Abstract
This study evaluates the potential of pseudocomplementary peptide nucleic acids (pcPNAs) for sequence-specific modification of enzyme activity towards double-stranded DNA (dsDNA). To this end, we analyze the ability of pcPNA–dsDNA complexes to site-selectively interfere with the action of four type IIs restriction enzymes. We have found that pcPNA–dsDNA complexes exhibit a different degree of DNA protection against cleaving/nicking activity of various isoschizomeric endonucleases under investigation (PleI, MlyI and N.BstNBI) depending on their type and mutual arrangement of PNA-binding and enzyme recognition/cleavage sites. We have also found that the pcPNA targeting to closely located PleI or BbsI recognition sites on dsDNA generates in some cases the nicking activity of these DNA cutters. At the same time, MlyI endonuclease, a PleI isoschizomer, does not exhibit any DNA nicking/cleavage activity, being completely blocked by the nearby pcPNA binding. Our results have general implications for effective pcPNA interference with the performance of DNA-processing proteins, thus being important for prospective applications of pcPNAs.
INTRODUCTION
There is significant interest in the discovery and development of small molecules (as compared with proteins) that target double-stranded DNA (dsDNA) in a site-selective manner but without serious sequence limitations. Such ligands may yield new robust biomolecular tools, sensitive DNA diagnostics and potent gene therapeutics. For these purposes, dsDNA–ligand complexes have to be sufficiently stable to compete with DNA-processing proteins. The resistance of a ligand to enzymatic degradation is also often essential to allow efficient DNA manipulation. Therefore, ligands are required, which bind dsDNA with (i) high affinity and (ii) high selectivity also characterized by (iii) sufficient biostability and (iv) lack of sequence restrictions.
Despite the fact that many natural and non-natural (synthetic and semi-synthetic) dsDNA-binding ligands with groove-specific or intercalative modes of complex formation have been identified (1–19), none of them meets all of these four key requirements simultaneously. The only group of ligands that actually comes close to reaching this goal are the minor groove-binding hairpin pyrrole-imidazole polyamides (6,17). Notwithstanding significant progress in this field, note that binding affinity of hairpin polyamides for single-base-mismatched sites is reduced ≤102 only, as compared with correct dsDNA targets. Although quite large, this degree of sequence specificity could be insufficient for some applications in vivo and in vitro, especially given the current trends in basic and applied studies to operate with entire genomes and crude samples/unpurified material. Therefore, the search for new ligands with alternative modes of binding to DNA duplexes is of paramount importance.
Peptide nucleic acids (PNAs) are nuclease/protease- resistant dsDNA-binding ligands that are capable of forming very stable and highly sequence-specific helix invasion complexes with target sites on duplex DNA (20–32). They have a unique ability to kinetically discriminate correct versus single-base-mismatched sites by a factor of approximately 103 (24–26), thus being promising for different applications ranging from molecular biotechnology to medicinal chemistry (33–40). Recently introduced pseudocomplementary (pc) PNAs are particularly promising for gene targeting as they exhibit rather mild sequence limitations for binding to designated dsDNA sites and also induce site-selective bending in DNA duplexes (29,41–43).
Along with ordinary Gs and Cs, pcPNAs carry, similar to the pc-type oligonucleotides (44), 2,6-diaminopurine (D) and 2-thiouracil (sU) nucleobases instead of all As and Ts, respectively. Whereas these modifications impede the base pairing between two mutually pseudocomplementary PNA oligomers, they do not prevent pcPNAs from binding to the corresponding sequences in DNA carrying normal nucleobases (29,41–43). As a result of such pseudocomplementarity between D/sU-modified PNA oligomers and common DNA strands, a pair of pcPNAs pries a dsDNA site open via formation of double-duplex invasion complexes (Fig. 1). This novel mode of PNA–DNA recognition significantly extends the sequence repertoire of possible DNA targets, since essentially any chosen mixed-base site on dsDNA can be targeted with pcPNAs, with the exception of very GC-rich (A + T content ≤40%) sites (29).
Figure 1.
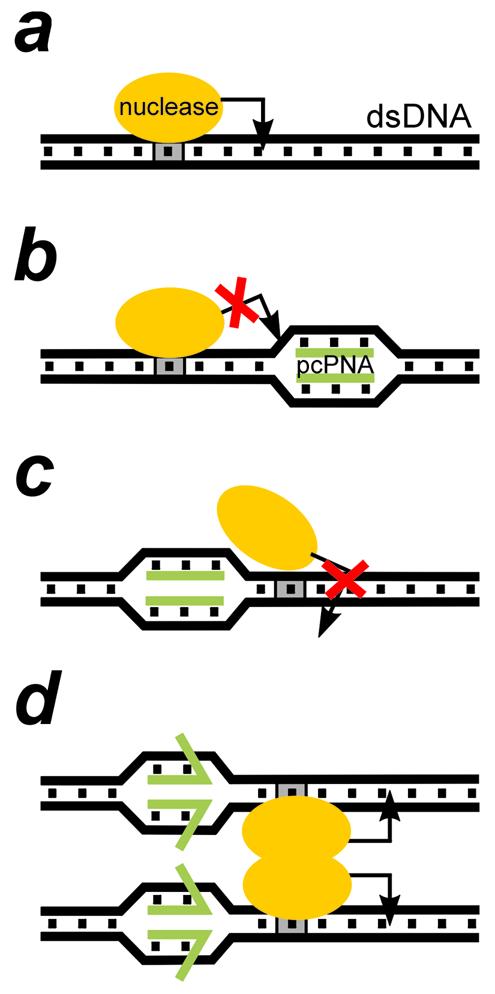
Schematics of the performance of the type IIs restriction endonucleases (the enzyme is shown as on orange oval) and its interference with the double-duplex invasion complex formed by a pair of pcPNAs (green bars) at different locations on dsDNA. (a) A type IIs restriction enzyme interacts with two discrete sites on dsDNA: the 4–7 bp non-palindromic recognition site (shown in gray) and the cleavage site, usually 1–20 bp away from the recognition site, indicated by an arrow (all enzymes used in this study cleave downstream of the recognition site). Certain nicking endonucleases (e.g. N.BstNBI studied here) behave like type IIs restriction enzymes and are closely related to them. (b) pcPNA binding downstream of the recognition site blocks the enzyme cleavage site. For smaller distances between the two sites, a PNA can also interfere with the enzyme binding (not shown). (c) PNA binding upstream in close proximity to the recognition site hinders enzyme binding, preventing DNA cleavage. (d) Dimerization of enzymes bound to two different recognition sites may be the driving force for partial displacement of pcPNAs from their complexes with dsDNA. In this case, restriction enzyme will efficiently cut dsDNA even if the ‘upstream’ PNA–DNA complex is located very close to the enzyme recognition site.
In order to assess the potential of pcPNAs for possible applications involving proteins, one needs to determine whether the pcPNA–dsDNA complexes are sufficiently stable to prevent DNA-processing proteins from binding to their recognition sites on dsDNA. To this end, Lohse et al. assayed the ability of pcPNA–dsDNA complexes to obstruct the activity of RNA polymerase (29). They found that pcPNA binding within the promoter region obstructs the initiation of transcription by T7 RNA polymerase. However, pcPNAs bound downstream of the promoter have no effect on the transcription elongation for both T3 and T7 RNA polymerases.
In another study, Izvolsky et al. showed that pcPNA binding to dsDNA efficiently blocks the access of common (type II) methylation/restriction enzymes to their recognition sites if the PNA-binding site completely covers or overlaps with the enzyme recognition site (41). The full protection against Dam methylase at low salt concentration and BclI restriction endonuclease at high salt concentration is achieved when the pcPNA-binding site embodies the enzyme-binding site. For Dam methylase, the limited (∼70%) protection is observed when the pcPNA and enzyme-binding sites overlap by 2 bp.
In the present work, we employ the type IIs restriction endonucleases and related DNA-nicking enzymes (45–47) as a test system to further examine pcPNA interference with protein activity on duplex DNA. Generally, these enzymes recognize an asymmetrical sequence and introduce a double-stranded or single-stranded break several bases away from their recognition site (Fig. 1). We have chosen this system because it provides us with a very simple and clear-cut method for detection of the protein-blocking potential of pcPNAs. Indeed, as a result of pcPNA protection, the chosen DNA sites will not be cut, which can be readily detected by regular gel electrophoresis. In addition, these enzymes make it possible to elucidate the interference of pcPNAs with protein activity on two levels, i.e. at the step of protein binding to its recognition site and at the step of DNA cleavage. Finally, competition of pcPNAs with restriction enzymes can be studied at physiologically relevant salt concentrations.
Specifically, we study here the interference of pcPNA–dsDNA complexes with the activity of four type IIs restriction enzymes, including three closely related isoschizomers. We deduce practical rules for effective blockage of the activity of DNA-processing proteins when pcPNAs bind in close proximity to the protein recognition site. Furthermore, a site-specific conversion of DNA cutters into DNA nickases by pcPNA targeting demonstrates the potential of pcPNAs to serve not only as selective blockers but also as modulators of protein activity on duplex DNA.
MATERIALS AND METHODS
Materials
The following pair of pcPNAs has been used in our study: H-Lys-GsUDGDsUCDCsU-LysNH2 (PNA 1495) and H-Lys-DGsUGDsUCsUDC-LysNH2 (PNA 1496). PNA concentration in stock solutions was calculated from their optical density measured at 260 nm using the following extinction coefficients: 11 700 (G), 10 200 (sU), 7600 (D) and 6600 (C) M–1cm–1. These PNAs were synthesized as previously described (29).
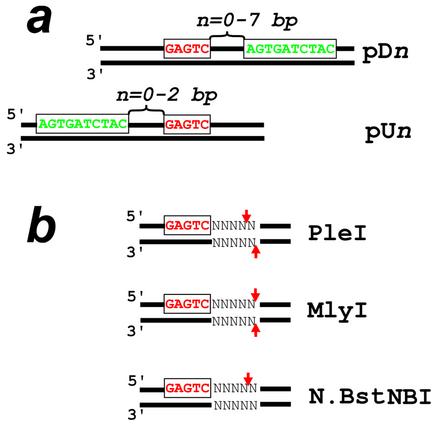
All restriction enzymes were obtained from New England Biolabs and used in the supplemented reaction buffers. Recombinant plasmids carrying the PNA-binding and enzyme recognition sites were obtained by cloning corresponding inserts into the BamHI–HindIII site of the pUC19 polylinker and introducing the plasmids into the Dam– strain of Escherichia coli (DM1™, Gibco-BRL). The plasmid inserts differ in the position of the PNA-binding site with respect to the recognition site of the restriction enzyme. According to our notation, the PNA-binding site is located n bp upstream (pUn series of plasmids) or downstream (pDn series of plasmids) of the enzyme recognition sequence (Fig. 2a). Note that plasmids carrying the recognition site for PleI, MlyI and N.BstNBI in the polylinker contain three more sites for these enzymes. The plasmid involved in the experiments with the restriction endonuclease BbsI has a single recognition site for this enzyme within the cloned insert.
Figure 2.
DNA constructs (a) used in this study with three isoschizomeric endonucleases (PleI, MlyI and N.BstNBI) and recognition/cleavage sites (b) of these enzymes. The enzyme recognition and PNA-binding sites are shown in red and green, respectively; arrows indicate the enzyme cleavage sites. PNA-binding sites are located downstream (pDn series of plasmids) or upstream (pUn series of plasmids) of the enzyme recognition site. The DNA constructs differ in the gap n between the two sites.
Analysis of the PNA-directed dsDNA protection from restriction enzymes
DNA–PNA complexes were obtained by incubating 20 nmol of PvuII-digested plasmids with both PNAs at a concentration of 0.8 µM in 40 µl of 10 mM TE buffer pH 7.4 for 2 h at 50°C. Such conditions provide the complete binding of pcPNAs to their target sites located within an ∼350 bp DNA fragment. To monitor the formation of DNA–PNA complexes, the electrophoretic mobility of which is significantly retarded with respect to naked DNA, the samples were resolved on an 8% polyacrylamide gel (PAAG) using 0.5× TBE as a running buffer (41,42). Unbound PNAs were removed by gel filtration through Sephadex G-50. About 1 nmol of the sample was introduced into a reaction buffer required for the enzyme activity and subjected to incubation with 3–10 U of the restriction enzyme under study for 40 min at 37°C (optimal time and temperature for quantitative digestion without PNA dissociation).
The DNA digestion by enzymes was stopped by addition of 10 mM EDTA (final concentration) and subsequent incubation at 65°C for 20 min. This treatment not only inactivates restriction enzymes but also induces pcPNAs dissociation from DNA targets. The final samples were resolved by electrophoresis in an 8% PAAG with 0.5× TBE as a running buffer. To enhance the retardation of nicked DNA fragments relative to intact ones, the samples were also run through an 8% PAAG containing 5 M urea (48). DNA was visualized by ethidium bromide staining and detected using a CCD camera coupled with the IS-1000 digital imaging system/software (Alpha Innotech Corporation). All images are presented as negatives; quantitative estimates of the extent of dsDNA cleavage/nicking were done by measuring the normalized intensities of the corresponding bands.
Analysis of the PNA-induced DNA-nicking activity of restriction enzymes
The scDNA assay. To check the PNA-induced DNA nicking, supercoiled (sc) plasmids were employed because of the well-known fact that the nicked, open circular dsDNA form has a different electrophoretic mobility in agarose gels as compared with covalently closed scDNA and linear dsDNA. These three DNA forms were resolved by electrophoresis in a 1% agarose gel using 1× TAE as a running buffer. The conversion of the initial scDNA sample into the open circular form clearly indicates dsDNA nicking.
The nick mapping assay. To localize the DNA-nicking position, 160 bp fluorescently labeled DNA fragments were generated by PCR from the pD5 template with Taq DNA polymerase (New England Biolabs) using one of the two primers tagged with Cy5 at the 5′ end. The PNA targeting and restriction digest of these fragments were performed as described above. The final samples were resolved on a 6% denaturing (7 M urea) PAAG run at 50°C using ALF Express DNA sequencer (Amersham Pharmacia Biotech) along with the four common dideoxy sequencing ladders. Sequencing ladders were obtained with the same PCR primers by using the AutoRead 200 sequencing kit (Amersham Pharmacia Biotech). The post-run data analysis was performed with an ALFwin Sequence Analyzer and DNA Fragment Analyzer software (Amersham Pharmacia Biotech).
RESULTS AND DISCUSSION
Blockage of DNA digestion with restriction and nicking enzymes by pcPNA–dsDNA complexes
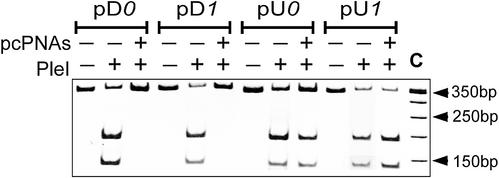
All three isoschizomeric enzymes used in this study, PleI, MlyI and N.BstNBI, recognize the same dsDNA sequence, 5′-GAGTC, and introduce either a double-stranded (PleI and MlyI) or a single-stranded (N.BstNBI) break 4–5 bp downstream of the enzyme recognition site (Fig. 2b). Figure 3 shows the results obtained with the PleI endonuclease using standard gel electrophoresis. One can see that pcPNA targeting upstream of the enzyme recognition site has little effect on the DNA cleavage activity of this enzyme. In contrast, the downstream PNA binding totally prevents digestion of the DNA target fragment. This is what one could expect since the PNA-binding site covers the enzyme’s cutting site thus obstructing DNA cleavage.
Figure 3.
Site-directed interference of pcPNA–dsDNA complexes with the DNA-cleaving activity of PleI analyzed by polyacrylamide gel electrophoresis. Restriction patterns with or without pcPNAs are compared for four DNA constructs indicated at the top of the gel. The 350 bp DNA target fragments were used in this experiment. Lane C is a 50 bp ladder.
Similar results have been obtained with the same set of plasmids in the case of pcPNA interference with the MlyI endonuclease: only the downstream pcPNA targeting is effective in inhibiting this restriction enzyme (data not shown). The expected complete blockage of the MlyI activity is also observed at larger, up to 5 bp downstream, distances between the PNA-binding and enzyme recognition sites. At the same time, uncommon enzymatic activity, i.e. the DNA-nicking activity at the remote DNA sites with degenerated enzyme recognition sequences, is detected in these cases with PleI restriction endonuclease (see Table 1 and next section). A partial protection of dsDNA against these enzymes is observed when the PNA-binding site is located 6 bp downstream of the enzyme recognition site. Both enzymes become insensitive to the presence of pcPNAs only when the distance between the corresponding sites reaches 7 bp.
Table 1. Effect of pcPNA–dsDNA complexes on the DNA-cleaving/nicking activity of isoschizomeric endonucleases depending on the relative location of corresponding binding/recognition sites and distances between them (pUn and pDn set of plasmids).
| Upstream PNA binding at various positions (bp) | Enzyme | Downstream PNA binding at various positions (bp) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 1 | 0 | 0 | 1 | 3 | 4 | 5 | 6 | 7 | |
| Cut | Cut | Cut | PleI | – | – | * | * | * | ** | Cut |
| Cut | Cut | Cut | MlyI | – | – | – | – | – | ** | Cut |
| Nick | – | – | N.BstNBI | – | – | – | – | – | Nick | Nick |
*Singly nicked DNA fragment at a site with the degenerate recognition sequence 5′-TAATC; a minute activity towards the bottom DNA strand is also present at this pseudo-site.
**Thirty percent of the fragment is cleaved, while the remaining 70% is singly nicked at the correct site.
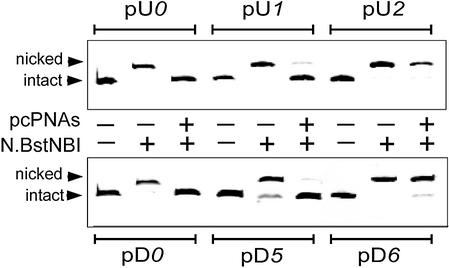
Figure 4 presents the results obtained with the N.BstNBI nicking enzyme using gel electrophoresis enhanced by the addition of urea to increase the retardation of nicked DNA (48). In this experiment, N.BstNBI is efficiently inhibited by the pcPNAs binding to dsDNA targets both upstream and downstream of the enzyme recognition site, in contrast to isoschizomeric common restriction enzymes. Note that the downstream PNA targeting strongly interferes with the activity of the N.BstNBI nickase within the 5 bp range from the enzyme recognition site, as was found in the case of MlyI. The DNA nickase becomes insensitive to pcPNAs in this case only when the gap between the corresponding sites reaches 6 bp. In contrast, the enzyme blockage by the upstream PNA targeting is effective at smaller distances between the PNA-binding and enzyme recognition sites: in the case of the 2 bp gap, N.BstNBI remains totally active after pcPNA binding.
Figure 4.
Site-directed interference of pcPNA–dsDNA complexes with the DNA-nicking activity of N.BstNBI monitored by polyacrylamide gel electrophoresis in the presence of urea. Introduction of a nick in the 350 bp DNA fragments leads to their retardation in gel electrophoresis. Upper panel: the upstream PNA targeting. Lower panel: the downstream PNA targeting.
The pcPNA-induced conversion of restriction enzymes into DNA nickases
In agreement with recently published results (43), Figure 5a shows that the PleI DNA cleavage activity vanishes upon binding of pcPNA 5 bp downstream of the enzyme recognition site. Obviously, the PNA binding denies the enzyme the access to its cleavage site. Instead, this DNA cutter exhibits PNA-induced DNA-nicking activity, as the PNA-targeted and enzyme-treated DNA fragment moves much more slowly in the gel with urea than the intact fragment (48). Analogous transformation of PleI into nickase is induced by the pcPNA targeting at 3 and 4 bp downstream of the enzyme recognition site (Fig. 5b). On the other hand, Figure 5 demonstrates that the PleI-related enzyme, MlyI, remains blocked, as expected, by targeting pcPNA 3–5 bp downstream of the enzyme-binding site without displaying any other activity.
Figure 5.
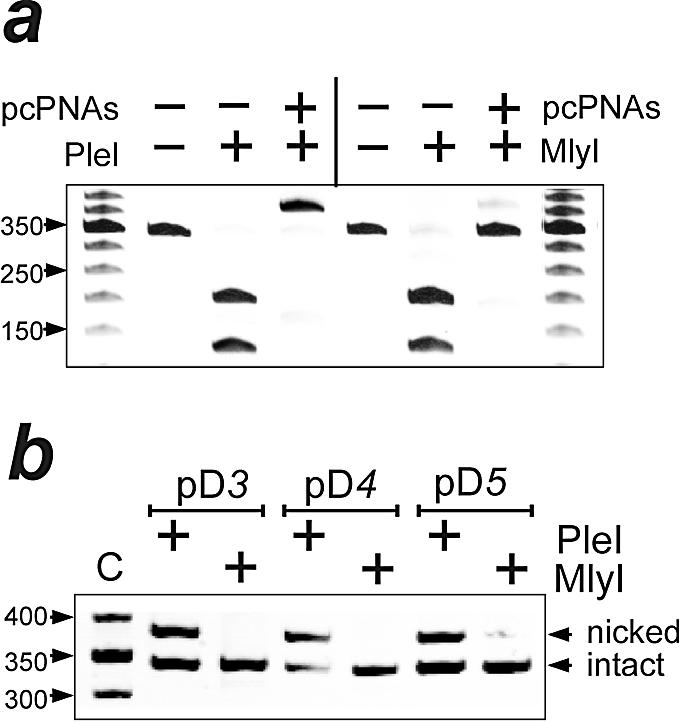
PNA-induced DNA nicking by PleI. Band patterns of PleI- and MlyI-treated 350 bp fragments of pDns are compared following gel electrophoresis in the presence of urea. (a) Binding of pcPNAs 5 bp downstream of the enzyme recognition site (pD5) offers full protection against restriction activity of both enzymes. However, the PleI-treated DNA fragment displays slower electrophoretic mobility, indicating the presence of a nick. (b) PNA-induced nicking activity is also detected for PleI, but not MlyI, in the case of a 3 and 4 bp gap between the two sites (pD3 and pD4). The 50 bp molecular weight ladder is shown in lane C.
The position of the PNA-induced PleI-generated nick was determined by a PCR-based mapping assay (see Materials and Methods for details and Fig. 6 as an example of using this assay with the BbsI enzyme). Using this assay, we located the single-stranded DNA break at a position 4 bp downstream of a site with a degenerated enzyme recognition sequence, 5′-TAATC. Such a pseudo-site was found on all DNA target fragments of the pDn plasmid set ∼20 bp away from the correct recognition sequence. Other pseudo-sites are also efficient in exhibiting the PNA-induced DNA-nicking activity of PleI, which is directed by site-specific DNA looping (43).
Figure 6.
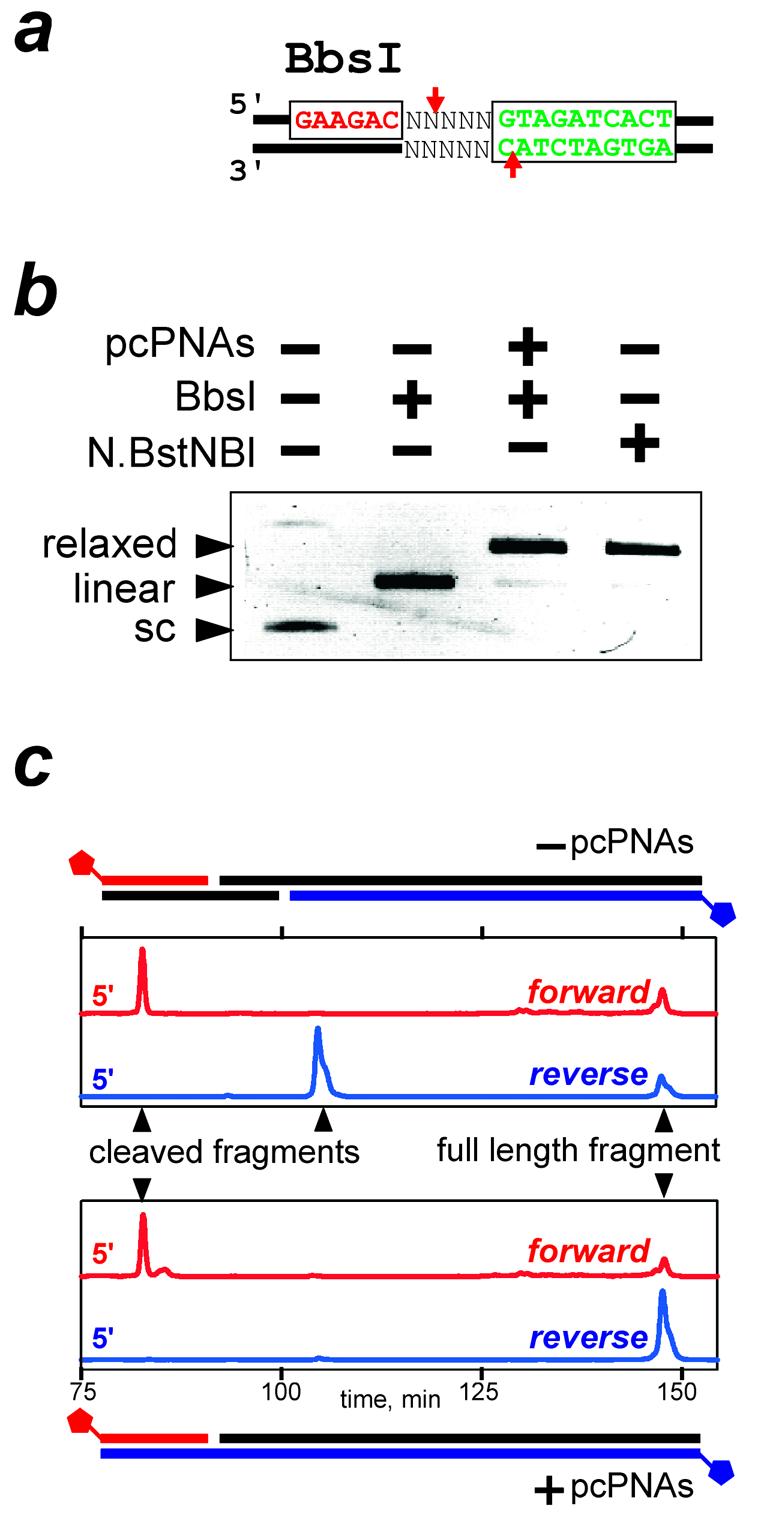
The PNA-induced DNA nicking by BbsI. (a) The DNA construct used in this study carries the BbsI recognition sequence (red) and pcPNA target (green); arrows indicate the enzyme cleavage sites. Note that the PNA-binding site is reversed, as compared with Figure 2. (b) Consistent with the DNA nicking, the PNA-bound supercoiled (sc) target plasmid is transformed into a relaxed (rather than linear) form by BbsI. (c) Mapping of the BbsI activity on fluorescently labeled dsDNA in the absence of pcPNAs (upper panel) and after the pcPNA binding to the DNA target (lower panel), as assessed by the time of DNA migration through the fluorescence detector of the DNA sequencer during denaturing polyacrylamide gel electrophoresis. PCR-generated DNA target fragments (peaks at 148 min) carry a single Cy5 tag on the 5′ end of either the forward (red trace) or reverse (blue trace) strand. In the absence of PNA, the appearance of shorter DNA fragments for both strands (peaks at 83 and 105 min) is consistent with the double-stranded cleavage of target DNA by BbsI. The PNA binding prevents cleavage of the reverse DNA strand, resulting in a nicked DNA product (peak at 83 min).
In the present work, we decided to test one more approach for the pcPNA-directed conversion of DNA cutters into nickases. To this end, we chose the BbsI endonuclease, which recognizes the 5′-GAAGAC sequence and cleaves dsDNA nearby, producing a 4 nt 5′ end overhang (Fig. 6a). As shown schematically in Figure 6a, we targeted the pcPNA pair to dsDNA in such a way that the PNA–DNA complex overlapped with the cleavage point of BbsI on only one (i.e. bottom) DNA strand. We assumed that in this case the PNA– DNA complex would not prevent the protein from binding to the recognition sequence and would allow the enzyme to cleave the top DNA strand, while the cleavage of the bottom DNA strand would be blocked. As a result, a single-stranded break, or nick, could be formed at a designated dsDNA site with the use of a common restriction enzyme. Such a strategy assumes the sequential, two-step mechanism of the dsDNA cleavage by restriction endonuclease, with DNA first being nicked and then further cut by enzyme to finally yield the double-stranded break (49–51).
Indeed, the binding of pcPNAs to the target site within scDNA and its subsequent digestion with the BbsI endonuclease yielded, similar to the action of N.BstNBI nickase, virtually complete conversion of scDNA into a relaxed (nicked) form, with only trace amounts of the linear form (Fig. 6b). Mapping of the PNA-induced DNA-nicking activity of BbsI in linear dsDNA by the PCR assay confirmed the effective enzymatic cleavage exclusively of the top DNA strand and at the expected position only (Fig. 6c). Hence, the site-directed interference of pcPNA–dsDNA complexes with the DNA-cleaving activity of type IIs restriction endonucleases makes it possible to efficiently produce the DNA nicks both near the correct enzyme recognition sites (the BbsI case) and at distantly located pseudo-sites (the PleI case). Artificial nicking systems designed in this way formally have a recognition specificity of ≥16 bp comprised of the enzyme recognition site(s) and the binding site for a pcPNA pair. The enzymes will, however, still create double-stranded cuts at the remaining target sites not protected by PNAs.
General implications for DNA-binding proteins
Table 1 summarizes our results on the interference of pcPNAs with the activity of the type IIs restriction enzymes. Obviously, the observed effects cannot be due to the direct inhibition of the enzymatic activity by pcPNAs. Indeed, unbound pcPNAs have been removed using gel filtration prior to introduction of the restriction enzyme to the reaction mixture. Also, all enzymes shown in Table 1 cleave DNA when the pcPNA target site is sufficiently far from the enzyme-binding/restriction sites, e.g. in pU2 and pD7. Our data can be explained only in terms of the protection of the binding sites and/or cleavage sites of the restriction enzymes by bound pcPNAs. As schematically shown in Figure 1, the pcPNAs binding both downstream (Fig. 1b) and upstream (Fig. 1c) of the enzyme recognition site should interfere with their action. Still, according to our data, the downstream PNA targeting is more effective since in this case the PNA–DNA complexes clash either with the process of enzyme binding or the subsequent DNA cleavage (or both).
Note that upstream pcPNA targeting close to the N.BstNBI recognition site prevents this enzyme from acting, whereas such targeting does not affect PleI and MlyI isoschizomers. In contrast to the N.BstNBI enzyme, the PleI and MlyI enzymes are capable of dimerization in the active state (43,49,52) that could be responsible for their different reaction to the PNA binding (Fig. 1d). However, it is most likely that the difference in the sensitivity of isoschizomeric enzymes to the proximal upstream PNA targeting may reflect the different shape of protein–DNA complexes: nicking enzyme N.BstNBI is somewhat larger than its PleI and MlyI isoschizomers (49,53) and, therefore, N.BstNBI binding to dsDNA could be more susceptible to closely located steric obstacles, such as a PNA–DNA complex.
Our results clearly demonstrate that the protection of dsDNA against cleavage by type IIs restriction enzymes by pcPNAs can proceed at two levels. First, enzyme inhibition occurs when the pcPNA target site is located in close proximity to the enzyme recognition site. Separation of the pcPNAs target site from the enzyme recognition site makes the protein binding possible but still hinders the DNA cleavage at a corresponding position if the PNA target site overlaps with or is adjacent to the cleavage site (see Table 1).
Our data indicate that a substantial distortion of the DNA duplex structure due to the pcPNA invasion is limited to the immediate vicinity of PNA–DNA complexes and does not propagate further than 1 bp from the pcPNA-binding site. This is consistent with our previous observations made for PNA double-duplex and triplex-invasion complexes (41,54).
The results of our study strongly support the notion that pcPNAs can generally be used as highly selective universal blockers and modulators of protein activity on duplex DNA. We believe that our present results, along with the previous findings with restriction/methylation enzymes and RNA polymerases (29,41,43), make it quite plausible to assume that binding to DNA of many other proteins involved in regulation and maintenance of DNA functions, such as transcription factors, histones and histone-like proteins, recombinases, helicases and ligases, can also be modulated by using pcPNAs. As compared with pyrimidine PNAs, which have been used successfully for similar purposes (54–58), pcPNAs represent the DNA-binding ligands that can form complexes with a much larger variety of DNA sequences (with the exception of GC-enriched sites), thus significantly extending the range of dsDNA targets suitable for these applications. Along with the single-base mismatch specificity of pcPNA binding (29,41,42), the single-base accuracy of the pcPNA-induced effects of interference with the DNA-processing enzymes enables the fine-tuning of DNA-recognizing/ modifying functions executed by proteins.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by an HFSP fellowship to E.P., PIF and SPRInG awards (Boston Univ.) to V.V.D., NIH grant GM59173 to M.D.F.K., and EU contract QLK3-CT-2000-00634 to P.E.N.
REFERENCES
- 1.Nielsen P.E. (1991) Sequence-selective DNA recognition by synthetic ligands. Bioconjug. Chem., 2, 1–12. [DOI] [PubMed] [Google Scholar]
- 2.Bailly C. and Henichart,J.P. (1991) DNA recognition by intercalator–minor-groove binder hybrid molecules. Bioconjug. Chem., 2, 379–393. [DOI] [PubMed] [Google Scholar]
- 3.Waring M.J. and Bailly,C. (1994) DNA recognition by intercalators and hybrid molecules. J. Mol. Recognit., 7, 109–122. [DOI] [PubMed] [Google Scholar]
- 4.Frank-Kamenetskii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 5.Sinden R.R., Pearson,C.E., Potaman,V.N. and Ussery,D.W. (1998) DNA: structure and function. Adv. Gen. Biol., 5A, 1–141. [Google Scholar]
- 6.Dervan P.B. and Burli,R.W. (1999) Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol., 3, 688–693. [DOI] [PubMed] [Google Scholar]
- 7.Gowers D.M. and Fox,K.R. (1999) Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res., 27, 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloomfield V.A., Crothers,D.M. and Tinoco,I. (1999) Nucleic Acids: Structures, Properties, and Functions, Chapter 12. University Science Books, Sausalito, CA. [Google Scholar]
- 9.Giovannangeli C. and Helene,C. (2000) Triplex-forming molecules for modulation of DNA information processing. Curr. Opin. Mol. Ther., 2, 288–296. [PubMed] [Google Scholar]
- 10.Winters T.A. (2000) Gene targeted agents: new opportunities for rational drug development. Curr. Opin. Mol. Ther., 2, 670–681. [PubMed] [Google Scholar]
- 11.Fox K.R. (2000) Targeting DNA with triplexes. Curr. Med. Chem., 7, 17–37. [DOI] [PubMed] [Google Scholar]
- 12.Neidle S. (2001) DNA minor-groove recognition by small molecules. Nat. Prod. Rep., 18, 291–309. [DOI] [PubMed] [Google Scholar]
- 13.Chaires J.B. and Waring,M.J., eds (2001) Drug–Nucleic Acid Interactions (Methods in Enzymology, Vol. 340). Academic Press, San Diego, CA. [Google Scholar]
- 14.Reddy B.S., Sharma,S.K. and Lown,J.W. (2001) Recent developments in sequence selective minor groove DNA effectors. Curr. Med. Chem., 8, 475–508. [DOI] [PubMed] [Google Scholar]
- 15.Knauert M.P. and Glazer,P.M. (2001) Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet., 10, 2243–2251. [DOI] [PubMed] [Google Scholar]
- 16.Neidle S. (2001) DNA minor-groove recognition by small molecules. Nat. Prod. Rep., 18, 291–309. [DOI] [PubMed] [Google Scholar]
- 17.Dervan P.B. (2001) Molecular recognition of DNA by small molecules. Bioorg. Med. Chem., 9, 2215–2235. [DOI] [PubMed] [Google Scholar]
- 18.Haq I. (2002) Thermodynamics of drug–DNA interactions. Arch. Biochem. Biophys., 403, 1–15. [DOI] [PubMed] [Google Scholar]
- 19.Boon E.M., Kisko,J.L. and Barton,J.K. (2002) Detection of DNA base mismatches using DNA intercalators. Methods Enzymol., 353, 506–522. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen P.E., Egholm,M., Berg,R.H. and Buchardt,O. (1991) Sequence selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science, 254, 1497–1500. [DOI] [PubMed] [Google Scholar]
- 21.Demidov V., Frank-Kamenetskii,M.D., Egholm,M., Buchardt,O. and Nielsen,P.E. (1993) Sequence selective double strand DNA cleavage by PNA targeting using nuclease S1. Nucleic Acids Res., 21, 2103–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demidov V.V., Potaman,V.N., Frank-Kamenetskii,M.D., Egholm,M., Buchardt,O., Sönnichsen,S.H. and Nielsen,P.E. (1994) Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol., 48, 1310–1313. [DOI] [PubMed] [Google Scholar]
- 23.Egholm M., Christensen,L., Dueholm,K.L., Buchardt,O., Coull,J. and Nielsen,P.E. (1995) Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res., 23, 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demidov V.V., Yavnilovich,M.V., Belotserkovskii,B.P., Frank-Kamenetskii,M.D. and Nielsen,P.E. (1995) Kinetics and mechanism of polyamide (‘peptide’) nucleic acid binding to duplex DNA. Proc. Natl Acad. Sci. USA, 92, 2637–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demidov V.V., Yavnilovich,M.V. and Frank-Kamenetskii,M.D. (1997) Kinetic analysis of specificity of duplex DNA targeting by homopyrimidine PNAs. Biophys. J., 72, 2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn H., Demidov,V.V., Frank-Kamenetskii,M.D. and Nielsen,P.E. (1998) Kinetic sequence discrimination of cationic bis-PNAs upon targeting of double-stranded DNA. Nucleic Acids Res., 26, 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efimov V.A., Choob,M.V., Buryakova,A.A., Kalinkina,A.L. and Chakhmakhcheva,O.G. (1998) Synthesis and evaluation of some properties of chimeric oligomers containing PNA and phosphono-PNA residues. Nucleic Acids Res., 26, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlmann E., Peyman,A., Breipohl,G. and Will,D.W. (1998) PNA: synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed., 37, 2796–2823. [DOI] [PubMed] [Google Scholar]
- 29.Lohse J., Dahl,O. and Nielsen,P.E. (1999) Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl Acad. Sci. USA, 96, 11804–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen P.E. (2001) Peptide nucleic acid targeting of double-stranded DNA. Methods Enzymol., 340, 329–340. [DOI] [PubMed] [Google Scholar]
- 31.Demidov V.V. and Frank-Kamenetskii,M.D. (2001) Sequence-specific targeting of duplex DNA by peptide nucleic acids via triplex strand invasion. Methods, 23, 108–122. [DOI] [PubMed] [Google Scholar]
- 32.Kristensen E. (2002) In vitro and in vivo studies on pharmacokinetics and metabolism of PNA constructs in rodents. In Nielsen,P.E. (ed.), Peptide Nucleic Acids: Methods and Protocols. Humana Press, Totowa, NJ, pp. 259–269. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen P.E. (1999) Applications of peptide nucleic acids. Curr. Opin. Biotechnol., 10, 71–75. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen P.E. and Egholm,M., eds (1999) Peptide Nucleic Acids: Protocols and Applications. Horizon Scientific Press, Wymondham, UK. [Google Scholar]
- 35.Ray A. and Nordén,B. (2000) Peptide nucleic acid (PNA): its medical and biotechnological applications and promise for the future. FASEB J., 14, 1041–1060. [DOI] [PubMed] [Google Scholar]
- 36.Dean D.A. (2000) Peptide nucleic acids: versatile tools for gene therapy strategies. Adv. Drug Deliv. Rev., 44, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demidov V.V. (2001) PD-loop technology: PNA openers at work. Expert Rev. Mol. Diagn., 1, 343–351. [DOI] [PubMed] [Google Scholar]
- 38.Braasch D.A. and Corey,D.R. (2002) Novel antisense and peptide nucleic acid strategies for controlling gene expression. Biochemistry, 41, 4503–4510. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen P.E., ed. (2002) Peptide Nucleic Acids: Methods and Protocols. Humana Press, Totowa, NJ.
- 40.Armitage B.A. (2003) The impact of nucleic acid secondary structure on PNA hybridization. Drug Discov. Today, 8, 222–228. [DOI] [PubMed] [Google Scholar]
- 41.Izvolsky K.I., Demidov,V.V., Nielsen,P.E. and Frank-Kamenetskii,M.D. (2000) Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry, 39, 10908–10913. [DOI] [PubMed] [Google Scholar]
- 42.Demidov V.V., Protozanova,E., Izvolsky,K.I., Price,C., Nielsen,P.E. and Frank-Kamenetskii,M.D. (2002) Kinetics and mechanism of the DNA double helix invasion by pseudocomplementary peptide nucleic acids. Proc. Natl Acad. Sci. USA, 99, 5953–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Protozanova E., Demidov,V.V., Soldatenkov,V., Chasovskikh,S. and Frank-Kamenetskii,M.D. (2002) Tailoring the activity of restriction endonuclease PleI by PNA-induced DNA looping. EMBO Rep., 3, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kutyavin I.V., Rhinehart,R.L., Lukhtanov,E.A., Gorn,V.V., Meyer,R.B.,Jr and Gamper,H.B.,Jr (1996) Oligonucleotides containing 2-aminoadenine and 2-thiothymine act as selectively binding complementary agents. Biochemistry, 35, 11170–11176. [DOI] [PubMed] [Google Scholar]
- 45.Roberts R.J. and Macelis,D. (2001) REBASE—restriction enzymes and methylases. Nucleic Acids Res., 29, 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts R.J., Belfort,M., Bestor,T., Bhagwat,A.S., Bickle,T.A., Bitinaite,J., Blumenthal,R.M., Degtyarev,S.Kh., Dryden,D.T., Dybvig,K., Firman,K., Gromova,E.S., Gumport,R.I., Halford,S.E., Hattman,S., Heitman,J., Hornby,D.P., Janulaitis,A. et al. (2003) A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res., 31, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhn H., Protozanova,E. and Demidov,V.V. (2002) Monitoring of single nicks in duplex DNA by gel electrophoretic mobility-shift assay. Electrophoresis, 23, 2384–2387. [DOI] [PubMed] [Google Scholar]
- 49.Higgins L.S., Besnier,C. and Kong,H. (2001) The nicking endonuclease N.BstNBI is closely related to type IIs restriction endonucleases MlyI and PleI. Nucleic Acids Res., 29, 2492–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potter B.V. and Eckstein,F (1984) Cleavage of phosphorothioate-substituted DNA by restriction endonucleases. J. Biol. Chem., 259, 14243–14248. [PubMed] [Google Scholar]
- 51.Snounou G. and Malcolm,A.D. (1984) Supercoiling and the mechanism of restriction endonucleases. Eur. J. Biochem., 138, 275–280. [DOI] [PubMed] [Google Scholar]
- 52.Bath A.J., Milsom,S.E., Gormley,N.A. and Halford,S.E. (2002) Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem., 277, 4024–4033. [DOI] [PubMed] [Google Scholar]
- 53.Snounou G. and Malcolm,A.D. (1984) Supercoiling and the mechanism of restriction endonucleases. Eur. J. Biochem., 138, 275–280. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen P.E., Egholm,M., Berg,R.H. and Buchardt,O. (1993) Sequence specific inhibition of restriction enzyme cleavage by PNA. Nucleic Acids Res., 21, 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen P.E., Egholm,M. and Buchardt,O. (1994) Sequence-specific transcription arrest by peptide nucleic acid bound to the DNA template strand. Gene, 149, 139–145. [DOI] [PubMed] [Google Scholar]
- 56.Veselkov A.G., Demidov,V.V., Frank-Kamenetskii,M.D. and Nielsen,P.E. (1996) PNA as a rare genome-cutter. Nature, 379, 214. [DOI] [PubMed] [Google Scholar]
- 57.Veselkov A.G., Demidov,V.V., Nielsen,P.E. and Frank-Kamenetskii,M.D. (1996) A new class of genome rare cutters. Nucleic Acids Res., 24, 2483–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhn H., Hu,Y., Frank-Kamenetskii,M.D. and Demidov,V.V. (2003) Artificial site-specific DNA-nicking system based on common restriction enzymes assisted by PNA openers. Biochemistry, 42, 4985–4992. [DOI] [PubMed] [Google Scholar]