Abstract
The catalytic subunit of telomerase (TERT) contains conserved reverse transcriptase-like motifs but N- and C-terminal regions unique to telomerases. Despite weak sequence conservation, the C terminus of TERTs from various organisms has been implicated in telomerase-specific functions, including telomerase activity, functional multimerization with other TERT molecules, enzyme processivity and telomere length maintenance. We studied hTERT proteins containing small C-terminal deletions or substitutions to identify and characterize hTERT domains mediating telomerase activity, hTERT multimerization and processivity. Using sequence alignment of five vertebrate TERTs and Arabidopsis thaliana TERT, we identified blocks of highly conserved amino acids that were required for human telomerase activity and functional hTERT complementation. We adapted the non-PCR-based telomerase elongation assay to characterize telomerase expressed and reconstituted in the in vitro transcription/translation rabbit reticulocyte lysate system. Using this assay, we found that the hTERT C terminus, like the C terminus of Saccharomyces cerevisiae TERT, contributes to successive nucleotide addition within a single 6-base telomeric repeat (type I processivity). Certain mutations in the hTERT C terminus also reduced the repetitive addition of multiple telomeric repeats (type II processivity). Our results suggest a functionally conserved role for the TERT C terminus in telomerase enzyme processivity.
INTRODUCTION
Telomeres are unique DNA–protein structures that cap the ends of linear chromosomes (1). Telomere integrity is essential for chromosome stability, and the maintenance of telomeric DNA is required for the long-term proliferation of eukaryotic cells (2). Several proteins have been identified that associate with telomeres and regulate various aspects of telomere structure, organization and length (1). In vertebrates, telomeres are composed of G-rich repeats synthesized by the ribonucleoprotein complex telomerase (3).
The human telomerase enzyme is minimally composed of a protein catalytic subunit, the telomerase reverse transcriptase (hTERT), and an RNA molecule, hTR. A short sequence within hTR serves as a template for de novo addition of telomere sequences (4). Comparative analysis of the amino acid sequence of TERT from different organisms revealed several conserved domains in TERT (5,6). The central region of TERT contains reverse transcriptase (RT)-like motifs and a telomerase-specific (T) motif essential for telomerase function in vitro and in vivo (7–10). TERT-specific motifs in the N terminus are important for enzymatic activity and are implicated in telomerase RNA binding and RNA template boundary definition (11–16). Recently, hTERT N-terminal amino acid residues that interact with hTR were defined (17). A DAT (dissociates activities of telomerase) region essential for telomere maintenance in vivo was mapped to the hTERT N terminus, and may be involved in the recruitment of the telomerase enzyme to telomeres (18). The N terminus of hTERT is also involved in functional and physical multimerization with other hTERT molecules, in telomerase ribonucleoprotein (RNP) formation in vivo, and in hTERT nucleolar localization (12,17,19–23).
The C terminus of TERT is not well conserved within the TERT family (24). However, the C-terminal domain is essential for human telomerase function in vivo. The addition of a hemagglutinin (HA) epitope tag at the C-terminal end impairs hTERT’s ability to maintain telomere length, increase the cellular life span of primary cells and immortalize transformed human cells (25,26). The expression of a hTERT C-terminal polypeptide in HeLa cells leads to telomere dysfunction, characterized by chromosome end-to-end fusions, and induces a senescence-like phenotype and apoptosis in the absence of telomere shortening (27). These studies suggest a role for the C-terminal domain in maintaining telomere length, integrity and structure (25–27). Recently, a DAT region essential for telomere maintenance in vivo was mapped to the hTERT C terminus (28). The Saccharomyces cerevisiae TERT C terminus also contributes to telomere length regulation, but is not required for cell survival (11). In addition, the C terminus of hTERT contains binding sites for the 14-3-3 and CRM1 proteins, which regulate the intracellular localization of telomerase (29).
The C terminus of the TERTs is also essential for telomerase catalytic function. Analysis of human and Tetrahymena TERT C-terminal truncations revealed that the C-terminal domain is required for in vitro enzymatic activity but probably not for binding to the telomerase RNA (12,15,16). Sequences at the junction of the RT motif E and the C terminus of hTERT play a role in functional and physical multimerization with other hTERT molecules (19,20). The processivity of human immunodeficiency virus type 1 (HIV-1) RT is mediated in part by residues in the C-terminal extension (24,30,31). Similarly, amino acids in the C-terminal domain of Est2p (S.cerevisiae TERT) contribute to telomerase activity, enzyme processivity and telomere length maintenance (24,32). These results reveal mechanistic similarities between HIV-1 RT and yeast telomerase, and support a role for enzyme processivity in telomere maintenance (24,32).
The TERT C terminus has been implicated in a number of telomerase-specific functions. However, the TERT amino acid residues and mechanisms mediating these functions have not been completely characterized for human telomerase. In the present study, human telomerases containing small deletions and substitutions were expressed in an in vitro transcription/translation system to investigate the regions of the hTERT C terminus involved in telomerase activity, multimerization with other hTERT molecules and enzyme processivity.
MATERIALS AND METHODS
Plasmid constructions
The construction of pET28b-hTERT, pET28a-GST-hTERT and pET28a-GST-hTERT D868N expression plasmids has been described previously (17,33). Internal C-terminal deletions and amino acid substitutions were generated using the Quick-Change site-directed mutagenesis protocol (Stratagene), the template pET28b-hTERT and appropriate pairs of primers. All C-terminal derivatives of hTERT were confirmed by restriction enzyme digestion and/or sequencing. The full-length hTR expression plasmid, phTR+1, has been described previously (34).
Sequence comparison
All TERT sequences used in the comparative analysis [NCBI BLAST (35)] were obtained from GenBank. The Rattus norvegicus TERT C-terminal amino acid sequence is a conceptual translation of the coding sequence available in the NCBI protein database.
In vitro transcription/translation
The T7-coupled transcription/translation rabbit reticulocyte lysate (RRL) system (Promega) was used as described previously (17). Full-length and C-terminal derivatives of hTERT were synthesized in RRL in the presence of purified hTR and [35S]methionine. hTR was synthesized and purified from FspI-linearized phTR+1 plasmid as previously described (34).
Telomerase activity assays
Telomerase activity was measured by two methods. First, a PCR-based telomerase repeat amplification protocol (TRAP) assay was performed on equal amounts of protein as described (36). An aliquot of 0.5–1 µl of RRL expressing hTERT or C-terminal variants in the presence of hTR was assayed for telomerase activity.
Secondly, a non-PCR-based telomerase elongation (also known as a standard, direct or conventional) assay was performed (37) with modifications. Equal amounts of hTERT or C-terminal variant proteins expressed in RRL in the presence of hTR (∼20 µl of RRL sample) were assayed for telomerase activity in a 40 µl final reaction volume using a gel-purified 5′-biotinylated (TTAGGG)4 as telomeric primer (Operon). Standard reaction conditions were 50 mM Tris–HCl pH 8.3, 50 mM KOAc, 1 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM spermidine, 1 µM telomere primer, 1.25 µM [α-32P]dGTP (800 Ci/mmol; NEN Life Science Products), 1 mM dATP and 1 mM dTTP. The reaction mix was incubated at 30°C for 2 h. The elongation step was terminated by adding 50 µl of RNase stop buffer (10 mM EDTA, 2 M NaCl, 0.1 mg/ml RNase A) and incubated at 37°C for 15 min. After addition of 50 µl of proteinase K solution (10 mM Tris– HCl pH 7.5, 0.3 mg/ml proteinase K) and a 15 min incubation at 37°C, a pre-washed Streptavidin MagnaSphere® Paramagnetic bead suspension (Promega) in 10 mM Tris–HCl pH 7.5, 1 M NaCl, 0.5 mM EDTA was added. The elongation products were immobilized on magnetic beads at room temperature for 30 min. Bead–elongation product complexes were washed three times with buffer A (10 mM Tris–HCl pH 7.5, 1 M NaCl), once with buffer B (10 mM Tris–HCl pH 7.5), resuspended in 10 µl of formamide dye (95% deionized formamide, 10 mM EDTA) and boiled for 30 min. Telomerase reaction products were analyzed by 8% polyacrylamide–urea gel electrophoresis. Gels were dried and exposed to X-ray film. Standard reaction assays were also performed using native telomerase extracted from telomerase-positive HL-60 cells (38) or with RRL and a 5′-end-labeled primer (TTAGGG)4.
Multimerization assay
Non-radiolabeled GST–hTERT proteins synthesized in RRL were immunopurified with a GST antibody (Amersham Pharmacia) pre-bound to protein A–Sepharose (Sigma) (17) and [35S]methionine-labeled His6-tagged hTERT, and C-terminal variants were immunopurified using Talon resin (Clontech) as described by the manufacturer. Equal amounts of partially purified 35S-labeled His6-tagged hTERT or C-terminal variants were added to the immobilized GST– hTERT proteins and co-immunoprecipitation was performed as previously described (17). Immunoprecipitated GST–hTERT and co-precipitated hTERT were resolved on an SDS–7.5% polyacrylamide gel and detected by western blot analysis and by visualization of 35S-labeled proteins, respectively.
Signals corresponding to GST–hTERT and 35S-labeled proteins were quantified by densitometric analysis of the autoradiographs (ImageQuant software, Molecular Dynamics). The efficiencies of co-immunoprecipitation of hTERT and C-terminal variant proteins were calculated by expressing the signal obtained from the co-precipitated 35S-labeled proteins as a percentage of the input signal. This co-immunoprecipitation value was then normalized to the levels of non-radiolabeled GST–hTERT detected in western blots, and the resulting values for each C-terminal variant were expressed as a percentage of the value obtained for wild-type hTERT. Data for individual mutants were statistically compared with the data for wild-type proteins using a paired two-tailed Student t-test.
Quantification of processivity
The signal of each telomerase elongation product was quantified by densitometric analysis of the autoradiographs or by phosphorimager analysis (ImageQuant software, Molecular Dynamics). Results of quantification using both methods were similar. The total counts for each signal were normalized to the amount of transcript by dividing each elongation product signal by the number of labeled dGTPs it contains.
The processivity (Pi) at each position within a single base pair repeat (type I) was calculated using the formula Pi = sum(Ti+1 + Ti+2 + …+ T6)/sum(Ti + Ti+1 + …+ T6) described in Peng et al. (2001) (24). T corresponds to the normalized signal at a given position, designated by i. The values obtained in independent experiments for equivalent positions within the first and second telomerase repeats (e.g. +1 is equivalent to +7) were averaged and statistically compared with average wild-type values using a paired two-tailed Student t-test. +1 and +7 refer to the second G in the human telomeric sequence TTAGGG. Similar processivity profiles were obtained for hTERT and C-terminal variants when equivalent telomerase elongation products within the first and second repeat were compared.
Repeat addition processivity (type II) was calculated using the normalized signals of the +6, +12, +18 and +24 telomerase elongation products (where products were detectable and quantifiable), and by applying the formula Pi = (Ti+6)/sum(Ti + Ti+6) described in Hardy et al. (2001) (39). For example, the normalized intensity of the second repeat addition products is divided by the sum of signals from the first and second repeat addition products. We obtained repeat processivity values for each pair of repeats separately. Repeat processivity values for each of the +6, +12, +18 and +24 elongation products from independent experiments were multiplied by 100, and then averaged. Data were statistically analyzed using a paired two-tailed Student t-test. Products included in this analysis were within the linear range of signal detection. Similar values for repeat addition processivity were obtained when first and second repeat addition products, second and third repeat addition products, or third and fourth repeat addition products were compared. As pulse–chase experiments were not performed, we cannot distinguish if long elongation products result from the continuous addition of multiple repeats onto a given telomeric primer, or from dissociation and subsequent rebinding and elongation.
RESULTS
BLAST alignment and mutagenesis
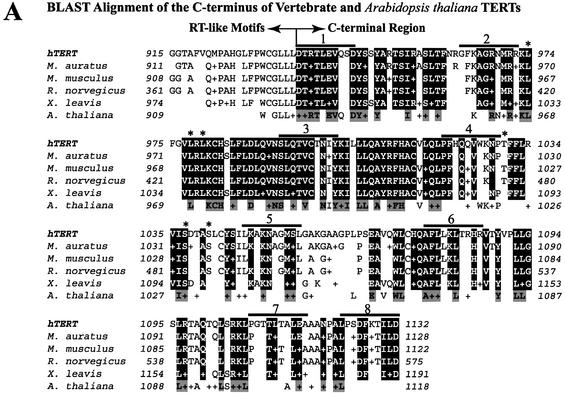
Alignment of C-terminal TERT sequences from diverse organisms indicates that this domain is not well conserved within the TERT family [data not shown; (24)]. We aligned five vertebrate TERT sequences and Arabidopsis thaliana TERT (5,7,40–46) using the NCBI BLAST program (35) to determine if portions of the C terminus were conserved among vertebrate and A.thaliana TERTs (Fig. 1A). The C terminus is defined in this report as beginning at hTERT residue 936. In contrast to the low degree of C terminus sequence homology among members of the entire TERT family, the C terminus of vertebrate and A.thaliana TERTs contained blocks of highly conserved residues. These residues are not conserved between hTERT and Est2p or between the TERTs and HIV-1 RT (data not shown).
Figure 1.
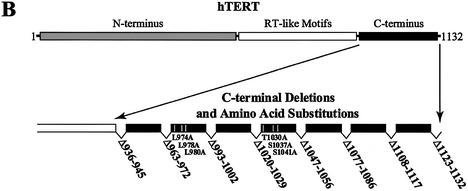
Map of the hTERT C terminus, location of C-terminal mutations, and sequence alignment of the C termini of vertebrate and A.thaliana TERTs. (A) Alignment of the C-terminal amino acid sequences of five vertebrate TERTs and A.thaliana TERT. Alignment of human, M.musculus, M.auratus, R.norvegicus, X.laevis and A.thaliana TERT sequences (5,7,40–46) was performed using the BLAST program. The symbol ‘+’ indicates non-identical conserved residues. Residues conserved in all vertebrate sequences are boxed in black. Residues boxed in gray are A.thaliana residues conserved with vertebrate sequences. Thick black lines above the protein sequence indicate the amino acids that are deleted in the hTERT variants. 1 = Δ936–945, 2 = Δ963–972, 3 = Δ993–1002, 4 = Δ1020–1029, 5 = Δ1047–1056, 6 = Δ1077–1086, 7 = Δ1108–1117, 8 = Δ1123–1132. The single amino acid substitutions L974A, L978A, L980A, T1030A, S1037A and S1041 are indicated by an asterisk. (B) A schematic illustration of hTERT. Mutations characterized in this study are indicated as broken (deletions) or white bars (substitutions) on the linear map of the hTERT C terminus. The numbering for each C-terminal variant indicates the amino acid positions of the deleted or substituted residues.
We generated a series of recombinant telomerases containing single amino acid substitutions or 10 amino acid deletions (Fig. 1B) to characterize the roles of the hTERT C terminus in telomerase activity, multimerization with other hTERT molecules and enzyme processivity. Deletions were regularly spaced at approximately 20 amino acid intervals. Single amino acid substitutions of the leucines to alanines at positions 974, 978 and 980 (L974A, L978A, L980A) in the CRM1-binding site (nuclear export signal) were generated (29). Single amino acid substitutions of the threonine to alanine at position 1030 and the serines to alanines at positions 1037 and 1041 (T1030A, S1037A, S1041A) in the 14-3-3-binding site were also generated (29). All C-terminal deletions and single amino acid substitutions affected highly conserved residues (the deletions removed between four and eight highly conserved residues). Thus, we identified vertebrate-conserved elements in the hTERT C terminus that may mediate functions specific to vertebrate telomerases. This mutagenic approach has been successfully employed to define various functions of hTERT N-terminal amino acid residues (17). We first addressed whether C-terminal vertebrate-conserved regions are implicated in human telomerase catalytic function.
Telomerase activity of hTERT C-terminal mutants detected by TRAP or by a conventional telomerase assay
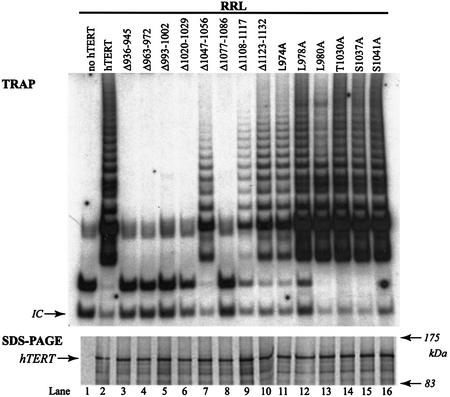
Wild-type hTERT or hTERT C-terminal mutants were expressed in vitro in RRL in the presence of in vitro-transcribed hTR. Wild-type hTERT and all mutant proteins were stably expressed in RRL (Fig. 2, bottom panel). The catalytic activities of reconstituted telomerases were assayed by the TRAP technique. The relative telomerase activities for mutant enzymes reconstituted in RRL are shown in Figure 2 (top panel). The Δ936–945, Δ963–972, Δ993–1002, Δ1020– 1029 and Δ1077–1086 C-terminal mutants were inactive. The Δ1047–1056, Δ1108–1117, Δ1123–1132, L974A, L978A, L980A, T1030A, S1037A and S1041A hTERT mutants reconstituted similar, or slightly reduced levels of activity compared with those reconstituted by wild-type hTERT. A previous study also reported that a triple alanine substitution T1030A, S1037A, S1041A in the 14-3-3-binding site does not alter telomerase activity, supporting the results we obtain with the variants containing single amino acid mutations at these positions (29). In addition, our results show that residues in the nuclear export signal-like (CRM1) motif in hTERT are not required for telomerase activity detected by TRAP. The loss of catalytic activity of certain of the C-terminal variants was not due to the inability of these mutants to interact with hTR (data not shown). Thus we identified multiple regions in the hTERT C terminus that are critical to the reconstitution of telomerase activity in vitro.
Figure 2.
Reconstitution of recombinant telomerases in an in vitro transcription/translation system and detection of telomerase activity using TRAP. Telomerase activity of C-terminal variants of hTERT expressed in RRL in the presence of hTR was detected using the PCR-based TRAP assay. Top panel: reconstituted telomerase activity of hTERT C-terminal mutants (Fig. 1). A PCR amplification control is indicated (IC). Bottom panel: expression of hTERT C-terminal mutants synthesized in RRL in the presence of [35S]methionine was detected by SDS–PAGE.
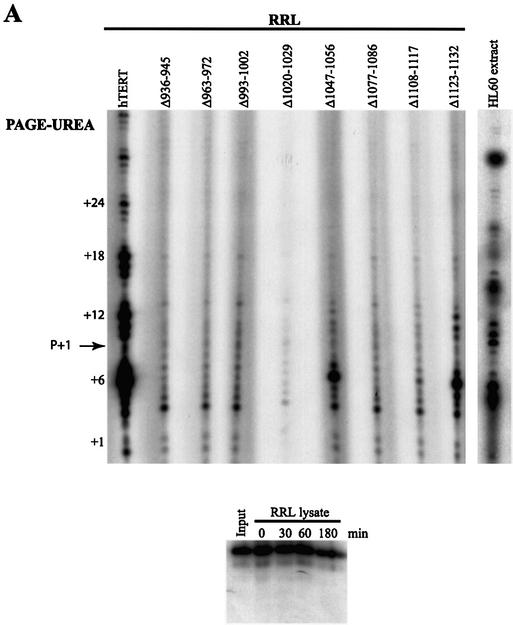
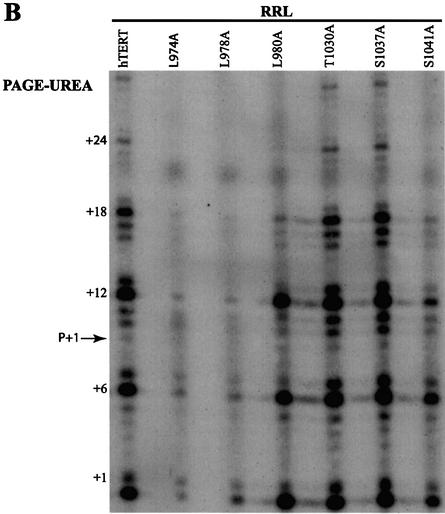
The C-terminal domain of Est2p has been implicated in enzyme processivity (24,32). To date, there have been no reports characterizing the role of hTERT in human telomerase processivity. We adapted the non-PCR-based telomerase elongation (also known as a standard, direct or conventional) assay (37) to characterize the processivity of human telomerases reconstituted in RRL. Wild-type telomerase reconstituted in RRL extended a telomeric (TTAGGG)4 oligonucleotide in vitro, generating a ladder of primer extension products with the 6-nt pausing pattern characteristic of human telomerase (47) (Fig. 3A, compare lanes labeled hTERT and HL-60). The generation of these products was hTERT and hTR dependent, and RNase and proteinase K sensitive (data not shown). The telomerase activities of all the deletion mutants, including those characterized as inactive by the TRAP technique, were weakly but reproducibly detected using the conventional assay (Fig. 3A). However, in contrast to the results obtained using the TRAP technique, no deletion mutant reconstituted telomerase activity as efficiently as wild-type hTERT (compare Figs 2 and 3A; hTERT, Δ1047–1056, Δ1108–1117, Δ1123–1132). In addition, many deletion mutants did not exhibit the 6-nt pausing pattern characteristic of wild-type human telomerase (Fig. 3A). Levels of telomerase activity were similar for wild-type, T1030A and S1037A hTERT mutants, whether detected by the conventional assay or by the TRAP technique (compare Figs 2 and 3B). However, the L980A and S1041A mutants reconstituted lower levels of activity than wild-type hTERT, whereas the L974A and L978A mutants reconstituted only weak levels of telomerase activity as detected by the conventional assay (Fig. 3B). Therefore, the conventional telomerase assay revealed catalytic defects that cannot be detected by the TRAP technique. Our results indicate that most vertebrate-conserved residues in the hTERT C terminus are required for catalytic activity, and that many of these residues are implicated in regulating overall levels of DNA synthesis.
Figure 3.
Detection of the catalytic activity of in vitro-reconstituted human telomerases using the direct primer extension telomerase assay. The telomerase activity of hTERT C-terminal variants expressed in RRL was detected using a non-PCR-based conventional assay. +6, +12, +18 and +24 refer to the first G in the telomeric sequence TTAGGG. The 3′-end-labeled biotinylated primer (TTAGGG)4 migrates as a 25mer [primer (P)+1], at the indicated position. (A) Upper panel: catalytic activity of RRL-expressed C-terminal deletion mutants and wild-type enzyme, and of native telomerase extracted from telomerase-positive HL-60 cells. Lower panel: a standard reaction assay was also performed with an [α-32P]-labeled (TTAGGG)4 primer added to the RRL for different periods of time. (B) Catalytic activity of C-terminal substitution mutants and wild-type enzyme.
Processivity defects of hTERT C-terminal mutants
Using the conventional telomerase elongation assay, short telomerase elongation products, representing the addition of the first few telomeric repeats onto the DNA substrate, were detectable for all mutant telomerases (Fig. 3). However, the deletion mutants reconstituted lower levels of activity than wild-type hTERT. The primer extension products generated by wild-type hTERT or C-terminal hTERT variants were quantified to determine if elongation defects were due to reduced enzyme processivity in addition to decreased levels of DNA synthesis. Telomerase processivity refers to the ability of the enzyme to (i) catalyze successive nucleotide additions within a single 6-base repeat (type I) and (ii) copy the RNA template repetitively, resulting in multiple 6-base repeats (type II). Type I processivity of yeast telomerase has been characterized, and is regulated by elements of the Est2p RT-like motifs and the C terminus (24). Type II processivity, or repeat addition processivity, is a feature of human and ciliate telomerases which can add hundreds of nucleotides to a DNA substrate in vitro (39,47,48). We quantified all elongation products generated by the reconstituted telomerases, including products that migrated below the input primer (TTAGGG)4. Products that migrate lower than the primer substrate may be generated by the addition of nucleotides onto input primer that has been cleaved by an endonuclease activity such as that previously characterized for ciliate and yeast telomerases (49,50). These lower products are not detected when [α-32P]-labeled primer is added to RRL, suggesting that they are not generated by random nucleases in RRL (Fig. 3A). +1 and +7 refer to the second G in the human telomeric sequence TTAGGG.
Quantification of the primer extension products [specifically, +6, +12, +18 and +24 telomerase elongation products (where applicable), see Materials and Methods] generated by all mutants revealed that deletion of many of the residues between amino acids 963 and 1117 caused defects in type II processivity (Fig. 3 and Table 1: Δ963–972, Δ1047–1056, Δ1077–1086, Δ1108–1117). The repeat addition processivity of these mutants ranged from 23 to 66% relative to wild type, and is comparable with the repeat addition processivity of recombinant Tetrahymena telomerases reconstituted with mutant telomerase RNA subunits that cause defects in enzyme processivity (18–73% relative to wild type) (39). Therefore, the results of these primer extension assays identify a possible type II processivity-modulating region in the C terminus of hTERT.
Table 1. Type II processivity of C-terminal hTERT mutants.
| Mutant | Average repeat addition processivity | Repeat addition processivity relative to wild type |
|---|---|---|
| hTERT | 29.85 ± 7.67 | 100 |
| Δ936–945 | 33.22 ± 8.24 | 111 |
| Δ963–972 | 14.05 ± 4.32* | 47 |
| Δ993–1002 | 25.19 ± 9.99 | 84 |
| Δ1020–1029 | n/a | n/a |
| Δ1047–1056 | 6.86 | 23 |
| Δ1077–1086 | 19.60 ± 9.12 | 66 |
| Δ1108–1117 | 12.81 ± 3.93* | 43 |
| Δ1123–1132 | 26.71 ± 12.54 | 89 |
| L974A | 23.41 ± 21.28 | 78 |
| L978A | 23.60 ± 8.96 | 83 |
| L980A | 21.65 ± 20.74 | 79 |
| T1030A | 25.12 ± 7.56 | 84 |
| S1037A | 23.81 ± 10.19 | 79 |
| S1041A | 29.94 ± 10.11 | 100 |
Processivity values were calculated as described in Materials and Methods. Type II processivity-defective mutants are indicated by bold typeface. Mutants with average processivity values that are significantly different compared with wild-type values (P < 0.05) are indicated by *. n/a indicates second-repeat products whose signal is too weak for quantification of type II processivity. Second-repeat products for mutant Δ1047–1056 were quantifiable only once; thus an average processivity value was not calculated, nor could statistical significance be calculated, despite the greatly reduced processivity.
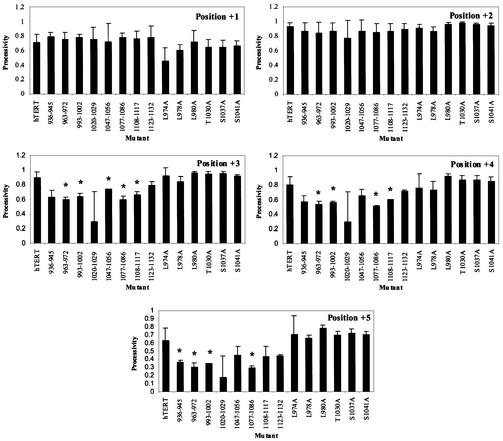
Analysis of type I processivity revealed that certain small deletions in the hTERT C terminus affected processivity at some positions within the telomeric repeat (Fig. 4). Processivity at position +3 was significantly reduced by 18–34% for the Δ963–972, Δ993–1002, Δ1047–1056, Δ1077– 1086 and Δ1108–1117 mutants compared with wild type. Processivity was reduced by 25–37% at position +4, and by 41–53% at position +5 for deletion mutants that had statistically significant enzyme processivity defects. The reduction in processivity observed for the C-terminal mutants is similar to the decreases reported for Est2p mutants defective in enyzme processivity (24). Deletion of the last 10 C-terminal amino acids, and substitution of residues in the 14-3-3- and CRM1-binding motifs did not affect type I processivity (Fig. 4). These results suggest that certain C-terminal regions characterized in this study contribute to both type I and type II enzyme processivity.
Figure 4.
Average type I processivity values for wild-type and C-terminal mutant telomerases reconstituted in RRL. Processivity values were calculated as described in Materials and Methods. Each plot depicts the processivity of wild-type and mutant enzymes at a specific position within the telomerase repeat product. +1 refers to the second G in the telomeric repeat TTAGGG, +2 to the third G, +3 to the first T, +4 to the second T and +5 to the A. Mutants with significantly different processivity values compared with wild type (P < 0.01) are indicated by *.
C-terminal mutations do not prevent the physical association of hTERT proteins
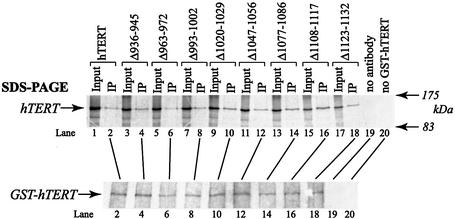
Previous studies indicate that human telomerase is a multimer and that an RT region adjacent to the C terminus of hTERT is implicated in multimerization (17–20,51). We and others have previously demonstrated that the physical association of hTERT molecules is hTR independent (17,18,20). Moreover, N-terminal hTERT mutations, including those that inactivate telomerase and abolish RNA binding, do not prevent the physical association of hTERT proteins (17). In an effort to determine the role of the C terminus in multimerization, we mixed immunopurified GST–hTERT protein with partially purified [35S]methionine-labeled hTERT or C-terminal mutants, and immunoprecipitated hTERT/GST–hTERT complexes. Levels of immunoprecipitated GST–hTERT were examined by western analysis, and co-precipitated [35S]methionine-labeled hTERTs were detected by SDS– PAGE and autoradiography (Fig. 5). All C-terminal mutant proteins co-precipitated with GST–hTERT. The association was specific because 35S-labeled proteins were not precipitated in the absence of GST–hTERT or GST antibody (lanes 19 and 20). When compared with wild-type hTERT, none of the mutant proteins exhibited defects in hTERT–hTERT protein interactions (Fig. 5). Quantification of GST–hTERT/hTERT interactions indicated that all mutant proteins co-precipitated with GST–hTERT more efficiently than did wild-type hTERT (data not shown). However, only the Δ936–945 mutant exhibited a statistically significant (P < 0.01) increased interaction with GST–hTERT (∼3-fold). In conclusion, none of the C-terminal mutants characterized in this study impaired the physical association of hTERT proteins.
Figure 5.
Physical association of C-terminal hTERT mutants with GST–hTERT in vitro. GST–hTERT and C-terminal mutants were independently synthesized in RRL in the absence of hTR. Immunopurified GST–hTERT was mixed with partially purified [35S]methionine-labeled hTERT or C-terminal mutants. Co-precipitated 35S-labeled hTERT and immunoprecipitated GST–hTERT proteins were detected by SDS–PAGE/autoradiography (top panel) and by western blot analysis (bottom panel), respectively. Five percent of input proteins and 50% of immunoprecipitated proteins were loaded (top panel). Control reactions were performed in the absence of antibody (lane 19) or GST–hTERT (lane 20).
Inactive C-terminal mutants cannot functionally complement an inactive RT domain mutant
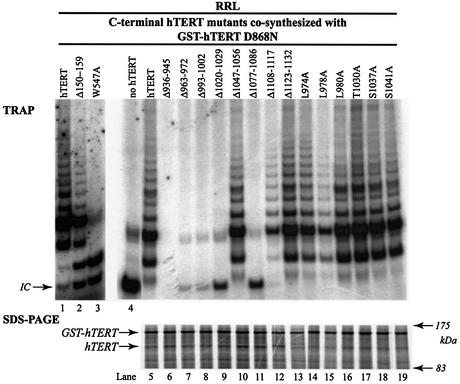
Previous studies indicate that separately inactive hTERT fragments can functionally multimerize in vitro and in vivo to reconstitute telomerase activity (12,17,19). Though N-terminal mutations do not prevent the physical association of hTERT proteins, some inactive N-terminal mutants fail to complement an inactive RT domain mutant to reconstitute activity (17). Therefore, we determined if inactive C-terminal mutants could complement an inactive RT domain mutant. The catalytically inactive point mutant GST–hTERT D868N (17) and C-terminal mutants were co-synthesized in RRL in the presence of hTR, and reactions were examined for telomerase activity and the expression of C-terminal mutants and GST–hTERT D868N (Fig. 6). Though all C-terminal mutants were expressed equally (Fig. 6, bottom panel), none of the inactive C-terminal mutants functionally complemented GST–hTERT D868N to reconstitute telomerase activity (Fig. 6, top panel). As previously reported, the inactive N-terminal hTERT mutant Δ150–159 functionally complemented GST–hTERT D868N to reconstitute telomerase activity (lane 2), whereas the inactive N-terminal mutant W547A did not functionally complement GST–hTERT D868N (17). Our results indicate that functional complementation requires the presence of intact RT motifs and catalytically essential C-terminal residues on the same hTERT molecule, supporting previously published results derived from the analysis of hTERT C-terminal truncations (19).
Figure 6.
Inactive C-terminal mutants cannot functionally complement an inactive RT domain mutant to reconstitute telomerase activity. GST–hTERT D868N and C-terminal mutants of hTERT were co-synthesized in RRL in the presence of hTR and [35S]methionine. Telomerase activity was measured by TRAP assay (top panel). The Δ150–159 mutant (lane 2) is an inactive N-terminal hTERT mutant that functionally complements GST–hTERT D868N to reconstitute telomerase activity (17). The W547A mutant is an inactive N-terminal hTERT mutant (lane 3) that does not functionally complement GST–hTERT D868N to reconstitute telomerase activity (17). GST–hTERT D868N and hTERT mutants were detected by SDS–PAGE/autoradiography (bottom panel).
DISCUSSION
In vitro and in vivo reconstitution of human telomerase has been used to map functions mediated by the N- and C-terminal extensions of hTERT (12,15,17–20,28). Our results obtained using the TRAP technique demonstrate an in vitro catalytic role for many vertebrate-conserved C-terminal residues. hTERT C-terminal mutations that altered the largest numbers of conserved residues impaired in vitro telomerase activity most severely (Figs 1 and 2). Despite the low degree of C terminus sequence homology among members of the entire TERT family, our data and those derived from previous studies of the C termini of human, Tetrahymena and yeast TERTs indicate that the TERT C-terminus is essential for telomerase catalytic activity (15,16,19,24,28).
Functional complementation requires a hTERT molecule containing both catalytically essential C-terminal residues and intact RT motifs
Inactive hTERT fragments can functionally complement each other to reconstitute telomerase activity, providing additional evidence that human telomerase exists as a dimer or multimer (17–20,51). The requirements for functional complementation include the association of hTR with only one active subunit of hTERT, intact hTR interaction sites in the N terminus of a hTERT molecule containing functional reverse transcriptase motifs, and catalytically important residues located C-terminal to amino acid 884 (17,19). This latter region includes the residues 914–928 that are implicated in hTERT oligomerization (20). A previous study reported that a truncated inactive version of hTERT encoding amino acids 1–884 cannot complement a motif A catalytic point mutant, indicating that an intact C terminus and RT motifs on the same hTERT molecule are required for functional multimerization (19). Our study, using hTERT variants containing small internal deletions at the C terminus, supports this latter conclusion. Our results also indicate that the catalytic and functional complementation defects of the C-terminal variants were not due to the inability of the mutant hTERTs to associate with hTR (data not shown). Thus, functional complementation requires a hTERT molecule containing catalytically essential C-terminal residues and intact RT motifs.
We also report that all of the hTERT C-terminal mutants can physically associate with wild-type GST–hTERT protein, suggesting that the catalytic and functional complementation defects of the C-terminal variants were not due to inhibition of dimerization or oligomerization. The mechanisms mediating the enhanced interactions of all C-terminal variants with GST–hTERT are unknown; perhaps regions implicated in negatively regulating hTERT association have been disrupted, or the conformation of the mutant proteins has been slightly altered. However, the association of the hTERT variants with hTR suggests that the proteins are not grossly misfolded (data not shown) (15,19). Deletion of hTERT residues 936–945 resulted in a statistically significant (P < 0.01) increase in hTERT interaction (∼3-fold). Arai and co-workers recently determined that amino acids 914–928 are essential for oligomerization (20). The proximity of residues 936–945 and 914–928 suggests that residues 936–945 could be part of a C-terminal region implicated in oligomerization. Thus C-terminal hTERT amino acids required for catalytic activity and functional complementation are not essential for the physical interaction of hTERT with another hTERT molecule or the telomerase RNA component. Recently, Arai and co-workers reported that the C terminus of hTERT can bind the N terminus of hTERT (20). As our experiments were performed using wild-type hTERT and C-terminal hTERT mutants, the C terminus of wild-type hTERT may bind the N terminus of the hTERT mutants, possibly masking defects in multimerization normally mediated by C-terminal hTERT residue interactions.
Role of the hTERT C terminus in DNA synthesis: TRAP versus the non-PCR-based elongation assay
The results we obtained using the standard telomerase assay varied from those obtained using the TRAP technique in two important aspects. First, the non-PCR-based assay is more sensitive than the TRAP technique for detecting differences in levels of reconstituted telomerase activity or DNA synthesis. Secondly, short telomerase elongation products, representing the addition of the first few telomeric repeats onto the DNA substrate, are detectable by the conventional telomerase assay but not by TRAP.
Using the TRAP technique, the Δ1047–1056, Δ1108–1117, Δ1123–1132, L974A, L978A, L980A and S1041A hTERT mutants reconstituted similar, or slightly reduced levels of activity compared with the levels reconstituted by wild-type hTERT, suggesting that these residues are not essential for catalysis. Our results obtained using the TRAP technique also demonstrate an in vitro catalytic role for many vertebrate-conserved C-terminal residues. hTERT C-terminal amino acid residues important for telomerase activity were recently identified at similar positions using telomerases reconstituted in vivo (28). Deletion (this study) or substitution (28) of hTERT residues at overlapping positions affected levels of telomerase activity similarly, suggesting that deletion of the hTERT residues does not grossly distort the overall structure of the protein.
Using the non-PCR-based telomerase elongation assay, we found that most of the vertebrate-conserved residues mutated in this study were required to reconstitute wild-type levels of in vitro catalytic activity (Fig. 3). Only the substitution of residues at positions 1030 and 1037, previously determined to be involved in regulatory interactions with 14-3-3 in vivo, did not affect levels of reconstituted telomerase activity assayed by the non-PCR-based technique. Interestingly, mutation of residues in the CRM1-interacting site of hTERT caused catalytic defects that were detected by the conventional assay, indicating that conserved residues in this motif may regulate the catalytic function of human telomerase in addition to modulating hTERT localization. Banik and colleagues (2002) recently reported that substitution of the last six hTERT residues 1127–1132, which constitute a biologically essential DAT domain, reduces but does not abolish telomerase activity, as detected by TRAP (28). In the present study, deletion of residues 1123–1132, which overlap with the DAT domain, nearly abolished telomerase activity detected by the conventional assay but did not affect telomerase activity detected by TRAP. These results suggest that catalytic defects could in part mediate the telomere maintenance defects of the DAT mutant (28). In fact, a correlation was recently found between telomerase activity and processivity mediated by the C terminus of S.cerevisiae TERT, and telomere length in vivo (24,32).
In the present study, we also report that mutants that are inactive when assayed by TRAP synthesize low levels of short elongation products detectable using the standard assay. It is likely that these short elongation products cannot be amplified by PCR due to the design of the TS and ACX primers, as the shortest products detectable by TRAP are 50 nt in length (52–54). Thus the standard assay allows the analysis of all elongation products generated by telomerase. Detailed mechanistic studies of S.cerevisiae and recombinant Tetrahymena telomerases have been possible through the use of direct telomerase assays (24,32,39,55). The non- PCR-based telomerase elongation assay that we report will now permit a detailed characterization of the human telomerase mechanism of action using recombinant telomerase reconstituted in RRL.
The hTERT C terminus is a determinant of human telomerase repeat addition processivity
Numerous studies have characterized the regulation of catalytic activity and processivity of endogenous telomerases (48,56–58). The processivity of endogenous human telomerase in vitro can be modulated by temperature, substrate (primer and dNTP) concentrations, primer sequence and G-quadruplex-interacting agents such as potassium ions (37,47,59). However, few studies have identified determinants within the TERT, telomerase RNA or associated protein components that regulate telomerase enzyme processivity. Detailed characterization of the telomerase mechanism of action has previously been most commonly reported using a non-PCR-based elongation assay, and this assay is the method of choice for mechanistic analyses of telomerase. Recently, a modified TRAP assay has also been reported for the analysis of processivity (specifically the generation of 9–10 repeats) and minimal template RNA length requirements of the human telomerase enzyme (53,54).
Studies of Euplotes crassus telomerase indicate that the developmentally regulated association of proteins with telomerase can influence repeat addition processivity (58, 60). Tetrahymena telomerase enzymes reconstituted in vitro or in vivo with mutant telomerase RNA subunits have type II processivity defects, implicating the RNA component in the regulation of enzyme processivity (61–63). Hardy et al. (2001) also reported the decreased repeat addition processivity of recombinant Tetrahymena telomerases reconstituted in RRL with mutant telomerase RNA subunits (39). Recently, nucleotide determinants adjacent to the template region of human and mouse telomerase RNAs were identified as regulators of enzyme processivity (64).
However, to date, there has been only one report characterizing the role of TERT in type II enzyme processivity. Bryan et al. (2000) identified a leucine at position 813 in motif C of Tetrahymena telomerase that affected the intrinsic processivity of the enzyme (65). Interestingly, in all retroviral RTs, the residue at this position is a tyrosine that is critical for catalytic activity. Substitution of the Tetrahymena telomerase leucine at position 813 with tyrosine resulted in increased repeat addition processivity (type II), creating an enzyme that resembles the highly processive classical RTs (65). Several of the C-terminal hTERT mutants that we characterized in the current study exhibited type II processivity defects, suggesting that a determinant of human telomerase repeat addition processivity resides in the hTERT C terminus, perhaps between residues 963 and 1117. The mechanism mediating defects in processivity remains to be determined. Defects in processivity could result from alignment of substrate at alternative template positions, defects in primer or dNTP binding or defects in primer extension or translocation. Human telomerase repeat addition processivity, like that of Tetrahymena telomerase, may also be mediated by other regions of hTERT, including the conserved RT motifs that are implicated in retroviral RT processivity.
Type I processivity: functional conservation between the C-terminal domains of human and yeast telomerase reverse transcriptase subunits
Processivity defects that map to the S.cerevisiae TERT C terminus have previously been characterized (24,32). C-terminal Est2p deletions affect type I processivity, and this region may be functionally analogous to the C-terminal extension of the HIV-1 RT (24). Interestingly, the deletion of the Est2p C terminus does not completely abolish telomerase activity as measured in a direct telomerase assay, though levels of DNA synthesis are reduced (24). Similar to the Est2p C-terminal mutants, many C-terminal hTERT mutants that we characterized in the current study exhibited significant type I processivity and overall DNA synthesis defects. Thus, despite weak conservation of C-terminal TERT sequences from vertebrates and yeast, the identification of C-terminal hTERT residues involved in modulating type I enzyme processivity suggests a functional conservation between the C-terminal domains of human and yeast telomerases. The mechanistic basis of this functional similarity will require careful characterization. Our results indicate that the hTERT C terminus plays essential roles in several human telomerase functions: catalytic activity, functional complementation with other hTERT molecules and enzyme processivity.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Maria A. Cerone and Dr Lea Harrington for comments on the manuscript, and Sophie Dupuis for excellent technical support. T.J.M. is the recipient of a Canadian Institutes of Health Research (CIHR) Doctoral Award. C.A. is the recipient of a Fonds de la Recherche en Santé du Québec Chercheur Boursier and a Boehringer Ingelheim (Canada) Inc. Young Investigator Award. This work was supported by grant MOP-14026 from the CIHR to C.A.
REFERENCES
- 1.Blackburn E.H. (2001) Switching and signaling at the telomere. Cell, 106, 661–673. [DOI] [PubMed] [Google Scholar]
- 2.Harrington L. and Robinson,M.O. (2002) Telomere dysfunction: multiple paths to the same end. Oncogene, 21, 592–597. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E. (1999) Telomerase. In Gestland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 609–635.
- 4.Feng J., Funk,W.D., Wang,S.-S., Weinrich,S.L., Avilion,A.A., Chiu,C.-P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J., Le,S., West,M.D., Harley,C.B., Andrews,W.H., Greider,C.W. and Villeponteau,B. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 6.O’Reilly M., Teichmann,S.A. and Rhodes,D. (1999) Telomerases. Curr. Opin. Struct. Biol., 9, 56–65. [DOI] [PubMed] [Google Scholar]
- 7.Harrington L., Zhou,W., McPhail,T., Oulton,R., Yeung,D.S.K., Mar,V., Bass,M.B. and Robinson,M.O. (1997) Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev., 11, 3109–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B., Taylor,R.D., Carlos,R., Andrews,W.H., Wright,W.E., Shay,J.W., Harley,C.B. and Morin,G.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 9.Counter C.M., Meyerson,M., Eaton,E.N. and Weinberg,R.A. (1997) The catalytic subunit of yeast telomerase. Proc. Natl Acad. Sci. USA, 94, 9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 11.Friedman K.L. and Cech,T.R. (1999) Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev., 13, 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (2000) Polymerization defects within human telomerase are distinct from telomerase RNA and Tep1 binding. Mol. Biol. Cell, 11, 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller M.C., Liu,J.K. and Collins,K. (2000) Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J., 19, 4412–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia J., Peng,Y., Mian,S. and Lue,N.F. (2000) Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol., 20, 5196–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachand F. and Autexier,C. (2001) Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA–protein interactions. Mol. Cell. Biol., 21, 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai C.K., Mitchell,J.R. and Collins,K. (2001) RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol., 21, 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriarty T., Huard,S., Dupuis,S. and Autexier,C. (2002) Functional multimerization of human telomerase requires an RNA interaction domain in the N-terminus of the catalytic subunit. Mol. Cell. Biol., 22, 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armbruster B.N., Banik,S.S.R., Guo,C., Smith,A.C. and Counter,C.M. (2001) N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol., 21, 7775–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (2001) Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol., 21, 6151–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arai K., Masutomi,K., Khurts,S., Kaneko,S., Kobayashi,K. and Murakami,S. (2002) Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem., 277, 8538–8544. [DOI] [PubMed] [Google Scholar]
- 21.Etheridge K., Banik,S., Armbruster,B., Zhu,Y., Terns,R., Terns,M. and Counter,C. (2002) The nucleolar localization domain of the catalytic subunit of human telomerase. J. Biol. Chem., 277, 24764–24770. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Chen,Y., Huang,H. and Weissman,S.M. (2002) Nucleolar localization of hTERT protein is associated with telomerase function. Exp. Cell Res., 277, 201–209. [DOI] [PubMed] [Google Scholar]
- 23.Bosoy D., Peng,Y., Mian,I.S. and Lue,N.F. (2002) Conserved N-terminal motifs of telomerase reverse transcriptase required for ribonucleoprotein assembly in vivo. J. Biol. Chem., 278, 3882–3890. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y., Mian,S.I. and Lue,N.F. (2001) Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell, 7, 1201–1211. [DOI] [PubMed] [Google Scholar]
- 25.Counter C.M., Hahn,W.C., Wei,W., Dickinson Caddle,S., Beijersbergen,R.L., Lansdorp,P.M., Sedivy,J.M. and Weinberg,R.A. (1998) Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc. Natl Acad. Sci. USA, 95, 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouellette M.M., Aisner,D.L., Savre-Train,I., Wright,W.E. and Shay,J.W. (1999) Telomerase activity does not always imply telomere maintenance. Biochem. Biophys. Res. Commun., 254, 795–803. [DOI] [PubMed] [Google Scholar]
- 27.Huang J.J., Lin,M.C., Bai,Y.X., Jing,D.D., Wong,B.C.Y., Han,S.W., Lin,J., Xu,B., Huang,C. and Kung,H. (2002) Ectopic expression of a COOH-terminal fragment of the human telomerase reverse transcriptase leads to telomere dysfunction and reduction of growth and tumorigenicity in HeLa cells. Cancer Res., 62, 3226–3232. [PubMed] [Google Scholar]
- 28.Banik S.S.R., Guo,C., Smith,A.C., Margolis,S.S., Richardson,D.A., Tirado,C.A. and Counter,C.M. (2002) C-terminal regions of the human telomerase catalytic subunit essential for in vivo enzyme activity. Mol. Cell. Biol., 22, 6234–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seimiya H., Sawada,H., Muramatsu,Y., Shimizu,M., Ohko,K., Yamane,K. and Tsuruo,T. (2000) Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J., 19, 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobo-Molina A., Ding,J., Nanni,R.G., Clark,A.D., Lu,K., Tantillo,C., Williams,R.L., Kamer,G., Ferris,A.L., Clark,P., Hizi,A., Hughes,S.H. and Arnold,E. (1993) Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl Acad. Sci. USA, 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bebenek K., Beard,W.A., Casas-Finet,J.R., Kim,H.-R., Darden,T.A., Wilson,S.H. and Kunkel,T.A. (1995) Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J. Biol. Chem., 270, 19516–19523. [DOI] [PubMed] [Google Scholar]
- 32.Hossain S., Singh,S.M. and Lue,N.F. (2002) Functional analysis of the C-terminal extension of telomerase reverse transcriptase: a ‘putative’ thumb domain. J. Biol. Chem., 277, 36174–36180. [DOI] [PubMed] [Google Scholar]
- 33.Bachand F. and Autexier,C. (1999) Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem., 274, 38027–38031. [DOI] [PubMed] [Google Scholar]
- 34.Autexier C., Pruzan,R., Funk,W.D. and Greider,C.W. (1996) Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J., 15, 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriarty T., Dupuis,S. and Autexier,C. (2002) Rapid up-regulation of telomerase activity in human leukemia HL-60 cells treated with the DNA-damaging drug etoposide. Leukemia, 16, 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun D., Lopez-Guajardo,C.C., Quada,J., Hurley,L.H. and Von Hoff,D.D. (1999) Regulation of catalytic activity and processivity of human telomerase. Biochemistry, 38, 4037–4044. [DOI] [PubMed] [Google Scholar]
- 38.Mayeda A. and Krainer,A.R. (1999) Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol., 118, 309. [DOI] [PubMed] [Google Scholar]
- 39.Hardy C.D., Schultz,C.S. and Collins,K. (2001) Requirements for the dGTP-dependent repeat addition processivity of recombinant Tetrahymena telomerase. J. Biol. Chem., 276, 4863–4871. [DOI] [PubMed] [Google Scholar]
- 40.Greenberg R.A., Allsopp,R.C., Chin,L., Morin,G.B. and DePinho,R.A. (1998) Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene, 16, 1723–1730. [DOI] [PubMed] [Google Scholar]
- 41.Guo W., Okamoto,M., Park,N.H., Lee,Y.M. and Park,N.H. (2001) Cloning and expression of hamster telomerase catalytic subunit cDNA. Int. J. Mol. Med., 8, 73–78. [DOI] [PubMed] [Google Scholar]
- 42.Kilian A., Bowtell,D.D.L., Abud,H.E., Hime,G.R., Venter,D.J., Keese,P.K., Duncan,E.L., Reddel,R.R. and Jefferson,R.A. (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet., 6, 2011–2019. [DOI] [PubMed] [Google Scholar]
- 43.Martin-Rivera L., Herrera,E., Albar,J.P. and Blasco,M.A. (1998) Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl Acad. Sci. USA, 95, 10471–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Dickinson,S.C., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q., Bacchetti,S., Haber,D.A. and Weinberg,R.A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 45.Fitzgerald M., Riha,K., Gao,F., Ren,S., McKnight,T. and Shippen,D. (1999) Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA, 96, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuramoto M., Ohsumi,K., Kishimoto,T. and Ishikawa,F. (2001) Identification and analyses of the Xenopus TERT gene that encodes the catalytic subunit of telomerase. Gene, 277, 101–110. [DOI] [PubMed] [Google Scholar]
- 47.Morin G.B. (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell, 59, 521–529. [DOI] [PubMed] [Google Scholar]
- 48.Greider C.W. (1991) Telomerase is processive. Mol. Cell. Biol., 11, 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins K. and Greider,C.W. (1993) Tetrahymena telomerase catalyzes nucleolytic cleavage and non-processive elongation. Genes Dev., 7, 1364–1376. [DOI] [PubMed] [Google Scholar]
- 50.Niu H., Xia,J. and Lue,N.F. (2000) Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol. Cell. Biol., 20, 6806–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenz C., Enenkel,B., Amacker,M., Kelleher,C., Damm,K. and Lingner,J. (2001) Human telomerase contains two cooperating telomerase RNA molecules. EMBO J., 20, 3526–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim N.W. and Wu,F. (1997) Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res., 25, 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gavory G., Farrow,M. and Balasubramanian,S. (2002) Minimum length requirement of the alignment domain of human telomerase RNA to sustain catalytic activity in vitro. Nucleic Acids Res., 30, 4470–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szatmari I. and Aradi,J. (2001) Telomeric repeat amplification, without shortening or lengthening of the telomerase products: a method to analyze the processivity of telomerase enzyme. Nucleic Acids Res., 29, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins K. and Gandhi,L. (1998) The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 95, 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohn M. and Blackburn,E.H. (1995) Telomerase in yeast. Science, 269, 396–400. [DOI] [PubMed] [Google Scholar]
- 57.Boswell-Fulton T. and Blackburn,E.H. (1998) Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol., 18, 4961–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greene E.C. and Shippen,D.E. (1998) Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes Dev., 12, 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maine I.P., Chen,S.-F. and Windle,B. (1999) Effect of dGTP concentration on human and CHO telomerase. Biochemistry, 38, 15325–15332. [DOI] [PubMed] [Google Scholar]
- 60.Bednenko J., Melek,M., Greene,E.C. and Shippen,D.E. (1997) Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-associated de novo telomere formation. EMBO J., 16, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Autexier C. and Greider,C.W. (1994) Functional reconstitution of wild type and mutant Tetrahymena telomerase. Genes Dev., 8, 563–575. [DOI] [PubMed] [Google Scholar]
- 62.Autexier C. and Greider,C.W. (1995) Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev., 15, 2227–2239. [DOI] [PubMed] [Google Scholar]
- 63.Ware T.L., Wang,H. and Blackburn,E.H. (2000) Three telomerases with completely non-telomeric template replacements are catalytically active. EMBO J., 19, 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J.-L. and Greider,C.W. (2003) Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J., 22, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000) A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem., 275, 24199–24207. [DOI] [PubMed] [Google Scholar]