Abstract
The Xrcc3 protein, which is required for the homologous recombinational repair of damaged DNA, forms a complex with the Rad51C protein in human cells. Mutations in either the Xrcc3 or Rad51C gene cause extreme sensitivity to DNA-damaging agents and generate the genomic instability frequently found in tumors. In the present study, we found that the Xrcc3 segment containing amino acid residues 63–346, Xrcc363–346, is the Rad51C-binding region. Biochemical analyses revealed that Xrcc363–346 forms a complex with Rad51C, and the Xrcc363–346– Rad51C complex possesses ssDNA and dsDNA binding abilities comparable to those of the full-length Xrcc3–Rad51C complex. Based on the structure of RecA, which is thought to be the ancestor of Xrcc3, six Xrcc3 point mutants were designed. Two-hybrid and biochemical analyses of the Xrcc3 point mutants revealed that Tyr139 and Phe249 are essential amino acid residues for Rad51C binding. Superposition of the Xrcc3 Tyr139 and Phe249 residues on the RecA structure suggested that Tyr139 may function to ensure proper folding and Phe249 may be important to constitute the Rad51C-binding interface in Xrcc3.
INTRODUCTION
Homologous recombinational repair (HRR) is one of the major pathways to repair double-strand breaks (DSBs), which are frequently caused by exposure to various DNA-damaging agents, including ionizing radiation, crosslinking reagents and oxidative stress (1–3). DSBs are also induced during chromosomal DNA replication (4). When cells are defective in HRR, unrepaired DSBs accumulate in chromosomes, resulting in genomic instability (5–7). In fact, single nucleotide polymorphisms (SNPs) and mutations in genes involved in HRR have been identified in tumor cells (8,9).
In the key step of the HRR pathway, the homologous-pairing step, a single-stranded DNA (ssDNA) tail derived from a DSB site invades the homologous double-stranded DNA (dsDNA) to form a heteroduplex. In Escherichia coli, the RecA protein catalyzes the homologous-pairing step (10,11). Two eukaryotic homologs of RecA, the Rad51 and Dmc1 proteins, which are conserved from yeast to human, have been identified (12–15), and have been shown to catalyze homologous pairing in vitro (16–20). The Dmc1 protein functions only in meiotic cells, but the Rad51 protein is expressed in both meiotic and mitotic cells (12–15). In addition, five Rad51 paralogs [Xrcc2 (21,22), Xrcc3 (22,23), Rad51B/hREC2/Rad51L1 (24–26), Rad51C/Rad51L2 (27) and Rad51D/Rad51L3 (26,28,29)], which share 20–30% amino acid identity with Rad51, have been identified in mammals.
The Xrcc2 and Xrcc3 genes were first identified as human genes that complement the DNA damage-sensitive hamster cell lines, irs1 and irs1SF, respectively (23,30–33), and were confirmed to be involved in HRR in vivo (34,35). Cells lacking Xrcc2 or Xrcc3 show extreme sensitivity to DNA crosslinking reagents, such as cisplatin, and ionizing radiation, and have significantly increased chromosomal missegregation (22,23, 36). Knockout experiments in the chicken DT40 cell lines showed that the Rad51B, Rad51C and Rad51D genes are also involved in the HRR pathway as well as the Xrcc2 and Xrcc3 genes (37,38).
The five Rad51 paralogs reportedly exist in two distinct complexes in human cells (39–42). One is composed of Rad51B, Rad51C, Rad51D and Xrcc2. This type of complex formation was predicted by the yeast two- and three-hybrid experiments (43). The Xrcc2–Rad51D and Rad51B–Rad51C subcomplexes were also purified as recombinant proteins (44,45). The Xrcc2–Rad51D complex has potential to catalyze homologous pairing between ssDNA fragment and superhelical dsDNA (44). Although Rad51B does not catalyze homologous pairing by itself (46,47), the Rad51B–Rad51C complex functions in the homologous-pairing step to antagonize the RPA-dependent repression in strand exchange by Rad51 (45). These results with the subunits of the Rad51B– Rad51C–Rad51D–Xrcc2 complex suggested that the complex functions in the homologous-pairing step of HRR.
Another Rad51-paralog complex found in human cells is composed of Xrcc3 and Rad51C (39–42). Interestingly, the two-hybrid screening with the human brain cDNA library showed that Xrcc2 and Xrcc3 independently interact with Rad51D and Rad51C, respectively, also suggesting that Xrcc2 and Xrcc3 exist in different complexes in brain cells (44,48). The purified Xrcc3–Rad51C complex binds to ssDNA and dsDNA (48,49), and has the potential to catalyze homologous pairing (48). Rad51C alone also promotes strand exchange between homologous single-stranded and double-stranded oligonucleotides, probably through its dsDNA destabilizing activity (47), suggesting that Rad51C is a catalytic center for this type of homologous pairing. On the other hand, no biochemical analysis of Xrcc3 itself has been reported yet. The Xrcc3 variant allele, Thr241Met, is found with significantly higher frequency in melanoma skin cancer and bladder cancer (50–52). However, no significant effect on the HRR pathway with the Xrcc3 variant was detected in the cells (53). Analyses of the domains and the amino acid residues required for the Rad51-paralog functions are important to understand the biological consequences of their SNPs and mutations.
In the present study, we performed a two-hybrid analysis with deletion mutants of Xrcc3 and Rad51C to identify the regions required for the Xrcc3–Rad51C interaction, and found that the N-terminal region (1–62 amino acids) of Xrcc3 is dispensable for Rad51C binding. On the other hand, the entire region of Rad51C, except for several residues in the N-terminus, is required for Xrcc3 binding. Biochemical analyses revealed that an Xrcc3 deletion mutant containing amino acid residues 63–346 (Xrcc363–346) forms a complex with Rad51C, and the Xrcc363–346–Rad51C complex can bind ssDNA and dsDNA as well as the full-length Xrcc3–Rad51C complex, although the DNA-binding ability of Rad51C alone is apparently lower than those of the complexes. A mutational analysis also showed that the Xrcc3 Phe249 residue, which is located on the solvent accessible surface when the Xrcc3 amino acids are superimposed on the RecA structure, is essential for Rad51C binding.
MATERIALS AND METHODS
Yeast two-hybrid analysis
The human Xrcc3 and Rad51C genes were cloned from human brain cDNA (purchased from Clontech) by the polymerase chain reaction (PCR). The DNA fragments encoding the Xrcc3 and Rad51C deletion mutants were designed based on secondary structure predictions, and were amplified by the PCR method. Mutations at Phe30, Tyr139, Phe180, Phe219, Phe223 and Phe249 of Xrcc3 were introduced by the site-directed mutagenesis method. The DNA fragments containing sequences derived from Xrcc3 were ligated into the NdeI site of the pAS2-1 vector (Clontech), and those containing sequences derived from Rad51C were ligated into either the SmaI or NcoI site of the pACT2 vector (Clontech). Plasmids pAS2-1 and pACT2 contain the GAL4 DNA-binding domain and the GAL4 activation domain, respectively, just upstream of their multiple cloning sites. The pAS2-1 vector containing the Xrcc3 sequence was introduced into the yeast strain AH109 and the pACT2 vector containing the Rad51C sequence was introduced into the yeast strain Y187. Two-hybrid interactions between Xrcc3 and Rad51C were tested by mating the AH109 and Y187 strains, according to the manufacturer’s protocol (Clontech Matchmaker GAL4 protocol). The interaction between Xrcc3 and Rad51C induced the expressions of the HIS3 and LacZ reporter genes, which were detected by yeast growth on a synthetic dextrose minimal medium (SD) plate without histidine and by β-galactosidase activity using a 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) filter assay, respectively.
Purification of the Xrcc363–346–Rad51C complex
The Xrcc3–Rad51C complex and Rad51C were purified as described previously (48). The DNA fragments containing Xrcc363–346 and Rad51C were inserted into the pET15b vector at the NcoI and NdeI sites, respectively. Then, the pET15b vector containing the Rad51C sequence was digested with BglII and BamHI, and the fragment with Rad51C was ligated into the BamHI site of the pET15b-Xrcc363–346 vector. The over-expression plasmid for Xrcc363–346 and Rad51C was introduced into E.coli JM109 (DE3) cells with the plasmid containing the genes for E.coli tRNAArg3 and tRNAArg4, which recognize the CGG and AGA/AGG codons, respectively. The cells were grown in 10 l of LB medium containing 100 µg/ml ampicillin and 34 µg/ml chloramphenicol at 30°C for 10 h, and then 200 µM isopropyl β-thiogalactopyranoside (IPTG) was added to induce protein expression. Xrcc363–346 and Rad51C were produced for 12 h at 18°C in the presence of IPTG, and the cells were harvested. The Rad51C protein contains a His6-tag and the Xrcc363–346 protein contains a FLAG-tag at the N-terminus. The cells producing Xrcc363–346 and Rad51C were resuspended in 20 ml of 20 mM sodium phosphate buffer (pH 8.5) containing 0.5 M NaCl, and were disrupted by sonication. The samples were centrifuged for 20 min at 30 000 g and the supernatants were incubated with 4 ml of Ni-NTA agarose (Qiagen) for 1 h at 4°C. Then, the Xrcc363–346 and Rad51C-bound resin was packed into a column and was washed with 300 ml of 20 mM Tris–HCl buffer (pH 8.5) containing 10 mM imidazole and 0.5 M NaCl. Subsequently, the resin was washed with 150 ml of 20 mM Tris–HCl buffer (pH 8.5) containing 10 mM imidazole. Xrcc363–346 and Rad51C were eluted by a gradient of imidazole from 10 to 400 mM in 20 mM Tris–HCl buffer (pH 8.5). The samples eluted from the Ni-column were applied to a heparin–Sepharose column (Amersham Biosciences) previously equilibrated with 20 mM Tris–HCl buffer (pH 8.5) containing 2 mM dithiothreitol and 10% glycerol. The proteins were eluted by a gradient of NaCl from 0 to 1200 mM and were dialyzed against 20 mM Tris–HCl buffer (pH 8.1) containing 5 mM dithiothreitol and 10% glycerol. The protein concentration was determined by the Bio-Rad protein assay kit (Bio-Rad), using bovine serum albumin (BSA) as the standard protein.
Preparation of closed circular double-stranded DNA
To avoid irreversible denaturation of the dsDNA, we prepared the plasmid DNA without any treatment that would potentially cause denaturation, such as alkaline treatment. The plasmid DNA (pGsat4; 3216 bp) containing the human α-satellite sequence was introduced into the E.coli DH5α strain and the cells were cultured for 12–16 h at 37°C. The cells were harvested, mildly disrupted with 0.5 mg/ml lysozyme and 0.1% sarkosyl, and centrifuged at 92 000 g for 1 h. The supernatant containing the pGsat4 plasmid DNA was extracted with phenol/chloroform three times, and the pGsat4 DNA in the aqueous phase was precipitated by ethanol. The pellet was dissolved in 1 ml of TE buffer and was treated with 0.15 mg/ml RNaseA at 37°C for 30 min. The pGsat4 DNA was purified by a 5–20% sucrose gradient centrifugation at 98 000 g for 18 h. DNA concentrations are expressed in moles of nucleotides.
Preparation of circular single-stranded DNA
The E.coli cells (JM109) containing pGsat4 DNA were cultured in LB medium until an A660 of 0.3 was achieved, and were then infected with the helper phage R408 at an multiplicity of infection (m.o.i.) of ∼20. After infection, the cells were continuously cultured for 6 h with vigorous agitation. Then, the supernatant was harvested and was treated with DNase I (10 U/ml) and RNase A (10 µg/ml) for 15 min at 37°C. The phage containing pGsat4 ssDNA was precipitated by adding 0.25 vol of phage precipitation solution, containing 3.75 M ammonium acetate (pH 7.5) and 20% polyethylene glycol 8000. The resulting pellet was resuspended in TE buffer, and the pGsat4 ssDNA was extracted with chloroform: isoamyl alcohol (24:1) followed by phenol:chloroform saturated with TE buffer. After several phenol:chloroform extractions, the aqueous phase was extracted with chloroform and the pGsat4 ssDNA was precipitated with 2 vol of ethanol in the presence of 3.25 M ammonium acetate. The resulting pellet was rinsed with 70% ethanol and was dried under vacuum. The pGsat4 ssDNA was dissolved in H2O, and its concentration is expressed in moles of nucleotides.
Single-stranded DNA binding assay
Single-stranded pGsat4 DNA (40 µM) was mixed with Xrcc3–Rad51C, Xrcc363–346–Rad51C or Rad51C in 10 µl of standard reaction buffer, containing 20 mM Tris–HCl (pH 8.1), 2 mM ATP, 1 mM dithiothreitol, 100 µg/ml BSA, 2 mM MgCl2 and 3% glycerol. Protein concentrations were 0.35, 0.7 and 1.1 µM. The reaction mixtures were incubated at 37°C for 10 min, and were analyzed by 0.8% agarose gel electrophoresis in 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA). The bands were visualized by ethidium bromide staining.
Double-stranded DNA binding assay
Superhelical pGsat4 DNA (15 µM; 3216 bp) was mixed with Xrcc3–Rad51C, Xrcc363–346–Rad51C or Rad51C in 10 µl of standard reaction buffer, containing 20 mM Tris–HCl (pH 8.1), 2 mM ATP, 1 mM dithiothreitol, 100 µg/ml BSA, 2 mM MgCl2 and 3% glycerol. Protein concentrations were 0.35, 0.7 and 1.1 µM. The reaction mixtures were incubated at 37°C for 10 min, and the samples were electrophoresed on a 0.8% agarose gel for 4 h at 3 V/cm in 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA). The bands were visualized by ethidium bromide staining.
Ni-bead pull-down assay
Co-expression vectors for Xrcc3 (or the Xrcc3 point mutants) and Rad51C were introduced into E.coli JM109 (DE3) cells with the plasmid containing the genes for E.coli tRNAArg3 and tRNAArg4. The freshly transformed cells were grown in 10 ml of LB medium at 30°C to an OD600 of 0.5, and protein expression was induced with 200 µM IPTG at 18°C for 12 h. The cell pellets were resuspended in 200 µl of 20 mM Tris–HCl buffer (pH 7.8) containing 0.5 M NaCl and 200 µg/ml lysozyme, and were lysed for 30 min on ice. The samples were centrifuged at 30 000 g for 10 min, and the supernatants (200 µl) were mixed with 40 µl of ProBond beads (50% slurry; Invitrogen) at 4°C for 1 h. Then, the beads were washed three times with 500 µl of 20 mM Tris–HCl buffer (pH 7.8) containing 0.5 M NaCl and the proteins bound to the beads were fractionated by 15–25% gradient SDS–PAGE. Bands were visualized by Coomassie brilliant blue staining.
Immuno-precipitation assay
Co-expression vectors for Xrcc3 (or the Xrcc3 point mutants) and Rad51C were introduced into E.coli BL21 CodonPlus cells (Stratagene). The freshly transformed cells were grown in 5 ml of LB medium at 30°C to an OD600 of 0.5, and protein expression was induced with 200 µM IPTG at 18°C for 12 h. The cell pellets were resuspended in 500 µl of 20 mM Tris–HCl buffer (pH 8.5) containing 0.5 M NaCl and 2 µg/ml lysozyme, and were disrupted by sonication. The samples were centrifuged at 17 000 g for 10 min, and the supernatants (100 µl) were mixed with 10 µl of anti-Rad51C antibody-conjugated rProtein A Sepharose Fast Flow (Amersham Biosciences) at 4°C for 2 h. Then, the beads were washed three times with 500 µl of phosphate buffer saline containing 1% NP-40. The proteins bound to the beads were eluted with 50 µl of ImmunoPure IgG elution buffer (Pierce), and were detected by the western blotting method using the anti-Rad51C or anti-Xrcc3 polyclonal antibody.
RESULTS
Deletion analysis of the Xrcc3 and Rad51C regions required for their interaction
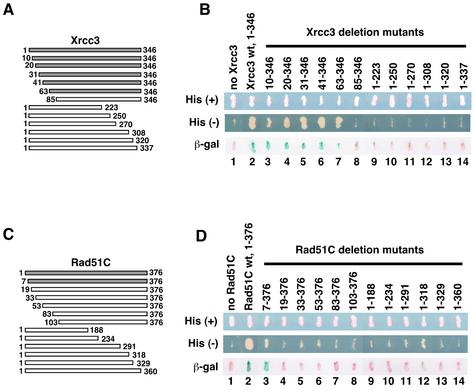
In order to study the Rad51C-interacting region of Xrcc3, we constructed 12 Xrcc3 deletion mutants, Xrcc310–346, Xrcc320–346, Xrcc331–346, Xrcc341–346, Xrcc363–346, Xrcc385–346, Xrcc31–223, Xrcc31–250, Xrcc31–270, Xrcc31–308, Xrcc31–320 and Xrcc31–337, which were composed of amino acid residues 10–346, 20–346, 31–346, 41–346, 63–346, 85–346, 1–223, 1–250, 1–270, 1–308, 1–320 and 1–337, respectively (Fig. 1A). These Xrcc3-deletion mutants were tested for their abilities to interact with Rad51C by the yeast two-hybrid analysis. In this system, the Xrcc3–Rad51C interaction induces the expressions of the HIS3 and LacZ reporter genes, which allow the yeast strain to grow in the absence of histidine (Fig. 1B, lane 2, middle row) and with blue color in the presence of X-Gal (Fig. 1B, lane 2, bottom row). As shown in Figure 1B, five N-terminally truncated Xrcc3 mutants, Xrcc310–346, Xrcc320–346, Xrcc331–346, Xrcc341–346 and Xrcc363–346, were able to interact with Rad51C as well as the full-length Xrcc3 (lanes 2–7); however, Xrcc385–346, which lacked the N-terminal 84 amino acid residues, did not interact with Rad51C (lane 8). These results indicate that the N-terminal 62 amino acid residues of Xrcc3 are not involved in Rad51C binding. In contrast, none of the C-terminally truncated Xrcc3 mutants interacted with Rad51C, and even Xrcc31–337, which lacked only nine amino acids from the C-terminal end, did not interact with Rad51C (Fig. 1B, lanes 9–14). Therefore, the C-terminal segment of residues 63–346 is the Rad51C-binding region of Xrcc3.
Figure 1.
Deletion analyses of the Xrcc3–Rad51C interaction. (A) Schematic representation of the Xrcc3 deletion mutants. Constructs that showed positive signals in the two-hybrid analysis are presented in gray bars. (B) The yeast two-hybrid results with the Xrcc3 deletion mutants. Yeast strains grown on SD plates with histidine (top row) and without histidine (middle row) are shown. The β-galactosidase activities, which were induced by the Xrcc3–Rad51C interaction, were detected by the X-Gal filter assay, and the results are shown in the bottom row. Lane 1 indicates a negative control with an empty vector instead of the Xrcc3 vector, and lane 2 indicates a positive control with the full-length Xrcc3 protein. Lanes 3–14 are experiments with the Xrcc3 deletion mutants containing amino acid residues 10–346, 20–346, 31–346, 41–346, 63–346, 85–346, 1–223, 1–250, 1–270, 1–308, 1–320 and 1–337, respectively. (C) Schematic representation of the Rad51C deletion mutants. Constructs with positive signals in the two-hybrid analysis are presented in gray bars. (D) The yeast two-hybrid results with the Rad51C deletion mutants. Yeast strains grown on SD plates with histidine (top row) and without histidine (middle row) are shown. The β-galactosidase activities, which were induced by the Xrcc3–Rad51C interaction, were detected by the X-Gal filter assay, and the results are shown in the bottom row. Lane 1 indicates a negative control with an empty vector instead of the Rad51C vector, and lane 2 indicates a positive control with the full-length Rad51C protein. Lanes 3–14 are experiments with the Rad51C deletion mutants containing amino acid residues 7–376, 19–376, 33–376, 53–376, 83–376, 103–376, 1–188, 1–234, 1–291, 1–318, 1–329 and 1–360, respectively.
Next, we analyzed the Xrcc3-interacting region of Rad51C. Based on the secondary structure prediction, six N-terminally and six C-terminally truncated Rad51C mutants were constructed (Fig. 1C), and were tested for their abilities to interact with Xrcc3 by the two-hybrid analysis. As shown in Figure 1D, only one Rad51C deletion mutant, which lacked seven amino acid residues from the N-terminal end, interacted with Xrcc3. Therefore, the entire region of Rad51C, except for several N-terminal residues, is required for the Xrcc3 binding. These Rad51C deletion mutants may be defective in the proper folding of the protein. It has been reported that the DNA damage-sensitive hamster cell lines, CL-V4B and irs3, have mutations in the Rad51C gene, and are probably missing exon 5 and exon 6, respectively (54,55). Exon 5 and exon 6 of the Rad51C gene encode the amino acid residues from Val236 and Val280, respectively. As shown in Figure 1D, Rad51C1–234 and Rad51C1–291, which lack the C-terminal regions from Lys235 and Arg292, respectively, were defective in the Xrcc3 interaction (lanes 10 and 11). The DNA damage-sensitive phenotypes of CL-V4B and irs3 may be due to a defective interaction between the Xrcc3 and Rad51C mutants.
The Xrcc3 deletion mutant, Xrcc363–346, forms a complex with Rad51C
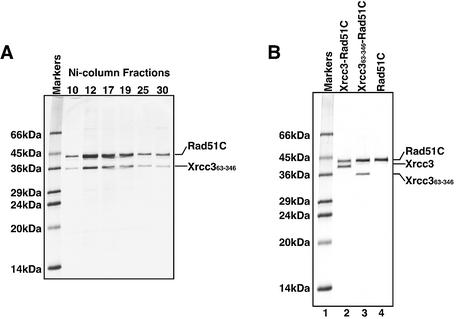
To determine whether Xrcc363–346 directly interacts with Rad51C, we co-expressed Xrcc363–346 with the His6- tagged Rad51C proteins in E.coli cells, and studied their interaction using Ni-nitrilotriacetate (Ni-NTA) agarose column chromatography. As shown in Figure 2A, Xrcc363–346, which did not contain a His6 tag, co-eluted with the His6-tagged Rad51C protein from the Ni-NTA agarose column. Both the Xrcc363–346 and Rad51C proteins were further co-purified by heparin–Sepharose column chromatography (Fig. 2B, lane 3). Therefore, these results indicate that Xrcc363–346 directly binds to Rad51C. Xrcc363–346 was not detected in the soluble fraction when it was expressed alone, suggesting that the complex formation with Rad51C is required for the proper folding of Xrcc363–346.
Figure 2.
Purification of the Xrcc363–346–Rad51C complex. (A) SDS–PAGE (15–25% gradient) of the Ni-column fractions of Xrcc363–346–Rad51C. Fractions (10 µl) containing Xrcc363–346–Rad51C were analyzed. Fraction numbers are indicated at the top of the gel. Bands were visualized by Coomassie brilliant blue staining. (B) SDS–PAGE (15–25% gradient) of the purified Xrcc3–Rad51C, Xrcc363–346–Rad51C and Rad51C proteins. Lane 1 is molecular weight markers. Lanes 2–4 indicate Xrcc3–Rad51C (0.5 µg), Xrcc363–346–Rad51C (0.5 µg) and Rad51C (0.5 µg), respectively. The proteins were visualized by Coomassie brilliant blue staining.
The DNA-binding activity of Xrcc363–346–Rad51C
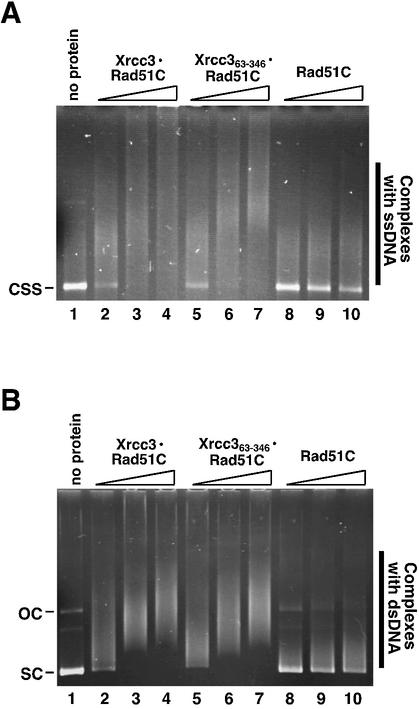
Next, we tested the DNA-binding ability of the purified Xrcc363–346–Rad51C complex. It was reported that the Xrcc3–Rad51C complex binds ssDNA and dsDNA (48,49), and that the DNA-binding ability of Rad51C alone is lower than that of Xrcc3–Rad51C (48). In the present study, pGsat4 circular ssDNA (3216 bases) and pGsat4 superhelical dsDNA (3216 bp) were used as substrates for DNA binding by Xrcc363–346–Rad51C. Consistent with previous observations (47,48), both Xrcc3–Rad51C and Rad51C alone bound to ssDNA and dsDNA, but the ssDNA- and dsDNA-binding abilities of Rad51C alone were significantly lower than that of Xrcc3–Rad51C (Fig. 3). Interestingly, Xrcc363–346–Rad51C bound to ssDNA and dsDNA as well as Xrcc3–Rad51C (Fig. 3). Therefore, Xrcc363–346 contains the functional domain, which is important in the DNA binding of Xrcc3–Rad51C. It should be noted that the ssDNA binding of Rad51C alone was clearly detected with short single-stranded oligonucleotides (47). However, Rad51C binding to the long circular ssDNA (3216 bases) was poorly observed (48) (Fig. 3). Xrcc3 may be required in cooperative binding of Rad51C to the long DNA.
Figure 3.
DNA-binding activity of Xrcc363–346–Rad51C. (A) ssDNA binding. Single-stranded pGsat4 DNA (40 µM) was mixed with the indicated amounts of Xrcc3–Rad51C, Xrcc363–346–Rad51C or Rad51C in 10 µl of the standard reaction buffer. The reaction mixtures were incubated at 37°C for 10 min and were analyzed by 0.8% agarose gel electrophoresis. The bands were visualized by ethidium bromide staining. (B) dsDNA binding. Superhelical pGsat4 DNA (15 µM; 3216 bp) was mixed with the indicated amounts of Xrcc3–Rad51C, Xrcc363–346–Rad51C and Rad51C in 10 µl of the standard reaction buffer. The reaction mixtures were incubated at 37°C for 10 min and the samples were analyzed by 0.8% agarose gel electrophoresis. The bands were visualized by ethidium bromide staining. Lane 1 indicates the experiment without protein. Lanes 2–4 indicate the experiments with Xrcc3–Rad51C. Lanes 5–7 indicate the experiments with Xrcc363–346–Rad51C. Lanes 8–10 indicate the experiments with Rad51C. Protein concentrations were 0.35 (lanes 2, 5 and 8), 0.7 (lanes 3, 6 and 9) and 1.1 µM (lanes 4, 7 and 10).
Design of mutations in Xrcc3
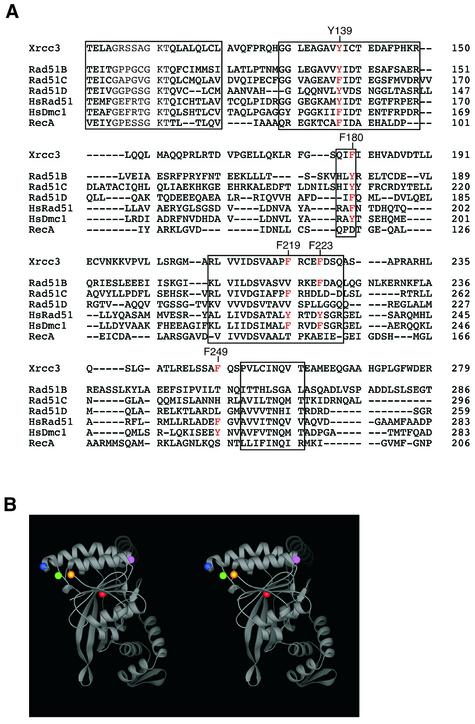
To identify the amino acid residues involved in the Xrcc3–Rad51C interaction, we constructed six mutant Xrcc3 genes, F30A, Y139A, F180A, F219A, F223A and F249A, each with an alanine replacement at positions Phe30, Tyr139, Phe180, Phe219, Phe223 and Phe249, respectively. Phenyl alanine and tyrosine were selected as amino acids for mutagenesis, because these aromatic residues were frequently found at the protein–protein interface. As shown in Figure 4A, Tyr139 of Xrcc3 is perfectly conserved as an aromatic residue among the RecA/Rad51-class proteins (the human Rad51B, Rad51C, Rad51D, Rad51 and Dmc1 proteins, and the E.coli RecA protein). Phe180 of Xrcc3 is also highly conserved, and Phe219, Phe223 and Phe249 of Xrcc3 are partially conserved among the RecA/Rad51-class proteins (Fig. 4A). When these Xrcc3 amino acid residues were superimposed on the crystal structure of RecA (56), Tyr139, corresponding to Phe92 in RecA, was located inside the RecA molecule (Fig. 4B, red sphere), where it probably forms the hydrophobic core of the protein. Phe180, Phe219, Phe223 and Phe249 of Xrcc3 were located on the molecular surface of the RecA structure (Fig. 4B, yellow, green, blue, and pink spheres, respectively). Therefore, these residues are considered as candidates that are directly involved in the interface between Xrcc3 and Rad51C. The Xrcc3 F30A mutant was designed as a negative control, which has an amino acid replacement outside the Rad51C-interacting region of Xrcc3 (amino acid residues 63–346). These six Xrcc3 mutants were tested for their interactions with Rad51C.
Figure 4.
Sequence alignments of the Rad51/RecA-class proteins, and locations of the Xrcc3 amino acid residues mutated in the present study. (A) The amino acid sequence of the human Xrcc3 protein is aligned with the human Rad51B, Rad51C, Rad51D, Rad51 and Dmc1 proteins, and the E.coli RecA protein. (B) Locations of the amino acid residues, Tyr139, Phe180, Phe219, Phe223 and Phe249, of Xrcc3 superimposed on the crystal structure of RecA (56). Red, orange, green, blue and pink spheres indicate the locations of Tyr139, Phe180, Phe219, Phe223 and Phe249, respectively.
Two-hybrid analysis of interactions between the Xrcc3 point mutants and Rad51C
First, we tested the two-hybrid interactions between the Xrcc3 point mutants and Rad51C. As expected, Xrcc3 F30A, which has a mutation outside the Rad51C-interacting region, positively interacted with Rad51C as well as the wild-type Xrcc3 (Fig. 5, lanes 2 and 3). The Xrcc3 F180A, F219A and F223A mutants were also positive in Rad51C binding (Fig. 5, lanes 5–7). These results indicate that Phe180, Phe219 and Phe223 of Xrcc3 are not involved in the Xrcc3–Rad51C interaction, although these residues are located on the solvent accessible surface in the model structure (Fig. 4B). In contrast, the Xrcc3 Y139A and F249A mutants did not show positive signals, suggesting that these mutants are defective in Rad51C binding (Fig. 5, lanes 4 and 8). Therefore, Tyr139 and Phe249 may be essential amino acid residues for Rad51C binding.
Figure 5.
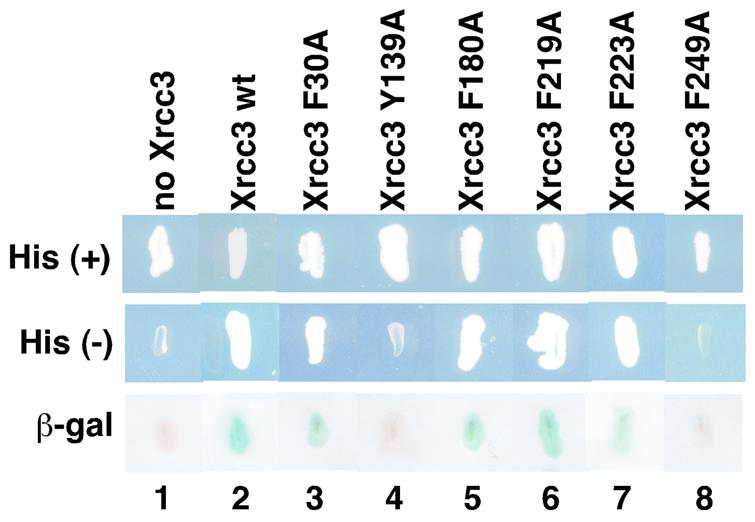
Mutational analysis of the Xrcc3–Rad51C interaction by the yeast two-hybrid system. Yeast strains grown on SD plates with histidine (top row) and without histidine (middle row) are shown. The β-galactosidase activities, which were induced by the Xrcc3–Rad51C interaction, were detected by the X-Gal filter assay, and the results are shown in the bottom row. Lane 1 indicates a negative control with an empty vector instead of the Xrcc3 vector and lane 2 indicates a positive control with the wild-type Xrcc3 protein. Lanes 3–8 indicate experiments with the Xrcc3 point mutants, F30A, Y139A, F180A, F219A, F223A and F249A, respectively.
Tyr139 and Phe249 of Xrcc3 are essential for Rad51C binding
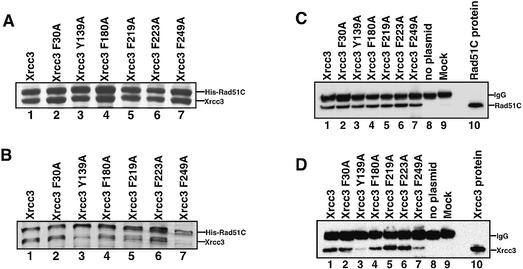
In order to confirm the results from the two-hybrid analysis with the Xrcc3 point mutants, we performed two independent assays, the Ni-bead pull-down and immuno-precipitation assays, with cell-free extracts containing the His6-tagged Rad51C protein and one of the FLAG-tagged Xrcc3 mutants. Cell-free extracts were prepared from the E.coli cells, in which each of the six FLAG-tagged Xrcc3 mutants was co-expressed with the His6-tagged Rad51C protein (Fig. 6A). In the Ni-bead pull-down assay, the Xrcc3 protein should coprecipitate with the His6-tagged Rad51C protein by the Ni-chelating beads (ProBond), if it was directly bound to Rad51C. As shown in Figure 6B, the wild-type Xrcc3 protein coprecipitated with the His6-tagged Rad51C protein (lane 1). Consistent with the results from the two-hybrid analysis, the Xrcc3 F30A, F180A, F219A and F223A mutants coprecipitated with the His6-tagged Rad51C protein as well as the wild-type Xrcc3 protein, indicating that these mutants were proficient in Rad51C binding (Fig. 6B, lanes 1, 2 and 4–6). On the other hand, the Xrcc3 Y139A and F249A mutants, which did not show an interaction with Rad51C in the two-hybrid analysis, exhibited a significant deficiency in Rad51C binding in the Ni-bead pull-down assay (Fig. 6B, lanes 3 and 7).
Figure 6.
Biochemical analyses of the Xrcc3–Rad51C interaction. (A) SDS–PAGE (15–25% gradient) of cell-free extracts from cells that co-expressed His6-tagged Rad51C and FLAG-tagged Xrcc3 (or the FLAG-tagged Xrcc3 point mutants). The cell-free extracts (3 μl of supernatant) were subjected to SDS–PAGE (15–25% gradient). Bands were visualized by Coomassie brilliant blue staining. Lane 1 indicates the positive control with the wild-type Xrcc3 protein. Lanes 2–7 indicate the experiments with the Xrcc3 point mutants, F30A, Y139A, F180A, F219A, F223A and F249A, respectively. (B) Protein fractions bound to the ProBond resin were analyzed by SDS–PAGE (15–25% gradient). Bands were visualized by Coomassie brilliant blue staining. Lane 1 indicates the positive control with the wild-type Xrcc3 protein. Lanes 2–7 indicate the experiments with the Xrcc3 point mutants, F30A, Y139A, F180A, F219A, F223A and F249A, respectively. (C and D) Immuno-precipitation analysis with the anti-Rad51C antibody. Protein fractions precipitated with the anti-Rad51C antibody-conjugated rProtein A Sepharose were detected by the anti-Rad51C antibody (C) and by the anti-Xrcc3 antibody (D). Lane 1 indicates the positive control with the wild-type Xrcc3 protein. Lanes 8 and 9 indicate the negative controls without the co-expression plasmid for Xrcc3–Rad51C and with rProtein A Sepharose treated with non-immune serum, respectively. Lanes 2–7 indicate the experiments with the Xrcc3 point mutants, F30A, Y139A, F180A, F219A, F223A and F249A, respectively. Purified Rad51C and Xrcc3 were applied as markers [lane 10 of (C) and lane 10 of (D), respectively].
Consistent results were obtained with the immuno- precipitation assay using a polyclonal antibody against Rad51C. As shown in Figure 6C, Rad51C was specifically precipitated with the anti-Rad51C antibody from the extracts expressing both Xrcc3 (or a Xrcc3 mutant) and Rad51C. When protein fractions precipitated with the anti-Rad51C antibody were probed with a polyclonal antibody against Xrcc3, the Xrcc3 F30A, F180A, F219A and F223A mutants were detected as well as the wild-type Xrcc3 protein. In contrast, the amounts of the Xrcc3 Y139A and F249A mutants that were coprecipitated with Rad51C by the anti-Rad51C antibody were significantly decreased (Fig. 6D, lanes 3 and 7). All of these results from the three independent experiments, the two-hybrid analysis, the Ni-bead pull-down assay and the immuno-precipitation assay, are perfectly consistent with each other. Therefore, we conclude that Tyr139 and Phe249 of Xrcc3 play essential roles in Rad51C binding.
DISCUSSION
In the present study, we identified the Xrcc3 segment from amino acid residues 63–346 as the Rad51C-interacting region. This region contains the Walker-type ATPase motifs, which are highly conserved among seven human Rad51-class proteins, Rad51, Dmc1, Rad51B, Rad51C, Rad51D, Xrcc2 and Xrcc3 (57). Recently, the crystal structure of the human Rad51 region (amino acid residues 97–339) was determined (58), which revealed that the structure of this Rad51 core region is quite similar to that of RecA. Therefore, Xrcc363–346 may also share a similar core region structure with those of the Rad51 and RecA proteins, although this region of Xrcc3 has the specific function to interact with Rad51C.
Mutational analyses of Xrcc3 identified Tyr139 and Phe249 as essential residues for Rad51C binding. Tyr139 of Xrcc3 was perfectly conserved among the Rad51/RecA-class proteins as an aromatic residue (Fig. 4A). When Tyr139 of Xrcc3 was superimposed on the RecA crystal structure, the residue was located deep inside the protein molecule (Fig. 4B, red sphere). Since the amino acid residues that directly interact with Rad51C must be located on the surface of the protein molecule, the Xrcc3 Y139A mutation may indirectly affect the Rad51 binding, probably through a perturbation of the Xrcc3 structure. Actually, in the RecA structure, Phe92, corresponding to Tyr139 of Xrcc3, forms a hydrophobic core with Leu77, Ile80, Leu99, Ile111 and Leu114 (56). So far, no mutations at the hydrophobic core of RecA has been reported. Interestingly, these hydrophobic residues are also conserved in Xrcc3, as Leu120, Pro127, Phe146, Leu170 and Leu173, respectively. The formation of the hydrophobic core around Tyr139 may be a key step in the proper folding of Xrcc3. The amino acid residue corresponding to Xrcc3 Tyr139 may be a central residue in the proper folding of the Rad51-class proteins.
In contrast, Phe249 of Xrcc3 was located on the surface of the protein molecule, when the residues were superimposed on the RecA structure (Fig. 4B, pink sphere). The mutation of Phe249 in Xrcc3 caused a significant decrease in the Rad51C-binding ability as evaluated by three independent assays, the two-hybrid analysis, the Ni-bead pull-down assay and the immuno-precipitation assay. Therefore, Phe249 of Xrcc3 is a strong candidate for one of the amino acid residues that constitute the interface for Rad51C binding. Interestingly, Phe249 of Xrcc3 is conserved in Rad51 as Phe259, which is an essential residue for Rad52 binding (59). The Rad51 Phe259 residue is located in the solvent accessible surface, when it is superimposed on the polymer structure of RecA (59). The Xrcc3 region around Phe249 is the protein-interacting region, which may have commonly evolved as a protein-binding surface among the eukaryotic Rad51-class proteins. Intriguingly, a variant Xrcc3 gene, T241M, is associated with an increased risk of cancer (52). The Xrcc3 T241M variant has methionine instead of threonine at position 241, which is located very close to Phe249, suggesting that the variant may perturb the proper Xrcc3–Rad51C interaction. An in vivo study of the Xrcc3 T241M variant revealed that it can complement the Xrcc3-deficent phenotypes in mammalian cells (53). This small modification on the Rad51C-interacting region in the Xrcc3 T241M variant may increase the risk of undesirable correction during DNA repair through homologous recombination.
In addition to the homologous-pairing step, Xrcc3 also reportedly functions in later stages of the HRR pathway, such as in the resolution of intermediates formed by homologous pairing (60). Among the Rad51 paralogs, Rad51B preferentially binds a synthetic Holliday junction, which mimics an intermediate formed by homologous pairing (46). Rad51B forms a stable complex with Rad51C, like Xrcc3. Rad51C may also function in the later stages of the HRR pathway with Xrcc3 and Rad51B. Further studies are required to understand these broad functional spectra of the Rad51 paralogs in the HRR pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Wataru Kagawa for assistance and discussions. This work was supported in part by the Bioarchitect Research Program (RIKEN), Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation (JST), and also by a Grant-in-Aid from the Ministry of Education, Sports, Culture, Science and Technology of Japan.
REFERENCES
- 1.Whitaker S.J. (1992) DNA damage by drugs and radiation: what is important and how is it measured? Eur. J. Cancer, 28, 273–276. [DOI] [PubMed] [Google Scholar]
- 2.Ward J.F. (1994) The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol., 66, 427–432. [DOI] [PubMed] [Google Scholar]
- 3.Caldecott K.W. (2001) Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays, 23, 447–455. [DOI] [PubMed] [Google Scholar]
- 4.Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 5.Thompson L.H. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson D.O. and Alt,F.W. (2001) DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene, 20, 5572–5579. [DOI] [PubMed] [Google Scholar]
- 7.van Gent D.C., Hoeijmarkers,J.H. and Kanaar,R. (2001) Chromosomal stability and the DNA double-stranded break connection. Nature Rev. Genet., 2, 196–206. [DOI] [PubMed] [Google Scholar]
- 8.Pierce A.J., Stark,J.M., Araujo,F.D., Moynahan,M.E., Berwick,M. and Jasin,M. (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol., 11, S52–S58. [DOI] [PubMed] [Google Scholar]
- 9.Thompson L.H. and Schild,D. (2002) Recombinational DNA repair and human disease. Mutat. Res., 509, 49–78. [DOI] [PubMed] [Google Scholar]
- 10.Shibata T., DasGupta,C., Cunningham,R.P. and Radding,C.M. (1979) Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc. Natl Acad. Sci. USA, 76, 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEntee K., Weinstock,G.M. and Lehman,I.R. (1979) Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc. Natl Acad. Sci. USA, 76, 2615–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- 14.Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 15.Habu T., Taki,T., West,A., Nishimune,Y. and Morita,T. (1996) The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res., 24, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 17.Baumann P., Benson,F.E. and West,S.C. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- 18.Gupta R.C., Bazemore,L.R., Golub,E.I. and Radding,C.M. (1997) Activities of human recombination protein Rad51. Proc. Natl Acad. Sci. USA, 94, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Golub,E.I., Gupta,R. and Radding,C.M. (1997) Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl Acad. Sci. USA, 94, 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong E.L., Shinohara,A. and Bishop,D.K. (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem., 276, 41906–41912. [DOI] [PubMed] [Google Scholar]
- 21.Cartwright R., Tambini,C.E., Simpson,P.J. and Thacker,J. (1998) The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res., 26, 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N., Lamerdin,J.E., Tebbs,R.S., Schild,D., Tucker,J., Shen,M.R., Brookman,K.W., Siciliano,M.J., Walter,C.A., Fan,W., Narayana,L.S., Zhou,Z., Adamson,A.W., Sorensen,K.J., Chen,D.J., Jones,N.J. and Thompson,L.H. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- 23.Tebbs R.S., Zhao,Y., Tucker,J.D., Scheerer,J.B., Siciliano,M.J., Hwang,M., Liu,N., Legerski,R.J. and Thompson,L.H. (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA, 92, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albala J.S., Thelen,M.P., Prange,C., Fan,W., Christensen,M., Thompson,L.H. and Lennon,G.G. (1997) Identification of a novel human RAD51 homolog, RAD51B. Genomics, 46, 476–479. [DOI] [PubMed] [Google Scholar]
- 25.Rice C.M., Smith,S.T., Bullrich,F., Havre,P. and Kmiec,E.B. (1997) Isolation of human and mouse genes based on homology to REC2, a recombinational repair gene from the fungus Ustilago maydis. Proc. Natl Acad. Sci. USA, 94, 7417–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cartwright R., Dunn,A.M., Simpson,P.J., Tambini,C.E. and Thacker,J. (1998) Isolation of novel human and mouse genes of the recA/RAD51 recombination-repair gene family. Nucleic Acids Res., 26, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dosanjh M.K., Collins,D.W., Wufang,F., Lennon,G.G., Albala,J.S., Shen,Z. and Schild,D. (1998) Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res., 26, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawabata M. and Saeki,K. (1999) Multiple alternative transcripts of the human homologue of the mouse TRAD/R51H3/RAD51D gene, a member of the rec A/RAD51 gene family. Biochem. Biophys. Res. Commun., 257, 156–162. [DOI] [PubMed] [Google Scholar]
- 29.Pittman D.L., Weinberg,L.R. and Schimenti,J.C. (1998) Identification, characterization and genetic mapping of Rad51d, a new mouse and human RAD51/RecA-related gene. Genomics, 49, 103–111. [DOI] [PubMed] [Google Scholar]
- 30.Jones N.J., Cox,R. and Thacker,J. (1987) Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res., 183, 279–286. [DOI] [PubMed] [Google Scholar]
- 31.Fuller L.F. and Painter,R.B. (1988) A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res., 193, 109–121. [DOI] [PubMed] [Google Scholar]
- 32.Jones N.J., Zhao,Y., Siciliano,M.J. and Thompson,L.H. (1995) Assignment of the XRCC2 human DNA repair gene to chromosome 7q36 by complementation analysis. Genomics, 26, 619–622. [DOI] [PubMed] [Google Scholar]
- 33.Thacker J., Tambini,C.E., Simpson,P.J., Tsui,L.C. and Scherer,S.W. (1995) Localization to chromosome 7q36.1 of the human XRCC2 gene, determining sensitivity to DNA-damaging agents. Hum. Mol. Genet., 4, 113–120. [DOI] [PubMed] [Google Scholar]
- 34.Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- 35.Pierce A.J., Johnson,R.D., Thompson,L.H. and Jasin,M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev., 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffin C.S., Simpson,P.J., Wilson,C.R. and Thacker,J. (2000) Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nature Cell Biol., 2, 757–761. [DOI] [PubMed] [Google Scholar]
- 37.Takata M., Sasaki,M.S., Sonoda,E., Fukushima,T., Morrison,C., Albala,J.S., Swagemakers,S.M.A., Kanaar,R., Thompson,L.H. and Takeda,S. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol., 20, 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takata M., Sakaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masson J.Y., Tarsounas,M.C., Stasiak,A.Z., Stasiak,A., Shah,R., McIIwraith,M.J., Benson,F.E. and West,S.C. (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev., 15, 3296–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiese C., Collins,D.W., Albala,J.S., Thompson,L.H., Kronenberg,A. and Schild,D. (2002) Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res., 30, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu N., Schild,D., Thelen,M.P. and Thompson,L.H. (2002) Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res., 30, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller K.A., Yoshikawa,D.M., McConnell,I.R., Clark,R., Schild,D. and Albala,J.S. (2002) RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem., 277, 8406–8411. [DOI] [PubMed] [Google Scholar]
- 43.Schild D., Lio,Y., Collins,D.W., Tsomondo,T. and Chen,D.J. (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem., 275, 16443–16889. [DOI] [PubMed] [Google Scholar]
- 44.Kurumizaka H., Ikawa,S., Nakada,M., Enomoto,R., Kagawa,W., Kinebuchi,T., Yamazoe,M., Yokoyama,S. and Shibata,T. (2002) Homologous pairing and ring and filament structure formation activities of the human Xrcc2-Rad51D complex. J. Biol. Chem., 277, 14315–14320. [DOI] [PubMed] [Google Scholar]
- 45.Sigurdsson S., Van Komen,S., Bussen,W., Schild,D., Albala,J.S. and Sung,P. (2001) Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev., 15, 3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoyama H., Kurumizaka,H., Ikawa,S., Yokoyama,S. and Shibata,T. (2003) Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem., 278, 2767–2772. [DOI] [PubMed] [Google Scholar]
- 47.Lio Y., Mazin,A.V., Kowalczykowski,S.C. and Chen,D.J. (2003) Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem., 278, 2469–2478. [DOI] [PubMed] [Google Scholar]
- 48.Kurumizaka H., Ikawa,S., Nakada,M., Eda,K., Kagawa,W., Takata,M., Takeda,S., Yokoyama,S. and Shibata,T. (2001) Homologous-pairing activity of the human DNA-repair proteins Xrcc3-Rad51C. Proc. Natl Acad. Sci. USA, 98, 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masson J.Y., Stasiak,A.Z., Stasiak,A., Benson,F.E. and West,S.C. (2001) Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl Acad. Sci. USA, 98, 8440–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winsey S.L., Haldar,N.A., Marsh,H.P., Bunce,M., Marshall,S.E., Harris,A.L., Wojnarowska,F. and Welsh,K.I. (2000) A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer. Cancer Res., 60, 5612–5616. [PubMed] [Google Scholar]
- 51.Matullo G., Guarrera,S., Carturan,S., Peluso,M., Malaveille,C., Davico,L., Piazza,A. and Vineis,P. (2001) DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int. J. Cancer, 92, 562–567. [DOI] [PubMed] [Google Scholar]
- 52.Kuschel B., Auranen,A., McBride,S., Movik,K.L., Antoniou,A., Lipscombe,J.M., Day,N.E., Easton,D.F., Ponder,B.A., Pharoah,P.D. and Dunning,A. (2002) Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum. Mol. Genet., 11, 1399–1407. [DOI] [PubMed] [Google Scholar]
- 53.Araujo F.D., Pierce,A.J., Stark,M. and Jasin,M. (2002) Variant XRCC3 implicated in cancer is functional in homology-directed repair of double-strand breaks. Oncogene, 21, 4176–4180. [DOI] [PubMed] [Google Scholar]
- 54.French C.A., Masson,J.Y., Griffin,C.S., O’Regan,P., West,S.C. and Thacker,J. (2002) Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem., 277, 19322–19330. [DOI] [PubMed] [Google Scholar]
- 55.Godthelp B.C., Wiegant,W.W., van Duijn-Goedhart,A., Scharer,O.D., van Buul,P.P.W., Kanaar,R. and Zdzienicka,M.Z. (2002) Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res., 30, 2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Story R.M., Weber,I.T. and Steitz,T.A. (1992) The structure of the E. coli recA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- 57.Thacker J. (1999) A surfeit of RAD51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- 58.Pellegrini L., Yu,D.S., Lo,T., Anand,S., Lee,M., Blundell,T.L. and Venkitaraman,A.R. (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature, 420, 287–293. [DOI] [PubMed] [Google Scholar]
- 59.Kurumizaka H., Aihara,H., Kagawa,W., Shibata,T. and Yokoyama,S. (1999) Human Rad51 amino acid residues required for Rad52 binding. J. Mol. Biol., 291, 537–548. [DOI] [PubMed] [Google Scholar]
- 60.Brenneman M.A., Wagener,B.M., Miller,C.A., Allen,C. and Nickoloff,J.A. (2002) XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell, 10, 387–395. [DOI] [PubMed] [Google Scholar]