Abstract
Spontaneous damage to DNA as a result of deamination, oxidation and depurination is greatly accelerated at high temperatures. Hyperthermophilic microorganisms constantly exposed to temperatures exceeding 80°C are endowed with powerful DNA repair mechanisms to maintain genome stability. Of particular interest is the processing of DNA lesions during replication, which can result in fixed mutations. The hyperthermophilic crenarchaeon Sulfolobus solfataricus has two functional DNA polymerases, PolB1 and PolY1. We have found that the replicative DNA polymerase PolB1 specifically recognizes the presence of the deaminated bases hypoxanthine and uracil in the template by stalling DNA polymerization 3–4 bases upstream of these lesions and strongly associates with oligonucleotides containing them. PolB1 also stops at 8-oxoguanine and is unable to bypass an abasic site in the template. PolY1 belongs to the family of lesion bypass DNA polymerases and readily bypasses hypoxanthine, uracil and 8-oxoguanine, but not an abasic site, in the template. The specific recognition of deaminated bases by PolB1 may represent an initial step in their repair while PolY1 may be involved in damage tolerance at the replication fork. Additionally, we reveal that the deaminated bases can be introduced into DNA enzymatically, since both PolB1 and PolY1 are able to incorporate the aberrant DNA precursors dUTP and dITP.
INTRODUCTION
Life at high temperatures represents a big challenge in respect to maintaining the integrity of genetic information stored in DNA. It has been demonstrated that the DNA molecule undergoes spontaneous deamination, depurination and oxidation at greatly increased rates at the high temperatures at which hyperthermophilic archaeal organisms such as Sulfolobus normally live (1). The lesions arising in DNA, particularly during chromosomal replication, must be quickly dealt with otherwise they impose genome instability leading to death or fixed mutations. To counteract increased DNA damage, hyperthermophiles are endowed with efficient DNA repair enzymes such as uracil DNA glycosylase b from Pyrobaculum aerophilum (Pa-UDGb), the representative of the fifth family of uracil-DNA glycosylases, which removes deamination products of both cytosine and adenine, i.e. uracil and hypoxanthine, respectively (2). It has been demonstrated that the archaeal B family replicative DNA polymerases such as Pfu also specifically recognize uracil in the template utilizing a special binding pocket in their N-terminal domain resulting in the termination of primer extension about 4 bases upstream from the lesion (3). Interestingly, the crenarchaea such as those from the genus Sulfolobus also possess another DNA polymerase, PolY1 (4,5), belonging to the Y family. The Y family DNA polymerases are specialized in the replication of aberrant DNA templates and are able to bypass certain DNA lesions in an error-free manner but show low fidelity on normal DNA template (6,7). The structural basis of lesion bypass by the Y family DNA polymerases has recently been investigated by crystallographic approaches utilizing PolY1 from two different strains of Sulfolobus (8–10). It has been suggested that the ability of Y family polymerases to bypass template lesions is due to fewer contacts with the primer–template and a more open catalytic site which lacks the α-helix (O-helix domain) directly involved in the geometrical selection of correct base pairs in other DNA polymerases (11,12).
In order to deepen our understanding of how DNA lesions are processed during progression of the replication fork in archaea we have tested interactions of two known DNA polymerases from the model crenarchaeon Sulfolobus solfataricus with selected heat-induced lesions present in the DNA template as well as in the nucleotide pool. Here we demonstrate that the progression of the replicative DNA polymerase PolB1 is arrested at the lesions while the DNA polymerase PolY1 bypasses them. We also extend the knowledge of specific ‘read-ahead’ recognition of uracil in the template (13) by PolB1 to another DNA deamination product, hypoxanthine.
MATERIALS AND METHODS
Enzymes and substrates
Sulfolobus solfataricus DNA polymerases B1 and Y1 were purified from Escherichia coli overexpressing strains as described previously (14,15). Rabbit polyclonal antiserum was raised against purified PolY1 by TaKaRa-Shuzo. Taq (rTAQ) and Pfu DNA polymerases were purchased from TaKaRa and Stratagene, respectively. FPLC grade dNTPs were purchased from Amersham Pharmacia Biotech. Template oligonucleotides containing modified bases, i.e. uracil, hypoxanthine, 8-hydroxyguanine (8-oxoG) and a synthetic abasic site (AP-site, tetrahydrofuran moiety) were synthesized by Nippon Gene Ltd (Toyama, Japan). All other oligonucleotides including Cy3-labeled ones were synthesized by BEX Corp. (Tokyo) and double purified by HPLC.
Quantitative western blot analysis
Sulfolobus solfataricus (strain P2) cells were grown aerobically at 80°C, pH 3.5, in 100 ml of Brock’s basal salt medium supplemented with glucose (2 g/l) (16). Growth was monitored spectrophotometrically at 600 nm and when the absorbance reached a value of 0.7 OD, 0.5 ml aliquots of the culture were withdrawn and centrifuged in Eppendorf tubes. Each pellet was resuspended in 100 µl of SDS–PAGE sample buffer (50 mM Tris–HCl, pH 6.8, 10% glycerol, 100 mM 2-mercaptoethanol, 0.6% SDS, 0.01% Bromophenol blue). Aliquots of the extract were subjected to SDS–PAGE together with samples of the purified DNA PolB1 and DNA PolY1. Then the gels were electroblotted onto PVDF membranes and western blot analyses were carried out with rabbit polyclonal antisera raised against DNA PolB1 and DNA PolY1. Antigen–antibody interactions were detected with horseradish peroxidase-conjugated secondary antibodies and the ECL+ kit (Amersham Pharmacia Biotech). Chemiluminescence was analyzed using a Chemi Doc 2000 System with the Quantity One software (Bio-Rad). For calculating the number of DNA PolB1 or PolY1 molecules, it was assumed that 1.1 × 109 cells/ml are present in a culture of Sulfolobus when the absorbance at 600 nm is 1 OD.
Primer extension assays
Standard DNA polymerase reactions (10 µl) were performed in 30 mM potassium phosphate buffer (pH 7.4), 7.5 mM MgCl2, 1.25 mM β-mercaptoethanol, 5% glycerol, 100 µM dNTPs and 50 nM annealed 5′-Cy3-primer/template. PolY1 and PolB1 were added at 50 nM concentration and Taq and Pfu DNA polymerases at 2.5 U/reaction, followed by incubation at 55°C for 10 min. Reactions were terminated by adding 1 vol of stop solution (98% formamide, 10 mM EDTA, 20 mg/ml blue dextran) and the products were resolved by electrophoresis in 15% denaturing polyacrylamide gel and visualized using a Molecular Imager FX (Bio-Rad). The template used for all reactions was 5′-GAAGGGATCCTTAAGACTGTAACCGGTCTTCGCGCG-3′ with lesions or altered bases individually placed at the underlined positions as specified in the relevant figures. The sequences of the primers used were 5′-CGCGCGAAGACCGG-3′ and 5′-CGCGCGA AGACCGGTTAC-3′.
DNA binding BIAcore assay
The analysis was carried out using a BIAcoreX instrument by injecting 90 µl of the appropriate DNA polymerase at five different concentrations (10, 50, 100, 250 and 500 nM) in standard HBS-EP buffer over a SA chip surface with captured oligonucleotide at a flow rate of 30 µl/min. The oligonucleotides used were basically the same as those in the primer extension assay except for being biotinylated at their 3′-termini to enable immobilization. The sequence was 5′-GAAGGGATCCTTAAGACTGTAACCGGTCTTC GCGCG-biotin-3′, where the underlined G was optionally substituted for uracil, hypoxanthine or 8-oxoG. The sequence of the optionally annealed primer was 5′-CGCGCG AAGACCGGTTA-3′. Prior to analysis ∼500 RU of the appropriate oligonucleotide was captured on the SA chip surface via biotin–streptavidin coupling. The chip was regenerated between injections by a pulse of 2 M NaCl to remove the remains of protein bound from a previous injection. Where applicable, the equilibrium dissociation constants (Kd) were calculated from the kinetic traces of every five injection set using the BIAevaluation Software version 3.0 (BIAcore, Uppsala, Sweden) employing global fitting according to the ‘1:1 binding with mass transfer’ predefined model.
RESULTS
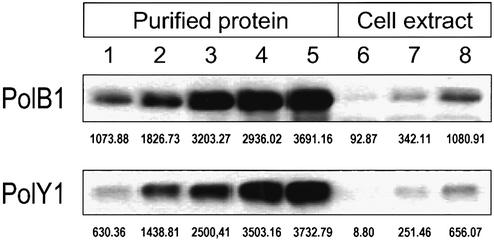
To approach the question of how DNA lesions inflicted by thermal stress affect DNA replication in hyperthermophiles, we have purified a new Y family DNA polymerase from S.solfataricus strain P2 which we term PolY1 (15). First we raised antiserum against the purified PolY1 in rabbits and compared its expression level to the level of PolB1 in vivo in growing S.solfataricus cells using quantitative western blot analysis (Fig. 1). We estimate that the number of molecules of PolB1 per cell at OD600 = 0.7 is around 1500, whereas for PolY1 it is about 4000.
Figure 1.
Expression of DNA polymerases B1 and Y1 in S.solfataricus cells. Quantitative western blots showing detection of the polymerases in cell lysates including standards with different amounts of purified proteins. The S.solfataricus cell extract was prepared as described in Materials and Methods. Lanes 1–5, purified proteins applied at 15, 30, 60, 120 and 240 ng/lane; lanes 6–8, S.solfataricus total cell extract applied at 5, 10 and 15 µl/lane. The chemiluminescence values shown beneath the lanes were expressed as the sum of the intensities of the pixels inside the volume boundary manually selected around each band per area of a single pixel, as described in Materials and Methods. The intensity values obtained for the amounts of 15, 30 and 60 ng (lanes 1–3) were used to construct a linear regression curve for each protein. From these titration curves, the protein concentration values were extrapolated for the lanes loaded with 15 µl of total cell extract.
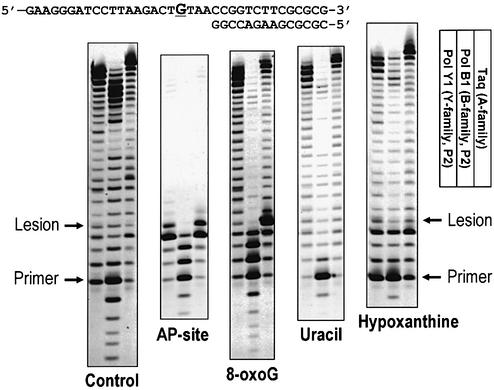
Next we investigated how PolY1 as well as the main chromosomal replicase PolB1 from the same organism perform primer extension and interact with several model DNA lesions in vitro (Fig. 2). The AP-site severely inhibited primer extension by both DNA polymerases from Sulfolobus as well as by the Taq DNA polymerase used as an A family thermostable polymerase control. The blockage of PolY1 by an AP-site may seem to conflict with the data of Boudsocq et al. (17), who reported efficient AP-site bypass with the same enzyme at 37°C. However, upon enhancing the image darkness we could observe limited AP-site bypass by PolY1 but not PolB1 under our experimental conditions. 8-oxoG, a major oxidative damage in DNA, also severely inhibited primer extension by PolB1 and most of the primers stopped 1 bp before the lesion. However, unlike the AP-site, 8-oxoG did not inhibit primer extension by PolY1. The primer was efficiently extended beyond the lesion and fully extended to the end of the template strand by PolY1. In the cases of uracil and hypoxanthine, the major products of primer extension by PolB1 corresponded to the position more than 1 bp upstream from the lesion sites. This premature halt of DNA polymerization upstream from the template deaminated bases resembles ‘read-ahead sensing’ of uracil by other archaeal B family DNA polymerases such as Pfu (13). It is apparent, however, that in the case of hypoxanthine there is significant primer extension upstream of the lesion and even to the full-size product. In contrast, the primer was efficiently extended beyond the lesions and elongated to the full size by PolY1 and Taq DNA polymerase.
Figure 2.
Comparison of DNA lesion bypass by thermostable DNA polymerases from the A, B and Y families. The side-by-side comparison of DNA polymerase abilities to bypass several heat-induced DNA lesions. The enzymes used were PolY1 and PolB1 from S.solfataricus and Taq DNA polymerase from T.aquaticus, shown schematically from left to right in the picture. The types of DNA lesions tested and associated controls (no lesion) are shown below the gel lines. The position of the lesions (or G in the control) is underlined within the full sequence context at the top. dNTPs were used at a concentration of 100 µM and the reactions were carried out at 55°C for 10 min as described in Materials and Methods.
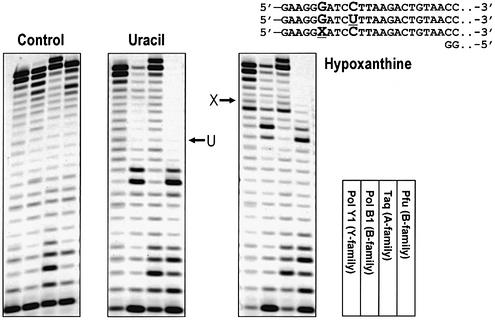
To determine the inhibitory distance from uracil and hypoxanthine, we have moved the primer start site further upstream from these lesions (Fig. 3). We also included the Pfu DNA polymerase as a positive control. PolB1 as well as Pfu paused 3–4 bp upstream from the lesions whereas PolY1 and Taq DNA polymerase bypassed the lesions efficiently. These results strongly suggest that PolB1 and Pfu recognize hypoxanthine as well as uracil by the read-ahead recognition mechanism. It should also be noted, however, that PolB1 was capable of extending a small fraction of primers to the full-length products, particularly in the case of hypoxanthine. This is in contrast to the clear stop of Pfu. The primer extension by Pfu mostly terminated at 4 bp upstream from the positions of uracil and hypoxanthine and no visible full-size extension was detectable. These results, together with the results shown in Figure 2, suggest that the read-ahead recognition of hypoxanthine by PolB1 is not so strict as for uracil and also that Pfu is more sensitive in sensing uracil and hypoxanthine in the template strand than PolB1.
Figure 3.
Recognition of deaminated bases in DNA by archaeal B family DNA polymerases by the read-ahead mechanism. The deaminated base products uracil (U) and hypoxanthine (X) were positioned further downstream from the 3′-end of the primer to determine the exact position of the replication block. Sequence details of the relevant parts of oligonucleotide substrates as well as the exact positions of the lesions on the gels are shown in the picture. The order of experimental lanes on the gels is schematically shown as well. The enzymes used were S.solfataricus PolY1 and PolB1 as well as the control DNA polymerases Taq from T.aquaticus and Pfu from Pyrococcus furiosus.
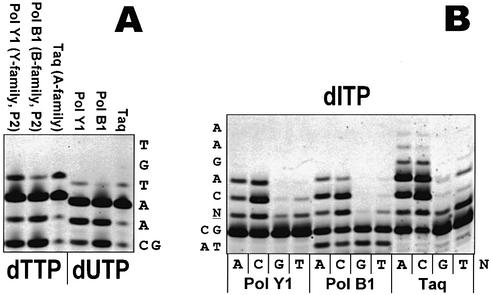
It has recently come to light that modified bases can be introduced into DNA enzymatically through the incorporation of modified dNTPs such as 8-oxo-dGTP or dUTP (18,19). We have previously demonstrated that PolY1 incorporates 8-oxo-dGTP and 2-OH-dATP into DNA in an erroneous manner (15). Here we show that both B and Y family DNA polymerases from S.solfataricus as well as the control Taq DNA polymerase from the aerobic gram-negative bacterium Thermus aquaticus efficiently insert dUTP and dITP into DNA (Fig. 4). dITP was more efficiently incorporated opposite template C and A compared with template G and T.
Figure 4.
Incorporation of deaminated dNTP derivatives into DNA by thermostable DNA polymerases. (A) Side-by-side comparison of incorporation of the normal base thymine (dTTP) and the erroneous uracil (dUTP) into DNA by the Y, B and A family DNA polymerases. The relevant substrate sequence is shown on the right next to the gel. (B) Incorporation and insertion specificity of the erroneous base hypoxanthine (dITP) into DNA by the Y, B and A family DNA polymerases. The relevant primer–template sequence is shown on the left next to the gel, where N stands for the variable base. Each of the four possible bases was tested at the variable base position as template for each enzyme in separate lanes as marked under the gel picture. The enzymes used were S.solfataricus PolY1 and PolB1 as well as the control DNA polymerase Taq from T.aquaticus.
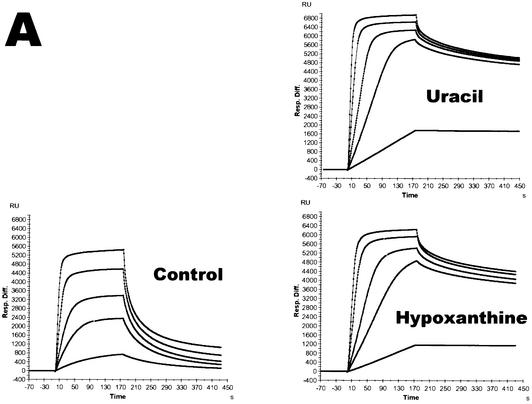
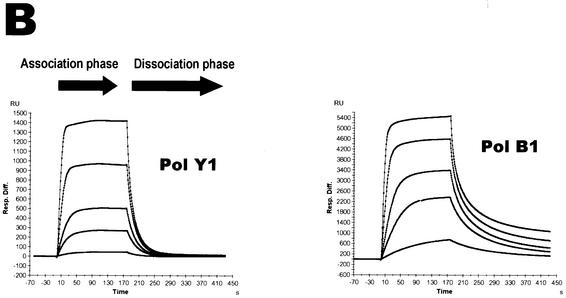
In order to further strengthen our findings on the specific recognition of deaminated bases in the template by PolB1, we have investigated the physical interactions of both PolB1 and PolY1 with template oligonucleotides containing lesions using the surface plasmon resonance technique. PolB1 showed greatly enhanced binding to oligonucleotides containing a single uracil or hypoxanthine (Fig. 5A). Because the binding kinetics of PolB1 to DNA did not fit well with the predefined 1:1 binding model, we were unable to accurately determine its Kd values. Nevertheless, based on the 1:1 model, we estimated the semi-quantitative Kd values of PolB1 to the oligonucleotides containing uracil or hypoxanthine as 2–4 nM (Table 1). In the cases of other lesions, i.e. the AP-site and 8-oxoG, we did not observe any differences in binding to the lesion-containing versus normal undamaged oligonucleotides. In contrast, PolY1 displayed no enhanced binding to the oligonucleotides containing any of the lesions. PolB1 has an affinity for normal undamaged single-stranded DNA (ssDNA) with a Kd of 55 nM, which is similar to the affinities for DNA of E.coli DNA polymerase I, rat DNA polymerase β or the previously characterized PolY1 from Sulfolobus strain P1 (5). PolY1 displayed generally weaker binding to undamaged ssDNA than PolB1, with a Kd of 456 nM (Fig. 5B). However, PolY1 displayed a slight increase in affinity for undamaged primed DNA which has partly double-stranded DNA sequences (Table 1). In summary, the strong binding of PolB1 to uracil and hypoxanthine in the template supports our conclusion that both bases are recognized by the uracil-binding pocket of the enzyme. This agrees with previous reports about the tight binding of archaeal B family DNA polymerases such as Pfu to uracil-containing DNA (3,13,20).
Figure 5.
Tight binding of PolB1 to ssDNA oligonucleotides containing deaminated bases. BIAcore sensograms showing the association and dissociation kinetics for binding of S.solfataricus DNA polymerases to immobilized oligonucleotides. (A) Demonstration of tight binding of PolB1 to oligonucleotides containing deaminated bases (either single uracil or single hypoxanthine as shown) compared to control oligonucleotide without lesions. (B) Comparison of PolY1 and PolB1 binding to control oligonucleotide. Binding to substrates was tested by injecting the appropriate protein independently five times at concentrations of 10, 50, 100, 250 and 500 nM.
Table 1. Comparison of the affinities of Sulfolobus DNA polymerases for oligonucleotides with or without DNA lesions immobilized on the BIAcore chip surface.
| Ligand | Kd (nM) | |
|---|---|---|
| PolB1a | PolY1 | |
| ssDNA containing uracil | 4 | 535 |
| ssDNA containing hypoxanthine | 2 | 325 |
| ssDNA containing abasic site | 15 | 483 |
| ssDNA containing 8-oxoG | 62 | 542 |
| ssDNA without lesions | 55 | 456 |
| Primed DNA | 81 | 158 |
The binding analysis was performed under the same conditions with five different analyte concentrations and fitted globally according to the ‘1:1 binding with mass transfer’ model using the BIAevaluation software.
aThe values for PolB1 are semi-quantitative as its binding kinetics did not fit well into the 1:1 model.
DISCUSSION
To examine the roles of DNA polymerases in the processing of DNA lesions in the hyperthermophilic archaeon S.solfataricus, we have purified the new Y family DNA polymerase PolY1 and its main replicase PolB1. Compared to its homolog from a related Sulfolobus strain characterized previously (5), PolY1 showed more robust synthetic activity requiring much lower enzyme concentrations to perform primer extensions. This is in agreement with the fact that PolY1 characterized by another group could substitute for Taq DNA polymerase in PCR reactions (17). Interestingly, the expression levels of PolB1 and PolY1 are 1500 and 4000 molecules/cell, respectively (Fig. 1). These numbers are noticeably higher than the ones reported for E.coli DNA Pol III (10–20 molecules/cell) or DNA Pol I (400 molecules/cell) (21), rather resembling the level of fully SOS-induced E.coli Pol IV (22). One possible explanation for this is that most of these molecules are engaged in repairing the massive lesions inflicted on the Sulfolobus chromosomal DNA by heat exposure. Because of the high expression level and the ability to bypass some of the lesions (Figs 2 and 3), we consider that PolY1 could play an important role in these DNA transactions within the cell. However, to establish whether its function is essential for cell growth, it is necessary to construct and test the viability of S.solfataricus mutant strains that are unable to express the corresponding gene or express structurally intact but inactive mutant protein.
We also examined the abilities of PolB1 and PolY1 to bypass various DNA lesions, i.e. the AP-site, 8-oxoG, uracil and hypoxanthine, using in vitro primer extension assays (Fig. 2). Hyperthermophiles such as S.solfataricus are strictly dependent on high temperature for optimal growth and thus face problems, such as heat-induced depurination, oxidation and deamination in DNA. The depurinated product, i.e. the AP-site, is a major cytotoxic lesion in DNA while deamination of cytosine and adenine results in the formation of uracil and hypoxanthine, respectively, which can induce transition type mutations. To examine the possible contribution of PolY1 to reducing the cytotoxic effects of the AP-site, we conducted a bypass DNA synthesis assay using template DNA containing an AP-site. However, the lesion efficiently inhibited primer extension by both PolB1 and PolY1. Because the bypass efficiency beyond the AP-site was only marginal even with PolY1, we suggest that PolY1 may not play an important role in the tolerance mechanism against the cytotoxic damage of the AP-site in this organism.
Unlike the AP-site, 8-oxoG displayed different inhibitory effects on PolB1 and PolY1: it severely inhibited primer extension by PolB1 but not by PolY1. Because of the ability of PolY1 to bypass 8-oxoG, it may take over the primer end from PolB1 and extend across this lesion during DNA replication. In this respect it resembles other Y family DNA polymerases, such as DNA polymerases η and κ, which readily bypass the 8-oxoG lesion (23–25). This raises the question of whether the bypass of 8-oxoG by PolY1 in vivo would be mutagenic. In our preliminary experiment PolY1 incorporated mainly A opposite 8-oxoG in the template (data not shown), which may indeed seem mutagenic. However, if one imagines that most of the 8-oxoG lesions are introduced into the template by the erroneous incorporation of 8-oxo-dGTP (15) in the first place, the insertion of A opposite the lesion during the second round of replication would fix the original error and therefore be error free. It is also possible that the base insertion specificity opposite 8-oxoG is further modulated by accessory factors such as proliferating cell nuclear antigen, replication factor C and RPA, normally participating in replication in vivo.
The ‘read-ahead’ mechanism is a unique function by which archaeal B family DNA polymerases such as Pfu detect uracil in the template strand during DNA synthesis. Pfu DNA polymerase stalls 4 bp upstream of a template uracil, thereby preventing the irreversible fixation of a G:C→A:T mutation. Thermophilic bacterial DNA polymerases, such as Taq DNA polymerase, or viral B family DNA polymerases, such as T4 DNA polymerase, can read through a template strand uracil without indication of a stall. Here, we demonstrate for the first time that not only uracil but also the deamination product of adenine, i.e. hypoxanthine (deoxyinosine), is recognized by the ‘read-ahead’ mechanism. Pfu stalled 3–4 bp before hypoxanthine as well as uracil in the template strand (Fig. 3). In addition, PolB1 also displayed a similar stalling behavior, although it extended a small fraction of primers to the full-length products, particularly in the case of hypoxanthine. Thus, we speculate that hypoxanthine fits into the uracil-binding pocket (3) in the N-terminal domain of Pfu as well as PolB1. This notion is supported by the strong binding of PolB1 to uracil and hypoxanthine in ssDNA (Table 1 and Fig. 5A). The precise determination of the structural aspects of deaminated base recognition by PolB1 and Pfu should await a co-crystallization study including the whole enzyme or the uracil-binding pocket domain and substrate. It remains to be experimentally confirmed whether the inhibition of B family archaeal DNA polymerases by deaminated DNA bases extends to other deamination products of, for example, guanine (i.e. xanthine), which was mentioned not to be inhibitory (13).
It is conceivable that the free nucleotide pool is more prone to damage than the relatively stable and closed double-stranded DNA. In the light of this the introduction of modified bases into DNA via erroneous incorporation of modified dNTPs during chromosomal replication may represent a significant source of DNA damage. In fact, in addition to the incorporation of 8-oxo-dGTP (15), we show here that the thermostable DNA polymerases also easily incorporate dUTP and dITP (Fig. 4). The insertion of dITP into DNA can be particularly harmful since hypoxanthine pairs with all four bases with varying efficiencies (Fig. 4B). To minimize the chances of such insertion, concentrations of both dUTP and dITP are kept low inside the cells by specialized pyrophosphatases (26–29). Despite efficient insertion, both uracil and hypoxanthine inhibit the B family archaeal DNA polymerases once introduced into the template. We have observed that PCR fails with B family archaeal DNA polymerases, but not Taq DNA polymerase, when uracil is incorporated into the template (data not shown). Primers containing deoxyinosine are also reported to fail in PCR if Pfu DNA polymerase is used (30).
Because the 3-dimensional structures of archaeal DNA polymerases from both the B and Y families have recently been determined (3,8–10), it is now possible to explore the structural basis of specific lesion recognition by computer simulation approaches backed by site-directed mutagenesis studies. Because PolY1 could readily bypass all tested lesions except the AP-site we speculate that a polymerase switch takes place upon lesion encounter by PolB1 at the replication fork in vivo. Polymerase switching is actually quite a common process during lagging strand synthesis. Alternatively, a specific repair enzyme such as DNA glycosylase may be recruited to remove certain lesions from the template. Evidently, further studies will be needed to address these issues. Additionally, we anticipate that the lesion-tolerant PolY1 could prove useful in biotechnology applications such as forensic PCR, where the conventional DNA polymerases fail due to excessive damage to the DNA template.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a grant-in-aid for International Collaborative Research from the Japan Health Science Foundation. A part of this study was financially supported by the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology. F.M.P. is the recipient of a grant from the European Union (contract no. QLK3-CT-2002-02071).
REFERENCES
- 1.Bruskov V.I., Malakhova,L.V., Masalimov,Z.K. and Chernikov,A.V. (2002) Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res., 30, 1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sartori A.A., Fitz-Gibbon,S., Yang,H., Miller,J.H. and Jiricny,J. (2002) A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J., 21, 3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fogg M.J., Pearl,L.H. and Connolly,B.A. (2002) Structural basis for uracil recognition by archaeal family B DNA polymerases. Nature Struct. Biol., 9, 922–927. [DOI] [PubMed] [Google Scholar]
- 4.Kulaeva O.I., Koonin,E.V., McDonald,J.P., Randall,S.K., Rabinovich,N., Connaughton,J.F., Levine,A.S. and Woodgate,R. (1996) Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res., 357, 245–253. [DOI] [PubMed] [Google Scholar]
- 5.Gruz P., Pisani,F.M., Shimizu,M., Yamada,M., Hayashi,I., Morikawa,K. and Nohmi,T. (2001) Synthetic activity of Sso DNA polymerase Y1, an archaeal DinB-like DNA polymerase, is stimulated by processivity factors proliferating cell nuclear antigen and replication factor C. J. Biol. Chem., 276, 47394–47401. [DOI] [PubMed] [Google Scholar]
- 6.Prakash S. and Prakash,L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev., 16, 1872–1883. [DOI] [PubMed] [Google Scholar]
- 7.Ohmori H., Friedberg,E.C., Fuchs,R.P.P., Goodman,M.F., Hanaoka,F., Hinkle,D.C., Kunkel,T.A., Lawrence,C.W., Livneh,Z., Nohmi,T. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 1–20. [DOI] [PubMed] [Google Scholar]
- 8.Ling H., Boudsocq,F., Woodgate,R. and Yang,W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell, 107, 91–102. [DOI] [PubMed] [Google Scholar]
- 9.Silvian L.F., Toth,E.A., Pham,P., Goodman,M.F. and Ellenberger,T. (2001) Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nature Struct. Biol., 8, 984–989. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B.L., Pata,J.D. and Steitz,T.A. (2001) Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell, 8, 427–437. [DOI] [PubMed] [Google Scholar]
- 11.Kokoska R.J., Bebenek,K., Boudsocq,F., Woodgate,R. and Kunkel,T.A. (2002) Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J. Biol. Chem., 277, 19633–19638. [DOI] [PubMed] [Google Scholar]
- 12.Potapova O., Grindley,N.D. and Joyce,C.M. (2002) The mutational specificity of the Dbh lesion bypass polymerase and its implications. J. Biol. Chem., 277, 28157–28166. [DOI] [PubMed] [Google Scholar]
- 13.Greagg M.A., Fogg,M.J., Panayotou,G., Evans,S.J., Connolly,B.A. and Pearl,L.H. (1999) A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA, 96, 9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisani F.M., De Felice,M. and Rossi,M. (1998) Amino acid residues involved in determining the processivity of the 3′-5′ exonuclease activity in a family B DNA polymerase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochemistry, 37, 15005–15012. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu M., Gruz,P., Kamiya,H., Kim,S.R., Pisani,F.M., Masutani,C., Kanke,Y., Harashima,H., Hanaoka,F. and Nohmi,T. (2003) Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep., 4, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brock T.D., Brock,K.M., Belly,R.T. and Weiss,R.L. (1972) Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol., 84, 54–68. [DOI] [PubMed] [Google Scholar]
- 17.Boudsocq F., Iwai,S., Hanaoka,F. and Woodgate,R. (2001) Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res., 29, 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colussi C., Parlanti,E., Degan,P., Aquilina,G., Barnes,D., Macpherson,P., Karran,P., Crescenzi,M., Dogliotti,E. and Bignami,M. (2002) The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol., 12, 912–918. [DOI] [PubMed] [Google Scholar]
- 19.Nilsen H., Rosewell,I., Robins,P., Skjelbred,C.F., Andersen,S., Slupphaug,G., Daly,G., Krokan,H.E., Lindahl,T. and Barnes,D.E. (2000) Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell, 5, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 20.Lasken R.S., Schuster,D.M. and Rashtchian,A. (1996) Archaebacterial DNA polymerases tightly bind uracil-containing DNA. J. Biol. Chem., 271, 17692–17696. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg A. and Baker,T.A. (1992) Prokaryotic DNA polymerases other than E. coli Pol I. In DNA Replication. W.H. Freeman and Co., New York, NY, pp. 165–196.
- 22.Kim S.R., Matsui,K., Yamada,M., Gruz,P. and Nohmi,T. (2001) Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics, 266, 207–215. [DOI] [PubMed] [Google Scholar]
- 23.Haracska L., Yu,S.L., Johnson,R.E., Prakash,L. and Prakash,S. (2000) Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nature Genet., 25, 458–461. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.S., Geacintov,N.E. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Yuan,F., Wu,X., Wang,M., Rechkoblit,O., Taylor,J.S., Geacintov,N.E. and Wang,Z. (2000) Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res., 28, 4138–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persson R., Cedergren-Zeppezauer,E.S. and Wilson,K.S. (2001) Homotrimeric dUTPases; structural solutions for specific recognition and hydrolysis of dUTP. Curr. Protein Pept. Sci., 2, 287–300. [DOI] [PubMed] [Google Scholar]
- 27.Chung J.H., Park,H.Y., Lee,J.H. and Jang,Y. (2002) Identification of the dITP- and XTP-hydrolyzing protein from Escherichia coli. J. Biochem. Mol. Biol., 35, 403–408. [DOI] [PubMed] [Google Scholar]
- 28.Chung J.H., Back,J.H., Park,Y.I. and Han,Y.S. (2001) Biochemical characterization of a novel hypoxanthine/xanthine dNTP pyrophosphatase from Methanococcus jannaschii. Nucleic Acids Res., 29, 3099–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S., McLennan,A.G., Ying,K., Wang,Z., Gu,S., Jin,H., Wu,C., Liu,W., Yuan,Y., Tang,R. et al. (2001) Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene. J. Biol. Chem., 276, 18695–18701. [DOI] [PubMed] [Google Scholar]
- 30.Knittel T. and Picard,D. (1993) PCR with degenerate primers containing deoxyinosine fails with Pfu DNA polymerase. PCR Methods Appl., 2, 346–347. [DOI] [PubMed] [Google Scholar]