotide was used at a series of concentrations. For addition of multiple dNTPs or rNTPs, the same reaction conditions were used except that all four nucleotides were present at concentrations of 100 or 500 µM each. To demonstrate the incorporation of ribonucleotides, alkali cleavage with piperidine was used to cleave at sites of ribonucleotide incorporation, as previously described (19).
RESULTS
Discrimination between dNTPs and rNTPs in single nucleotide incorporation
To investigate the selectivity of wild-type Dbh for deoxynucleotides over ribonucleotides, incorporation of a single dGTP or rGTP opposite a template C, in DNA duplex A (Table 1), was measured under single-turnover conditions. Measurements were carried out at 37°C because this temperature provided a good compromise between a detectable level of rNTP incorporation, and a dNTP incorporation reaction that was slow enough for the convenience of manual quenching. Pre-steady-state kinetic constants for nucleotide binding (Kd) and the maximum rate of incorporation (kpol) of dGTP and rGTP were determined from plots of the first-order rate constants (kobsd) as a function of nucleotide concentration (Table 1). The results for dGTP are consistent with our previous data for Dbh and demonstrate the low efficiency of nucleotide incorporation by this enzyme, approximately six orders of magnitude lower than for Klenow fragment (7). rGTP incorporation by wild-type Dbh was ∼1000-fold slower than dGTP incorporation, though the Kd values for both substrates were similar. The extremely slow rate of rGTP addition by wild-type Dbh translated to a high level of discrimination (3400-fold) against ribonucleotides at the polymerase active site.
Table 1. Pre-steady-state kinetic constants for dNTP and rNTP incorporation by wild-type and F12A Dbha.
| Protein/DNA | Kd (mM) | kpol (s–1) | kpol/Kd | Kd (mM) | kpol (s–1) | kpol/Kd | Selectivityb |
|---|---|---|---|---|---|---|---|
| DNA duplex A (non-frameshift) | 5′-TCGAAGCGACGGG | ||||||
| 3′-AGCTTCGCTGCCCCTTTCCATCA | |||||||
| dGTP |
rGTP |
dG/rG | |||||
| Wild-type | 0.37 ± 0.19 | (2.3 ± 0.9) × 10–2 | 62 | 0.77 ± 0.36 | (1.4 ± 0.3) × 10–5 | 1.8 × 10–2 | 3400 |
| F12A | 0.95 ± 0.1 | (2.1 ± 0.5) × 10–3 | 2.2 | 1.3 ± 0.2 | (7.8 ± 1.4) × 10–4 | 0.6 | 3.7 |
| DNA duplex B (frameshift) | 5′-GCATTTTCTGCTCCGG | ||||||
| 3′-CGTAAAAGACGAGGCCCGCC | |||||||
| dCTP |
rCTP |
dC/rC | |||||
| Wild-type | 0.46 ± 0.21 | (1.9 ± 1.1) × 10–2 | 41 | 0.58 ± 0.49 | (3.1 ± 2.9) × 10–6 | 5.3 × 10–3 | 7700 |
| F12A | 1.6 ± 0.6 | (4.0 ± 2.0) × 10–4 | 0.25 | 3.3 ± 1.0 | (1.6 ± 0.5) × 10–4 | 0.05 | 5.0 |
aKinetic parameters are the average of at least two independent determinations and are reported as mean ± SD. The relatively large standard deviations in the case of rNTP incorporations by wild-type Dbh are a consequence of the extremely slow rates of these reactions.
bSelectivity was calculated as the ratio of kpol/Kd values (dNTP/rNTP).
Structural studies predict that a highly conserved active-site side chain of family Y polymerases (F12 in Dbh) may function to prevent rNTP incorporation by steric exclusion of the 2′-OH (4–6). To test this hypothesis, we reduced the size of this side chain by making the F12A mutation, and expressed and purified the mutant Dbh derivative. We then measured the incorporation of dNTPs and rNTPs by the F12A mutant protein under single-turnover conditions, as described above for wild-type Dbh. A typical plot of the pre-steady-state data is shown in Figure 1, and the kinetic parameters are listed in Table 1. The maximum rate of dGTP incorporation (kpol) by F12A Dbh into DNA duplex A (Table 1) was 10-fold slower than for wild-type Dbh. However, the rate of rGTP incorporation by F12A Dbh was only 2–3-fold slower than incorporation of dGTP, so that, overall, F12A Dbh incorporated a single rNTP ∼50-fold faster than the wild-type protein.
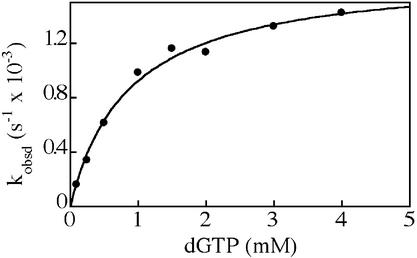
Figure 1.
Kinetics of dGTP incorporation by F12A Dbh. The first-order rate constants (kobsd) for dGTP addition to the DNA duplex A (Table 1) were plotted against dGTP concentration. The solid line represents the best fit of the data to a hyperbolic equation, giving a dissociation constant (Kd) of 0.87 ± 0.11 mM for binding of dGTP to the complex of F12A Dbh with primer/template, and a maximum incorporation rate (kpol) of (1.7 ± 0.1) × 10–3 s–1.
Because Dbh originates from a thermophile, we carried out a similar, though less extensive, set of measurements at 50°C (Fig. 2). Although the rate of dGTP incorporation by wild-type Dbh at 50°C was too fast to measure manually, the results were essentially identical to those described above: wild-type Dbh discriminated strongly against rNTPs, such that rGTP addition was barely detectable, whereas the F12A mutant Dbh incorporated dGTP and rGTP at similar rates.
Figure 2.
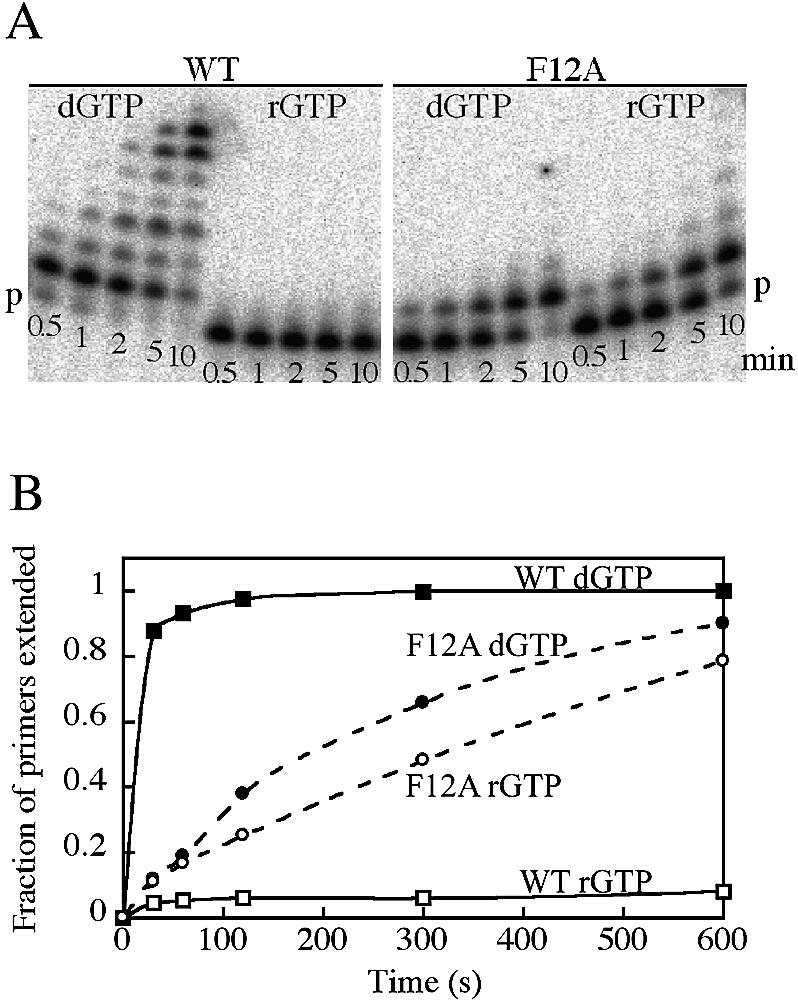
Incorporation of dGTP and rGTP by wild-type and F12A Dbh at 50°C, using DNA duplex A (Table 1). Reactions were carried out with 3 mM dGTP or rGTP using the conditions described in Materials and Methods. (A) Polyacrylamide-urea denaturing gel of samples removed and quenched at the indicated times (min). ‘p’ marks the position of the unextended primer. In the addition of dGTP by wild-type Dbh (WT), some primers have incorporated multiple G residues, either by misinsertion or slippage. (B) Quantitation of the gel shown in (A). The extent of reaction is measured as the fraction of primers that have been extended by at least one nucleotide.
rNTP discrimination in frameshift insertion
Error specificity studies of Dbh and other DinB polymerases indicate that these enzymes have a high propensity for generating single-base deletion mutations by skipping over a template pyrimidine and inserting dCTP opposite a G that is the 5′ neighbor of the skipped base (7–9). We have shown that the kinetics of frameshift insertion are very similar to non-frameshift incorporation, so that the frameshift incorporation reaction can readily be studied in vitro (7). To investigate whether the incoming nucleotide occupies a similar position within the Dbh active site in normal (non-frameshift) and frameshift insertion, we compared dCTP and rCTP incorporation by wild-type and F12A Dbh within a frameshift ‘hot spot’ DNA sequence (DNA duplex B in Table 1). The kinetic parameters (Table 1) were measured under single-turnover conditions at room temperature (22°C); this temperature was chosen because dCTP frameshift incorporation by wild-type Dbh was too rapid at higher temperatures to obtain reliable rate information using manual sampling. For both wild-type and F12A Dbh, the selectivity against ribonucleotide incorporation within a frameshift context was very similar to that during normal nucleotide incorporation. Wild-type Dbh selected against rCTP incorporation by several orders of magnitude, while F12A Dbh demonstrated almost no discrimination. No nucleotide incorporation by either wild-type or F12A Dbh was observed with a control DNA duplex that removed the potential for frameshifting by replacing the template 5′ G with a T (data not shown), confirming that the previously measured rates corresponded to bona fide frameshift incorporations of dCTP or rCTP, and not to C-dCTP/rCTP misinsertion events.
Addition of multiple rNTPs
Since F12A Dbh incorporated single ribonucleotides almost as efficiently as deoxyribonucleotides, we investigated the incorporation of successive rNTPs. When all four rNTPs were present under the same conditions used for the single-turnover assay, F12A Dbh readily added successive rNTPs to the DNA substrate, copying the entire 10 nt single-stranded template and generating a hybrid DNA/RNA product (Fig. 3). The rates of formation of the full-length product by F12A Dbh were very similar regardless of whether dNTPs or rNTPs were supplied. Although addition of the first ribonucleotide was slower (consistent with our kinetic data), subsequent rNTP addition appeared more processive than dNTP addition, so that fewer intermediate products were visible during RNA synthesis (Fig. 3). In contrast, the E710A steric gate mutant of Klenow fragment added the initial ribonucleotide much more rapidly than F12A Dbh but paused substantially after addition of approximately five ribonucleotides. As a result, there was an accumulation of products extended by five to seven residues, but very little full-length material, even after prolonged incubation. The contrast between E710A Klenow fragment and F12A Dbh is much more dramatic than it appears in Figure 3 because the higher specific activity of E710A Klenow fragment translates to a larger number of polymerase–DNA encounters in a given time interval. As expected from the results of the single rNTP addition experiments, wild-type Dbh gave almost no detectable product in the presence of four rNTPs (data not shown), but was capable of fully extending the DNA duplex substrate with all four dNTPs (Fig. 3). Primer extension by both wild-type and F12A Dbh gave products 1–2 nt longer than the expected full-length product (* in Fig. 3). This could result either from slippage during synthesis or from non-templated addition beyond the terminal base pair, as has been documented for a number of DNA polymerases (28). The possibility that minor dNTP contaminants might have contributed to the products observed during successive rNTP addition is ruled out by the difference in gel mobilities of the dNTP and rNTP extended primers. In addition, alkali cleavage of the products of rNTP addition regenerated the original DNA primer (data not shown).
Figure 3.
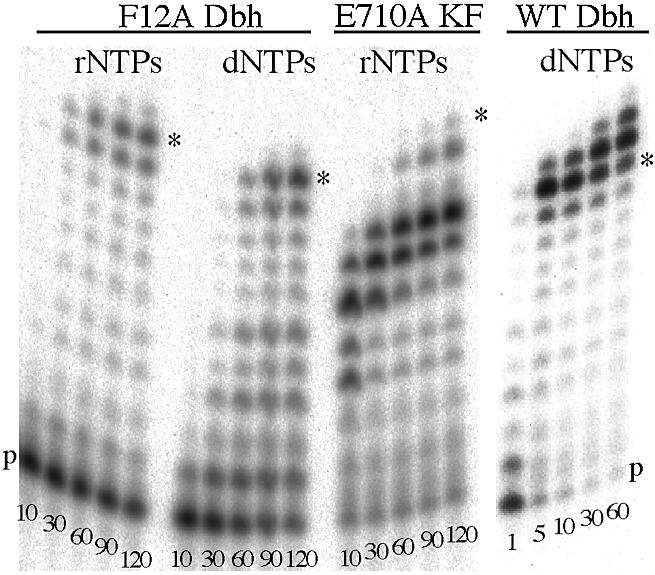
Addition of successive rNTPs or dNTPs to DNA duplex A (Table 1) by wild-type (WT) and F12A Dbh and E710A Klenow fragment (KF). Reactions were carried out as described in Materials and Methods, using all four dNTPs or rNTPs at 100 µM (F12A Dbh and E710A KF) or 500 µM (WT Dbh). Samples were removed and quenched at the indicated times (min), and fractionated on a polyacrylamide-urea denaturing gel. ‘p’ marks the position of unextended primer and an asterisk indicates the position of full-length product, corresponding to the addition of 10 nt. In the F12A reactions, faint bands larger than the expected fully extended primer were visible, presumably due to slippage or non-templated addition. These bands were more noticeable in the reaction of WT Dbh, due to the overall faster reaction rate.
DISCUSSION
Sugar recognition by Dbh
Structures of bypass polymerases suggest a more accessible active site, with fewer contacts between the protein and its substrates, compared with the classical DNA polymerases (4–6,10), and this is certainly consistent with the weak binding of dNTPs to Dbh noted in this study and our previous work (7). Despite these obvious differences in the binding of the incoming dNTP, the ability to recognize the sugar structure and discriminate between dNTPs and rNTPs is preserved even in family Y polymerases. Studies of DNA polymerases from families A, B, and now Y, and of reverse transcriptases, show that the wild-type enzymes discriminate against ribonucleotide incorporation by several thousand fold (Table 2). It is unclear whether differences in the magnitude of the dNTP/rNTP selectivity from enzyme to enzyme are significant, or merely reflect differences in experimental design and choice of nucleotide base. Certainly, the dGTP/rGTP selectivity of Dbh is similar in magnitude to the dCTP/rCTP selectivity of Klenow fragment, even though the family A DNA polymerases differ from the family Y DNA polymerases in having a much more snug binding pocket for the incoming nucleotide (13,14).
Table 2. Discrimination against rNTPs in four DNA polymerase families.
| DNA polymerase | dNTP/rNTP selectivitya | Wild-type/mutant selectivity ratiob | Reference | |
|---|---|---|---|---|
| Pol I (family A) | ||||
| KF | Wild-type | 3400 (C) | (19) | |
| E710A | 4.3 | 790 | ||
| Wild-type | 1 700 000 (T/U) | |||
| E710A | 33 | 50 000 | ||
| Pol α (family B) | ||||
| RB69 | Wild-type | 64 000 (C) | (22) | |
| Y416A | 19 | 3400 | ||
| φ29 | Wild-type | 4 400 000 (T/U) | (23) | |
| Y254V | 2300 | 1900 | ||
| Vent | Wild-type | (25) | ||
| Y412V | ||||
| Reverse transcriptase | ||||
| MMLV | Wild-type | 16 000 (T/U) | (20) | |
| F155V | 24 | 650 | ||
| HIV-1 | Wild-type | 130 000 (A) | (24) | |
| Y115A | 4.8 | 27 000 | ||
| Y115V | 170 | 765 | ||
| Family Y | ||||
| Dbh | Wild-type | 3400 (G) | This work | |
| F12A | 3.7 | 920 |
aSelectivity was calculated as the ratio of incorporation efficiencies (kpol/Kd), for Klenow fragment (KF), RB69 DNA polymerase and Dbh, and the ratio of Vmax/Km (or kcat/Km) values for φ29 DNA polymerase and the reverse transcriptases. Quantitative kinetic data were not reported for Vent DNA polymerase, but the overall trend was the same as for the other polymerases listed. The nucleotide used in each investigation is noted in parentheses.
bLoss of discrimination due to the side chain mutation was assessed as the ratio of selectivity values (dNTP/rNTP)wild type/(dNTP/rNTP)mutant.
Not only does Dbh have the same high level of rNTP discrimination as seen in other polymerases, but it also appears to use the same mechanism, namely steric exclusion of a ribonucleotide 2′-OH. As in other DNA polymerases (Table 2), mutation of the active-site side chain (F12) that is predicted to clash with the 2′-OH of an incoming ribonucleotide, produces a mutant Dbh derivative which shows almost no selectivity against ribonucleotides. In the family Y DNA polymerases, as in family B and reverse transcriptases, the relevant side chain is aromatic, whereas in family A a glutamate serves the same purpose; in each family the steric gate side chain is invariant or very highly conserved (21). Replacement of the side chain by alanine leads, in every case examined, to a dramatic decrease in dNTP/rNTP selectivity, whereas replacement by an intermediate-sized side chain causes a smaller decrease in selectivity (Table 2).
Kinetic aspects of sugar recognition
The three DNA polymerases for which single-turnover kinetics of rNTP incorporation have been studied (Dbh, Klenow fragment and RB69 DNA polymerase) show some interesting differences in their individual kinetic parameters. For wild-type Dbh, discrimination against rNTPs results entirely from changes in kpol; nucleotide binding is essentially the same for dNTPs and rNTPs. Similarly, for Klenow fragment, the discrimination is largely the result of kpol (∼103-fold), with a small (2-fold) effect on Kd (19). In contrast, in RB69 DNA polymerase, rNTP discrimination is attributable to almost equal effects on kpol and Kd (22). Moreover, in Klenow fragment and RB69 DNA polymerases, the effect of mutations in the steric gating residue is seen in Kd as well as in kpol. Thus, although different DNA polymerase families use a similar strategy to prevent rNTP incorporation, there are differences in the extent to which the discrimination operates at the ground state or the transition state for incorporation.
Kinetic studies of DNA polymerases (including an example from the Y family) suggest a common reaction pathway for dNTP incorporation in which a slow non-covalent step precedes the more rapid phosphoryl transfer step (29,30). Although it has been widely assumed that the rate-limiting non-covalent step corresponds to the closing of the fingers subdomain inferred from cocrystal structures of the classical polymerases, recent studies indicate that the closing of the fingers subdomain is more likely an early rapid step immediately following nucleotide binding (31,32; V. Purohit, N. D. F. Grindley and C. M. Joyce, submitted for publication). Therefore ground state nucleotide binding (Kd) should reflect interactions seen in the closed ternary complex, as has indeed been observed (32). Below, we discuss data on rNTP discrimination in the context of ternary complex cocrystal structures, which we believe provide the most satisfactory model for ground state nucleotide binding and for the subsequent transition state(s).
Structural comparison of sugar recognition strategies
Examination of the arrangement of protein side chains within the nucleotide binding pockets of classical and lesion bypass DNA polymerase families reveals some common features, suggesting that analogous selectivity strategies may operate in diverse polymerase families (Fig. 4). The steric gating amino acid (purple in Fig. 4) is at the C-terminus of the sequence motif A of Delarue et al. (33), which corresponds to a secondary structure element that is part of the common polymerase fold within the palm subdomain. In all four DNA polymerase families for which ternary complex structures are available, the incoming dNTP makes similar interactions with the steric gate residue and its immediate neighbors. The backbone NH of the steric gate residue is hydrogen-bonded to the dNTP 3′-OH, when present. (In the Klentaq and Dpo4 structures a dideoxy nucleotide was used and therefore the 3′-OH is absent; however, the distance between C3′ and the backbone nitrogen is essentially identical in all four structures.) The residue N-terminal to the steric gate is a highly conserved hydrophobic amino acid that forms part of the hydrophobic packing around the bound nucleotide; its backbone NH is hydrogen-bonded to a non-bridging oxygen within the triphosphate moiety. This is the β-phosphate in the three structures with a bound dNTP, but the α-phosphate of the dNDP of Dpo4.1 (suggesting the phosphates of the nucleotide may be mis-positioned in the latter structure). The net result of these interactions is to place the steric gate side chain in a consistent position relative to the nucleotide in all four polymerase families.
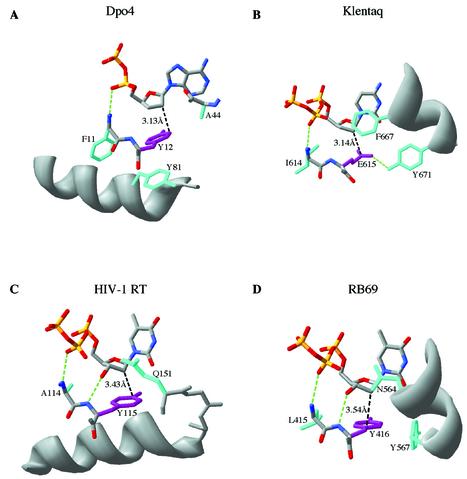
Figure 4.
Comparison of the structural determinants of sugar selectivity in four DNA polymerase families. Amino acid side chains close to the incoming dNTP in polymerase–DNA–dNTP ternary complexes are illustrated. In each case the steric gate residue that blocks the 2′-OH of an incoming rNTP is shown in purple, and the closest approach of this side chain to the C2′ position is indicated in black (Å). Other side chains (most of which are highly conserved in their respective polymerase families) that appear to play a role in maintaining the geometry of the dNTP binding pocket are shown in cyan. Important hydrogen bonds are illustrated in green. The polymerases are: (A) Dpo4, PDB file 1JX4 (4); (B) Klentaq, PDB file 3KTQ (14); (C) HIV-1 reverse transcriptase, PDB file 1RTD (16); (D) RB69 DNA polymerase, PDB file 1QSS (15). Note that in the Dpo4 structure (A) the nucleotide shown is a nucleoside diphosphate instead of a nucleoside triphosphate, due to hydrolysis during crystallization; moreover, the interaction between the α-phosphate and the polypeptide backbone resembles the interaction involving the β-phosphate in the other three illustrated structures. This figure was made using Swiss PDB viewer (version 3.7b2) (38).
In addition to the steric gate residue itself, an important component of the steric exclusion of rNTPs is the ability of other active-site side chains to hold the steric gate and the 2′-OH in a consistent juxtaposition so that specificity is enforced (19). In the family A polymerases, exemplified by Klentaq (Fig. 4B), the C-terminal residues of the O-helix play a crucial positioning role, with the invariant Y671 hydrogen-bonded to the E615 steric gate, and F667 (conserved as Phe or Tyr in family A) (26) constraining the position of the base and sugar of the incoming nucleotide (13,14). Indeed, mutation of the equivalent residue (F762) to Ala in Klenow fragment decreased dNTP/rNTP selectivity (19). In family B, exemplified by RB69 DNA polymerase (Fig. 4D), a highly conserved Asn (N564) interacts with the base and sugar, while the invariant Tyr (Y567) is in van der Waal’s contact with the Y416 steric gating side chain (15,34). The N564 and Y567 side chains are analogous to the Phe and Tyr side chains described above in the family A polymerases in that both pairs of side chains define the final turn of similarly positioned helices in the fingers subdomain, and are aligned with one another in the conserved sequence motif B of Delarue et al. (33). Although the steric gating residue in family B polymerases is always Tyr (never Phe), mutations to Phe had little or no effect on rNTP incorporation (22,23,25), suggesting that selectivity does not require interactions with the Tyr OH in order to position the side chain correctly. In reverse transcriptases (Fig. 4C), a highly conserved Gln side chain is positioned close to the base and sugar of the bound dNTP. The Gln is part of a conserved sequence motif (35) at the N-terminus of a helix that forms the base of the dNTP binding pocket and supports the Tyr or Phe steric gate side chain. In the bypass polymerase structure (Fig. 4A), an analogous helix, including an almost invariant Tyr (Y81 in Dpo4), supports the steric gate side chain. In contrast to the other polymerase structures, the fingers subdomain of Dpo4 presents a relatively featureless surface, with the backbone of A44 closest to the dNTP base and sugar (6). This side chain is not particularly highly conserved though small side chains predominate in this region (J. Pata, personal communication). Nevertheless, the high dNTP/rNTP selectivity of Dbh suggests that these minimal protein contacts must constrain the dNTP sufficiently to enforce steric selection against rNTPs.
Successive rNTP addition by F12A Dbh
The F12A mutant derivative of Dbh differs from the steric gate mutants of classical DNA polymerases in its much greater ability to incorporate multiple ribonucleotides, so that it operates equally efficiently as either a DNA or an RNA polymerase. Although F12A Dbh adds the first rNTP ∼2-fold more slowly than dNTP addition, incorporation of subsequent rNTPs appears to be as fast as dNTP addition and may also be slightly more processive, resulting in efficient copying of a 10 nt template overhang. In contrast, the steric gate mutants of other DNA polymerases tend to stall after addition of two to six ribonucleotides (19,20,24,25,36). This is nicely illustrated by E710A Klenow fragment in Figure 3; although the efficiency (kpol/Kd) of the first rNTP addition (and of normal dNTP addition) is 105-fold greater than that of F12A Dbh, the Dbh mutant synthesized a substantial amount of full-length product under conditions where the Klenow fragment mutant synthesized hardly any. The limited ribonucleotide incorporation by mutants of the classical DNA-dependent DNA polymerases appears to correlate with the template–primer duplex conformations seen in ternary complexes of these polymerases. Although the last few primer–terminal base pairs are frequently seen in an A-like conformation (13,14,16,37), which would be compatible with an RNA–DNA hybrid product, the rest of the duplex is B-DNA. Moreover, the numerous interactions between the protein and the duplex probably serve to enforce the observed conformation so that an RNA–DNA hybrid product would not be well tolerated further from the active site. In contrast, the comparatively open duplex binding site of the bypass polymerases may play a more passive role, not enforcing any particular conformation. Thus, a bypass polymerase could bind a DNA duplex as B-form, as seen in the Dpo4 ternary complex (4), and yet not discriminate against an A-form RNA–DNA hybrid, as implied by our biochemical data. Examination of the Dbh and Dpo4 structures suggests that an A-form duplex could be accommodated with a small adjustment of the thumb and a somewhat larger reorientation of the C-terminal domain (J. Pata, personal communication). [It is worth noting that the C-terminal domain is seen in different positions in two crystal structures (4,5), and that its attachment to the rest of the Dbh molecule is protease-sensitive (4,6)]. The ability of bypass polymerases to tolerate a variety of DNA geometries would be consistent with the need to accommodate bulky, helix-distorting modifications during lesion bypass.
Nucleotide binding during frameshift insertion by Dbh
Discrimination against rNTP incorporation depends on the positioning of the steric gate side chain relative to the incoming nucleotide, and can therefore serve as a diagnostic for changes in the geometry of nucleotide binding. When Dbh adds a nucleotide such that the first available templating base is skipped over (frameshift insertion), the dNTP/rNTP selectivity and the contribution of the F12 side chain to that selectivity were in close agreement with the selectivities measured for non-frameshift incorporation. We therefore conclude that nucleotide positioning during frameshift insertion does not differ greatly from normal (non-frameshift) nucleotide incorporation.
Contrary to this conclusion, the ternary complex of Dpo4 in an apparently frameshifted conformation (Dpo4.2) would predict less stringent rNTP discrimination during frameshift insertion since the sugar C2′ position is ∼1.3 Å further from F12 than it is in the Dpo4.1 complex (Fig. 5) (4). However, the Dpo4.2 complex may not be a good representation of the situation during frameshift insertion because it is demonstrably not a catalytically competent configuration. Although the primer terminus in the Dpo4.2 complex has a 3′-hydroxyl, no reaction has taken place with the incoming dNTP. The Dpo4.2 complex may have failed to achieve a catalytically competent configuration because the template–primer sequence does not conform to the favored sequence for –1 deletion formation by polymerases of the DinB group (7–9); the skipped base is a purine instead of the preferred pyrimidine, and the incoming nucleotide is dGTP rather than the preferred dCTP. Additionally, there are some puzzling features of the Dpo4.1 and Dpo4.2 structures that raise questions as to the inferences that can be drawn from these complexes: the differences in metal coordination (Fig. 5), the loss of the γ-phosphate in the Dpo4.1 complex, the differences in phosphate positions between Dpo4.1, Dpo4.2 and other DNA polymerase ternary complexes (see Fig. 4) and the presence of a molecule of ethylene glycol opposite the skipped template base in the Dpo4.2 complex (4). Based on our rNTP discrimination data, we predict that the ternary complex in frameshift insertion will have the dNTP in a position similar to that seen during normal (non-frameshift) insertion and that the substantial structural rearrangements necessary to accommodate the skipped base will be seen on the template side of the binding site.
Figure 5.
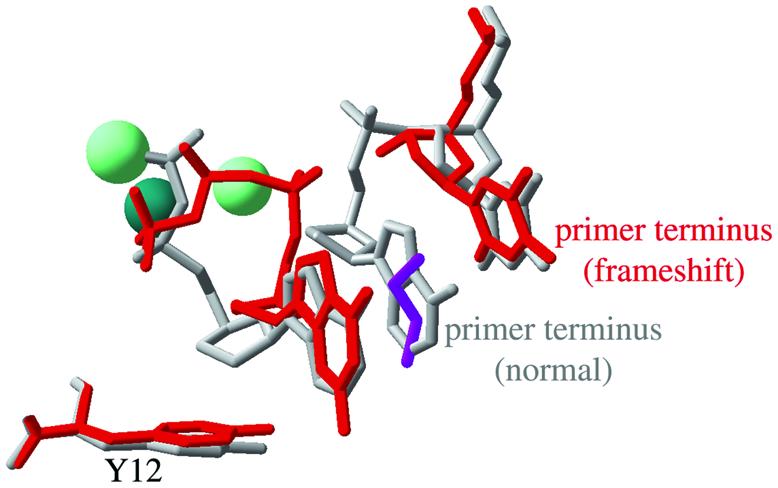
Comparison of the positions of the incoming nucleotide and the Tyr12 steric gate side chain in the Dpo4.1 and Dpo4.2 complexes. The Dpo4.1 complex, in which the incoming nucleotide (ddADP) is paired opposite the first unpaired template base, is illustrated in gray. The Dpo4.2 complex, in which an incoming ddGTP is paired opposite the base 5′ to the first unpaired template base (an apparently frameshifted conformation) is shown in red. Also shown are the primer terminus and the preceding nucleotide from the Dpo4.1 complex, and the primer terminus from the Dpo4.2 complex. The Dpo4.2 complex contains three metal ions at the polymerase active site: two Mg2+ (pale green) and one Ca2+ (dark green). The Dpo4.1 complex contains only the Ca2+ ion in essentially the same position as illustrated for Dpo4.2. The Dpo4.2 complex also contains a molecule of ethylene glycol (purple) that occupies the large space between the primer terminus and the incoming nucleotide. This figure was made by superimposing the alpha carbons from the PDB structure files 1JX4 (Dpo4.1) and 1JXL (Dpo4.2) (4) using Swiss PDB viewer (version 3.7b2) (38).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Olga Potapova for excellent technical support, and Janice Pata and Bo-Lu Zhou for providing the Dbh expression strain. We also thank Janice Pata for her thoughtful discussions regarding Dbh structure, for an unpublished alignment of bypass polymerase sequences and for comments on this manuscript. This work was supported by NIH Grant GM-28550.
REFERENCES
- 1.Goodman M.F. (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem., 71, 17–50. [DOI] [PubMed] [Google Scholar]
- 2.Ohmori H., Friedberg,E.C., Fuchs,R.P.P., Goodman,M.F., Hanaoka,F., Hinkle,D., Kunkel,T.A., Lawrence,C.W., Livneh,Z., Nohmi,T. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- 3.Kulaeva O.I., Koonin,E.V., McDonald,J.P., Randall,S.K., Rabinovich,N., Connaughton,J.F., Levine,A.S. and Woodgate,R. (1996) Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res., 357, 245–253. [DOI] [PubMed] [Google Scholar]
- 4.Ling H., Boudsocq,F., Woodgate,R. and Yang,W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell, 107, 91–102. [DOI] [PubMed] [Google Scholar]
- 5.Silvian L.F., Toth,E.A., Pham,P., Goodman,M.F. and Ellenberger,T. (2001) Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nature Struct. Biol., 8, 984–989. [DOI] [PubMed] [Google Scholar]
- 6.Zhou B.L., Pata,J.D. and Steitz,T.A. (2001) Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell, 8, 427–437. [DOI] [PubMed] [Google Scholar]
- 7.Potapova O., Grindley,N.D.F. and Joyce,C.M. (2002) The mutational specificity of the Dbh lesion bypass polymerase and its implications. J. Biol. Chem., 277, 28157–28166. [DOI] [PubMed] [Google Scholar]
- 8.Kokoska R.J., Bebenek,K., Boudsocq,F., Woodgate,R. and Kunkel,T.A. (2002) Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J. Biol. Chem., 277, 19633–19638. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi S., Valentine,M.R., Pham,P., O’Donnell,M. and Goodman,M.F. (2002) Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem., 277, 34198–34207. [DOI] [PubMed] [Google Scholar]
- 10.Trincao J., Johnson,R.E., Escalante,C.R., Prakash,S., Prakash,L. and Aggarwal,A.K. (2001) Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell, 8, 417–426. [DOI] [PubMed] [Google Scholar]
- 11.Joyce C.M. and Steitz,T.A. (1994) Function and structure relationships in DNA polymerases. Annu. Rev. Biochem., 63, 777–822. [DOI] [PubMed] [Google Scholar]
- 12.Brautigam C.A. and Steitz,T.A. (1998) Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol., 8, 54–63. [DOI] [PubMed] [Google Scholar]
- 13.Doublié S., Tabor,S., Long,A., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 14.Li Y. and Waksman,G. (2001) Crystal structures of a ddATP-, ddTTP-, ddCTP- and ddGTP-trapped ternary complex of Klentaq1: insights into nucleotide incorporation and selectivity. Protein Sci., 10, 1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin M.C., Wang,J. and Steitz,T.A. (2001) Structure of the replicating complex of a pol α family DNA polymerase. Cell, 105, 657–667. [DOI] [PubMed] [Google Scholar]
- 16.Huang H., Chopra,R., Verdine,G.L. and Harrison,S.C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- 17.Yang W. (2003) Damage repair DNA polymerases Y. Curr. Opin. Struct. Biol., 13, 23–30. [DOI] [PubMed] [Google Scholar]
- 18.Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd Edn. W.H. Freeman and Company, New York, NY. [Google Scholar]
- 19.Astatke M., Ng,K., Grindley,N.D.F. and Joyce,C.M. (1998) A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl Acad. Sci. USA, 95, 3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G., Orlova,M., Georgiadis,M.M., Hendrickson,W.A. and Goff,S.P. (1997) Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl Acad. Sci. USA, 94, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce C.M. (1997) Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl Acad. Sci. USA, 94, 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang G., Franklin,M., Li,J., Lin,T.C. and Konigsberg,W. (2002) A conserved Tyr residue is required for sugar selectivity in a Pol α DNA polymerase. Biochemistry, 41, 10256–10261. [DOI] [PubMed] [Google Scholar]
- 23.Bonnin A., Lázaro,J.M., Blanco,L. and Salas,M. (1999) A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase. J. Mol. Biol., 290, 241–251. [DOI] [PubMed] [Google Scholar]
- 24.Cases-González C.E., Gutiérrez-Rivas,M. and Menéndez-Arias,L. (2000) Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem., 275, 19759–19767. [DOI] [PubMed] [Google Scholar]
- 25.Gardner A.F. and Jack,W.E. (1999) Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res., 27, 2545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Astatke M., Grindley,N.D.F. and Joyce,C.M. (1998) How E. coli DNA polymerase I (Klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J. Mol. Biol., 278, 147–165. [DOI] [PubMed] [Google Scholar]
- 27.Joyce C.M. and Derbyshire,V. (1995) Purification of E. coli DNA polymerase I and Klenow fragment. Methods Enzymol., 262, 3–13. [DOI] [PubMed] [Google Scholar]
- 28.Clark J.M. (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res., 16, 9677–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benkovic S.J. and Cameron,C.E. (1995) Kinetic analysis of nucleotide incorporation and misincorporation by Klenow fragment of Escherichia coli DNA polymerase I. Methods Enzymol., 262, 257–269. [DOI] [PubMed] [Google Scholar]
- 30.Washington M.T., Prakash,L. and Prakash,S. (2001) Yeast DNA Polymerase η utilizes an induced-fit mechanism of nucleotide incorporation. Cell, 107, 917–927. [DOI] [PubMed] [Google Scholar]
- 31.Arndt J.W., Gong,W., Zhong,X., Showalter,A.K., Liu,J., Dunlap,C.A., Lin,Z., Paxson,C., Tsai,M.-D. and Chan,M.K. (2001) Insight into the catalytic mechanism of DNA polymerase β: structures of intermediate complexes. Biochemistry, 40, 5368–5375. [DOI] [PubMed] [Google Scholar]
- 32.Vande Berg B.J., Beard,W.A. and Wilson,S.H. (2001) DNA structure and aspartate 276 influence nucleotide binding to human DNA polymerase β. Implication for the identity of the rate-limiting conformational change. J. Biol. Chem., 276, 3408–3416. [DOI] [PubMed] [Google Scholar]
- 33.Delarue M., Poch,O., Tordo,N., Moras,D. and Argos,P. (1990) An attempt to unify the structure of polymerases. Protein Eng., 3, 461–467. [DOI] [PubMed] [Google Scholar]
- 34.Filée J., Forterre,P., Sen-Lin,T. and Laurent,J. (2002) Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol., 54, 763–773. [DOI] [PubMed] [Google Scholar]
- 35.Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia G., Chen,L., Sera,T., Fa,M., Schultz,P.G. and Romesberg,F.E. (2002) Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl Acad. Sci. USA, 99, 6597–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson S.J., Taylor,J.S. and Beese,L.S. (2003) Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl Acad. Sci. USA, 100, 3895–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]