Abstract
Polyadenylation plays an important role in RNA degradation in bacterial cells. In Escherichia coli, exoribonucleases, mostly RNase II and polynucleotide phosphorylase, antagonize the synthesis of poly(A) tails by poly(A) polymerase I (PAP I). In accordance with earlier observations showing that only a small fraction of bacterial RNA is polyadenylated, we demonstrate here that ∼10% of rpsO mRNA harbors short oligo(A) tails ranging from one to five A residues in wild-type cells. We also compared the length, frequency and distribution of poly(A) tails at the 3′-end of rpsO transcripts in vivo in the presence and absence of Hfq, a host factor that in vitro stimulates the activity of PAP I, and found that Hfq affects all three parameters. In the hfq+ strain the average length of oligo(A) tails and frequency of polyadenylated transcripts was higher than in the hfq– strain and a smaller proportion of tails was found at the 3′ end of transcripts terminated at the Rho- independent terminator. Our data led us to the conclusion that Hfq is involved in the recognition of 3′ RNA extremities by PAP I.
INTRODUCTION
The occurrence of polyadenylation in prokaryotes was hypothesized in 1974 when several adenylate residues were discovered at the 3′ hydroxyl end of T7 early messengers (1). One year later three separate laboratories described low levels of poly(A) RNA in bacteria (reviewed in 2). In spite of the discovery of a poly(A) polymerase (PAP I) in Escherichia coli in 1962 (3) and extensive studies on the catalytic properties of this protein (3,4), no function was associated with this activity in bacteria until recently.
It is now accepted that PAP I adds poly(A) extensions to the 3′ ends of many kinds of RNA, including mRNA, precursor and mature forms of rRNAs and tRNAs, regulatory RNAs and small RNAs with unknown functions (5–13). In some cases, polyadenylation was demonstrated to facilitate degradation by 3′→5′ exoribonucleases. This is the case for RNA molecules whose 3′ ends are sequestered in stable stem–loop structures which cannot be readily attacked by these enzymes. Addition of a 3′ poly(A) extension offers a toehold for polynucleotide phosphorylase (PNPase) to bind the RNA and initiate its degradation (14–17). In contrast, RNase II, which is also able to remove these tails, fails to pass through secondary structures which protect the messenger (17–19). It is generally accepted that repeated steps of poly(A) addition and exonuclease digestion are needed to overcome the resistance of structured RNAs to exonucleolytic decay (15,20).
On the basis of in vitro experiments, it was proposed that PAP I preferentially recognizes single-stranded RNA extremities (21,22). In contrast, in vivo poly(A) tails have been mapped to both single-stranded and folded 3′ termini distributed at several positions within mRNA molecules, suggesting that PAP I shows little or no sequence specificity for the addition of poly(A) tails (20). Also, PAP I polyadenylates tightly folded mRNA fragments (15,23) and non-coding small RNAs (RNA I, Cop A and Sok) that are mostly degraded by poly(A)-dependent RNases (6,24–26). The discrepancy between PAP I specificity in vitro and in vivo could be explained if cofactors such as unwinding proteins or RNA chaperones were involved in the recognition of 3′ extremities and adenylation by PAP I. Host factor I (Hfq), which stimulates poly(A) tail elongation both in vivo and in vitro (27) and exhibits chaperone activity in vitro (28), may play this role. In the case of Qβ RNA, Hfq affects the folding of the 3′ extremity of the molecule and promotes its replication by the replicase (29). We therefore decided to examine whether Hfq affects the occurrence of poly(A) sites and the extent of polyadenylation in vivo.
Degradation of rpsO mRNA, encoding ribosomal protein S15, is initiated by an RNase E cleavage between the coding sequence and the transcriptional terminator. The body of the messenger is then further degraded by the combined actions of PAP I, PNPase and RNase II. Poly(A) tails were detected at the termini of nascent terminated mRNA and at the end of decay intermediates produced by endonucleolytic and exonucleolytic digestion (20). In this report, we demonstrate that ∼10% of rpsO mRNA 3′ extremities harbor oligo(A) tails ranging from one to five A residues in wild-type cells (4 out of 39 clones). To analyze polyadenylation independently of the exonucleolytic nibbling which removes poly(A) tails and of RNase E, which has been shown to initiate the rapid degradation of the transcript (30) and to cleave poly(A) tails from RNA in vitro (31), we mapped the position and determined the frequency and length of poly(A) tails in rpsO mRNA in cells deficient for RNase II, PNPase and RNase E, and found that Hfq affects all three parameters.
MATERIALS AND METHODS
Strains, plasmid and growth conditions
Strains MG1693, SK5704 (rne-1ts pnp-7 rnb-500ts) and IBPC922 [rne-1ts pnp-7 rnb-500ts hfq1::Ω(KmR, BclI)] and the pFB1 plasmid carrying the rpsO locus have been described (27,32). These strains and their transformed derivatives were grown at 30°C in LB medium supplemented with thymine (40 µg/ml) and ampicillin (100 µg/ml) when required. Aliquots were prepared from exponential growing cells at 30°C (wild-type cells) or shifted to 44°C for 15 min to inactivate the thermosensitive enzymes RNase E and RNase II (RT–PCR analysis). To stop transcription, rifampicin (500 µg/ml) was added at the time of the shift (t0) (northern blotting analysis).
RNA preparation and northern blotting
RNA preparation and northern blotting were as described (27). Cells transformed with pFB1 were used for RT–PCR analysis while non-transformed strains were used for northern blotting experiments. Directed cleavage of RNAs by RNase H was performed as described (17).
RT–PCR cloning
Oligonucleotides used for 3′ end and poly(A) tail detection and characterization were the oligoribonucleotides RIBOLI (5′-pUGGUGGUGGAUCCCGGGAUC-3′) and RIBOFED (5′-pGCUUGGUGGUGGAUCCCG-3′), respectively, and the oligodeoxyribonucleotides DEOXYLI (5′-GATCCCGGGATCCACCACCA-3′) and DEOXYFED (5′-CGGGATCCACCACCAAGC-3′), respectively, as reverse primers. Selective amplification of rpsO cDNA was performed using the oligo 5′-GCAAACGACACCGGTTCTAC as forward primer. Bases in italics represent partial HindIII restriction sites and bases in bold the BamHI site used in the subsequent cloning procedure.
An aliquot of 2.5 µg total RNA was ligated to 100 pmol oligo RIBOLI or RIBOFED using 20 U T4 RNA ligase in the reaction buffer and incubation conditions described in Li et al. (33). After ethanol precipitation, an aliquot of the ligation was annealed to the DEOXY oligo complementary to the linker. cDNA synthesis was performed by incubating the annealing mix with 10 U AMV reverse transcriptase in 50 mM Tris–HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 100 mM DTT and 100 mM each dNTP at 42°C. The cDNA was subjected directly to PCR amplification with the same DEOXY oligo and the rpsO forward primer, under conditions suggested by the manufacturer. PCR products were digested with PstI and BamHI or HindIII and cloned into vector pT3T718U digested with the same enzymes. Polyadenylated clones are counted as those clones that possess extra A residues at the 3′ end of the RNA. Since we cannot distinguish between two encoded A residues at the 3′ end, the addition of one A after an encoded A or two A residues polymerized by PAP I, we assume that we underestimate the number of polyadenylation sites and size of the poly(A) tails.
RESULTS
About 10% of rpsO mRNAs harbor an oligo(A) tail in wild-type cells
It is already known that the relative amount of RNA harboring poly(A) tails is low in bacteria. In order to determine precisely the fraction of rpsO transcripts harboring poly(A) extensions and the length of these tails, we analyzed the 3′ ends of this particular mRNA species in wild-type cells in vivo. To this end, we end-ligated the RIBOLI oligoribonucleotide to RNA extracted from strain MG1693 transformed with pFB1 and performed an RT–PCR using the DEOXYLI and rpsO forward oligonucleotides complementary to the end-ligated oligoribonucleotide and rpsO mRNA, respectively, so as to selectively amplify rpsO mRNA 3′ ends (7). The PCR fragments were cloned and sequenced.
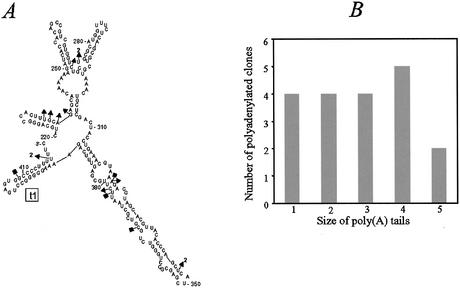
Most of the 39 3′ ends analyzed by this method corresponded to transcripts lacking the transcription terminator hairpin and only four of them (10%) harbored very short oligo(A) extensions ranging from one to three A residues (Fig. 1A, square symbols). Because of the low frequency of poly(A)-tailed rpsO transcripts, we modified the above RT–PCR protocol so as to select for mRNA harboring at least two consecutive A residues at their 3′ ends. For this purpose, we ligated the RIBOFED oligoribonucleotide terminating with 5′-GCUU… to total RNA. The junction between this oligoribonucleotide and RNAs terminating with two A residues created a HindIII restriction site on the RT–PCR products which permitted selective cloning of amplified DNA fragments containing adenylated rpsO mRNAs (12). Strikingly, 34 out of 49 3′ ends analyzed (69%) contained at least two terminal A residues corresponding to encoded nucleotides, strongly reinforcing the notion that the relative frequency of RNA harboring post-transcriptionally added poly(A) extensions is very low. On the other hand, the other 15 ends contained non-encoded A residues in stretches ranging from one to five A residues in length, confirming that oligo(A) tails are very short in the wild-type strain (Fig. 1B). Moreover, the identification of poly(A) extensions both upstream (two clones at position 347) and downstream of hairpin structures (two clones at position 417) suggests that PAP I can polymerize AMP downstream of both single-stranded and double-stranded sequences (Fig. 1A).
Figure 1.
rpsO mRNA polyadenylation in wild-type cells. (A) Predicted structure of the rpsO mRNA 3′ region from position 320 (PstI cloning site) to position 420 corresponding to the 3′ end of the transcript terminated at the Rho-independent transcription terminator t1. Residue 1 is the first residue transcribed from the natural P1 promoter (40). This 201 nt long region was folded with mfold (http://www.bioinfo.rpi.edu/applications/mfold). The locations of rpsO 3′ extremities harboring poly(A) tails in RNAs isolated from wild-type strain MG1693/pFB1 are indicated on this secondary structure. Positions were deduced from RT–PCR analysis without (squares) (4 clones out of 39) and with selection of RNA 3′ extremities harboring at least two A residues at their 3′ ends (triangles) (13 clones in the rpsO sequence out of 49). Number 2 in bold indicates that two clones harboured poly(A) at the same position. Extremities terminated by two encoded A residues are not shown in this diagram. (B) Clones harboring post-transcriptionally added oligo(A) tails shown in (A) are plotted as a function of tail length. Two polyadenylated extremities located at the upstream RNase III site of the rpsO–pnp intercistronic region were included in this analysis.
Hfq affects the frequency of oligoadenylated rpsO transcripts
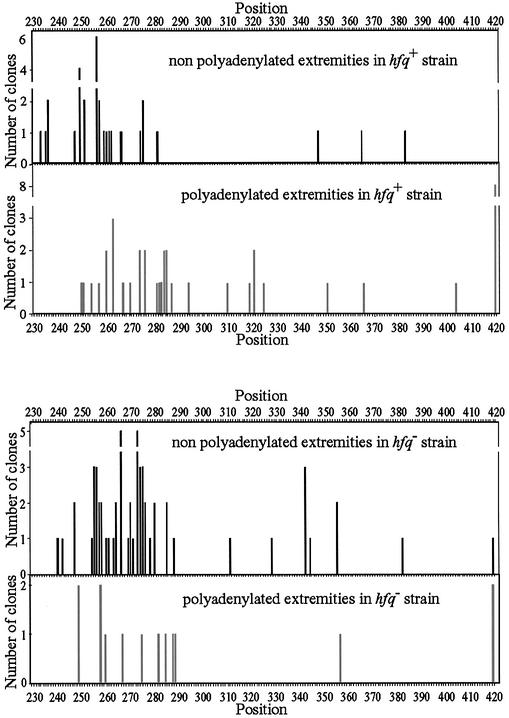
The current model of polyadenylation in bacteria postulates that the length of poly(A) tails results from a dynamic equilibrium between the opposite activities of PAP I and 3′→5′ exonucleases, namely PNPase and RNase II, which shorten or completely remove single-stranded nucleotides from the 3′ end of RNAs. To investigate the role of Hfq in poly(A) synthesis, we compared the frequency of tails in isogenic hfq+ and hfq– strains deficient for PNPase, RNase II and RNase E. RNase E inactivation has the advantage of increasing the intracellular concentration of the rpsO transcript. RT–PCR of rpsO transcripts was performed using an oligoribonucleotide ligated to the 3′ end of total RNA as described above (7). Strikingly, sequencing of the amplified rpsO cDNAs showed that 57% (41 clones out of 72) of 3′ mRNA extremities were adenylated in an hfq+ strain compared to only 20% (14 clones out of 70) of 3′ mRNA extremities in the strain lacking Hfq (Fig. 2). A χ2 test of the data indicates that differences in the number of adenylated and non-adenylated clones in hfq+ and hfq– strains is highly significant (χ2 = 20.4, P << 0.001). We concluded that Hfq enhances the polyadenylation status of RNA transcripts.
Figure 2.
Location of both adenylated and non-adenylated 3′ extremities generated in strains deficient or proficient for Hfq. Distribution of non- adenylated and adenylated rpsO 3′ extremities in SK5704 (rne-1ts pnp-7 rnb-500ts) (upper panels) and IBPC922 (rne-1ts pnp-7 rnb-500ts hfq-1::Ω) (lower panels) 15 min after the temperature shift which inactivates thermosensitive RNase II and RNase E. Both strains carried the pFB1 plasmid overproducing the rpsO mRNA. Positions were deduced from the RT–PCR analysis during which all RNA 3′ extremities harboring (grey) or not (black) non-encoded nucleotides were mapped. The locations and the number of independent clones corresponding to non-adenylated 3′ ends (31 clones from strain SK5704, 56 clones from strain IBPC922) and adenylated 3′ ends (41 clones from strain SK5704, 14 clones from strain IBPC922) are presented according to their positions in the rpsO sequence. Position 420 corresponds to rpsO mRNA terminated at the Rho-independent transcription terminator (Fig. 1A).
Hfq decreases polyadenylation at the terminus of the primary transcript
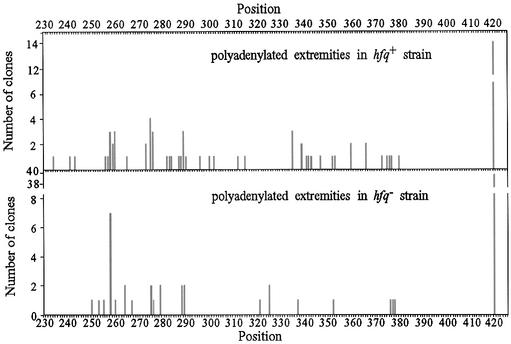
Since the number of oligoadenylated 3′ extremities isolated from the hfq– strain was not sufficient to allow comparison between hfq+ and hfq– strains as to the location and length of poly(A) tails, we repeated the RT–PCR experiment with the oligoribonucleotide allowing the selective cloning of RNAs harboring at least two adjacent A residues at their 3′ end. We isolated 71 and 70 oligoadenylated extremities from the hfq+ and hfq– strains, respectively, in the triple mutant background and found polyadenylation at different sites in the hfq+ and hfq– strains (Fig. 3). In the hfq– strain we observed that 39 out of 70 clones were located at position 420, as compared to 14 out of 71 clones located at the same position in the hfq+ strain. This poly(A) site is isolated from the others and corresponds to the 3′ end of the primary transcript. A χ2 test of the data (χ2 = 14.6, P << 0.001) indicates that this difference in the number of adenylated clones located at the end of the primary transcript in hfq+ and hfq– strains is highly significant.
Figure 3.
Sites of rpsO poly(A) tails isolated from strains deficient or proficient for Hfq. Amplified rpsO cDNA clones from strains SK5704 (rne-1ts pnp-7 rnb-500ts) (upper panel) and IBPC922 (rne-1ts pnp-7 rnb-500ts hfq-1::Ω) (lower panel) containing at least two A residues at the junction with the oligoribonucleotide were cloned selectively. The diagram shows 3′ extremities containing non-encoded riboadenylates on the sequence of the rpsO mRNAs isolated from the hfq+ and hfq– cells (15 min after the temperature shift which inactivates thermosensitive RNase II and RNase E). Position 420 corresponds to rpsO mRNA terminated at the Rho-independent transcription terminator (Fig. 1A).
The large fraction of mRNA–oligo(A) junctions mapping downstream of transcription terminator t1 in the hfq– strain [56% of oligo(A) tails] might reflect a preferential polyadenylation of primary transcripts, harboring a 3′ terminal stem–loop structure, in the absence of Hfq. The fact that this fraction drops to 20% in cells containing Hfq suggests that Hfq may facilitate adenylation of the 3′ end of molecules truncated within the coding sequence of rpsO resulting from both endo- and exonucleolytic cleavage of the primary transcript. Alternatively, Hfq might destabilize the terminated-polyadenylated transcripts in an rne-pnp-rnb independent way.
Oligo(A) tails are shorter when Hfq is inactivated
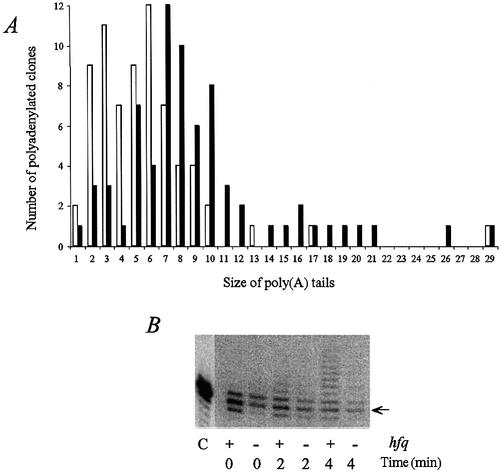
Comparison of oligoadenylated 3′ extremities isolated from hfq+ and hfq– strains (71 and 70 clones, respectively) lacking exoribonucleases and RNase E suggested that Hfq also affects the length of poly(A) tails. Indeed, the number average lengths calculated from data reported in the histogram in Figure 4A are 5.6 ± 0.9 and 9.1 ± 1.2 in the hfq– and the hfq+ strains, respectively. If tails longer than 14, which seldom occur, are excluded, the number average lengths fall slightly to 5.1 ± 0.6 in the hfq– and 7.3 ± 0.6 in the hfq+ strains. These results show that in the absence of RNase II, PNPase and RNase E, tails are longer in an hfq+ strain than in the mutant.
Figure 4.
Length of rpsO poly(A) tails isolated from strains deficient or proficient for Hfq. (A) Clones containing post-transcriptionally added tails isolated from hfq+ (black) and hfq– (open symbol) cells, described in Figure 3, are plotted as a function of poly(A) tail length. (B) Rate of poly(A) tail elongation by PAP I, downstream of the rpsO transcription terminator in SK5704 (rne-1ts pnp-7 rnb-500ts) and IBPC922 (rne-1ts pnp-7 rnb-500ts hfq-1::Ω) visualized on a northern blot. Cells grown at 30°C to an OD650 nm = 0.4 were shifted to 44°C at t0 to inactivate RNase E and RNase II. Rifampicin was added at the time of the shift and total RNAs were prepared from bacteria withdrawn at different times after the shift, indicated in minutes at the bottom of the autoradiograph. Aliquots of 5 µg total RNA extracted from strains containing (+) or not (–) the Hfq protein and run off transcript (c) were treated with RNase H in the presence of the chimeric rpsO oligonucleotide described in Marujo et al. (17) and analyzed on a northern blot, probed for rpsO mRNA. The arrow indicates the normal 3′ terminus of the rpsO primary transcript.
To test whether this difference could reflect the rate of addition of A residues, we analyzed the length of the tails at different times after inactivation of RNase II and RNase E in cells lacking PNPase, at single nucleotide resolution. This was done by northern blotting of RNA fragments corresponding to the 3′ end of rpsO transcripts obtained by RNase H-directed cleavage 50 nt upstream of the transcription termination site (17). Figure 4B shows that the rpsO transcript may gain up to three additional nucleotides at its 3′ extremity (presumably A residues) 4 min after the inactivation of RNase II and RNase E in the strain lacking Hfq compared to seven nucleotides in cells containing Hfq. This elongation suggests that Hfq stimulates the rate of polyadenylation at the end of the primary transcript immediately after the inactivation of RNase E and RNase II in the absence of PNPase. However, we cannot exclude the possibility that Hfq may protect rpsO RNA tailed at different positions from degradation by ribonucleolytic activities different from RNase II, PNPase and RNase E.
DISCUSSION
A minor fraction of E.coli rpsO mRNA is adenylated in wild-type cells
The RT–PCR analysis of RNA isolated from wild-type cells strongly reinforces the idea that the fraction of mRNAs harboring a poly(A) tail is very low in E.coli; indeed ∼10% of rpsO mRNAs are adenylated (4 clones out of 39). More strikingly, no tail longer than five A residues was found even when selecting for adenylated 3′ extremities harboring at least two terminal A residues. Our results strongly indicate that mRNAs (like mature stable RNAs, their precursors and regulator RNAs) exhibit very short oligo(A) tails at their 3′ ends. RNAI, Sok RNA, CopA RNA, MS2 genomic RNA, SraL, CI RNA of phage P4, precursors and mature forms of rRNA and tRNAtrpts harbor 3′ extensions which do not exceed seven A residues (6–8,10,12,13,25,26). No tail longer than 29 A residues was found when RNase E together with RNase II and PNPase, the two 3′→5′ exoribonucleases which shorten poly(A) tails, were inactive and the vast majority of the tails that were sequenced were homopolymeric; only 5 out of the 176 tails analyzed contained a C or a G residue, probably incorporated by PAP I (21,34). It is curious that the long poly(A) tails such as the 45–55 A residue long ones that were observed in the wild-type E.coli strain, containing PAP I and exoribonucleases (9), were not detected in this study. It is possible that only a few preferentially adenylated RNA species (that do not include the rpsO mRNA) can acquire poly(A) tails of such length. On the other hand, one could also imagine that these long tails correspond to a very small fraction of all RNA species that have undergone many successive steps of elongation without being attacked by exoribonucleases. This latter hypothesis may explain why an rpsO mRNA harboring 69 A residues was characterized in an exonuclease-deficient strain by a method selecting for long transcripts (32). However, the probability of isolating RNAs with stretches of A residues this long is presumably very low. On the other hand, our failure to detect the long heteropolymeric tails containing G, C and U residues that have been detected by others at the 3′ end of rpsO transcripts in wild-type cells suggests that these 3′ extensions are only synthesized when PNPase is overproduced in strains lacking PAP I (35).
Hfq affects rpsO mRNA 3′ end formation and its polyadenylation
In a previous work, we showed that Hfq stimulates processive synthesis of poly(A) by PAP I in vitro (27). Here, we demonstrate that Hfq is also a cofactor of poly(A) metabolism in vivo that affects the fraction of polyadenylated molecules, the distribution of adenylation sites and the length of poly(A) tails. In particular, the higher fraction of polyadenylated molecules found in the presence of Hfq suggests that this protein facilitates the recognition of 3′ ends by PAP I (Fig. 2). The scattering of RNA extremities with poly(A) extensions in the presence of Hfq, compared to the preferential adenylation of the primary transcript (position 420) in its absence, suggest that this cofactor improves the polyadenylation of unstructured 3′ extremities, which probably result from cleavages internal to rpsO mRNA. Moreover, these data indicate that the 3′ end of the primary transcript (typical of RNAs terminated at Rho-independent terminators) harboring a U6C sequence downstream of a stable hairpin is very efficiently adenylated by PAP I. This result is consistent with in vitro experiments showing that the addition of a few single-stranded nucleotides facilitates the adenylation by PAP I of RNAs harboring a terminal stable stem–loop (21,22). Reduction of polyadenylation at this site in the presence of Hfq suggests either that PAP I could be titrated by RNA extremities resulting from ribonucleolytic cleavage which become more accessible to PAP I in the presence of Hfq or that this protein masks the U6C terminal motif of the primary transcript. The specificity of eukaryotic Lsm and Sm proteins, belonging to the same family as Hfq, for U-rich motifs and the affinity of Hfq for an oligo(U) polyribonucleotide are consistent with this latter hypothesis (36,37; M. Folichon, in preparation). Altogether, these data indicate that Hfq reduces the preference that PAP I seems to exhibit towards the 3′ ends of primary trancripts released at Rho-independent terminators or harboring 3′ terminal hairpins. On the other hand, it is also conceivable that Hfq protects poly(A) from degradation by 3′→5′ exonucleases remaining in cells deficient for PNPase and RNase II. In this case, the high affinity of Hfq for oligoadenylated RNA may explain why the fraction of polyadenylated molecules is higher when Hfq is present in the cell. Finally, it must be pointed out that the primer anchor method used in these experiments favors the identification of clones containing unstructured mRNA extremities; indeed, ligation of the primer is much less efficient downstream of stable 3′ hairpins (E. Hajnsdorf, unpublished results). This probably explains why only 20% of the oligo(A) tails identified above are located downstream of t1, compared to 52% when reverse transcription was initiated from oligo(dT18) hybridized to poly(A) (20). This implies that the relative number of clones containing RNA extremities mapping downstream of t1 or in the coding sequence likely do not reflect the relative concentrations of full-length and truncated transcripts. However, in spite of this bias, the primer anchor method allows a comparison of the distribution of adenylated and tail-less mRNA extremities and length of tails in cells containing different sets of enzymes.
We confirm here that Hfq stimulates poly(A) addition. The number average length of tails is greater in an exonuclease-deficient strain containing Hfq and longer tails were found in this strain than in the isogenic hfq– strain. The fact that the length of poly(A) was analyzed after a heat shock which inactivates thermosensitive RNase E and RNase II of a PNPase-deficient strain suggests that this difference reflects a stimulation of poly(A) elongation by Hfq occurring after the inactivation of exonucleases. Consistent with this idea, we found that longer poly(A) tails are synthesized in the presence of Hfq after inactivation of RNase II and RNase E than in its absence (Fig. 4B). The fact that PAP I is not among the few polypeptides whose synthesis was reported to be modified by Hfq argues against an effect of this protein on PAP I synthesis (38). These data are in agreement with our earlier observation that rpsO transcripts are more rapidly elongated in the presence of Hfq in vitro (27). However, experiments presented here show that PAP I, when stimulated by Hfq, synthesizes tails whose number average length is 9.1 A residues in vivo (the longest ones are 29 A residues in length), while earlier in vitro experiments demonstrated that PAP I can processively synthesize very long tails of nearly 1000 nt in the presence of Hfq (27). This clearly demonstrates that other factors, including exoribonucleases remaining in cells lacking RNase II and PNPase, prevent the appearance of such tails. On the other hand, the poor expression of PAP I (39), its very low intracellular concentration (34) and the failure to detect such long tails in the cell raises the possibility that the Hfq-promoted processivity of poly(A) elongation may only affect a small fraction of bacterial RNA or become more efficient under particular physiological conditions.
The current model of poly(A) metabolism in E.coli postulates that the tails synthesized by PAP I are removed by 3′→5′ exoribonucleases. We clearly demonstrate here that poly(A) tails resulting from this dynamic equilibrium are rare and short in spite of the fact that all accessible RNA extremities that are not masked by proteins, aminoacylated or engaged in strong secondary structures can probably be adenylated by PAP I assisted by Hfq.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. Kushner for bacterial strains MG1693 and SK5704 and M.E. Winkler for strain TX2808 used for P1 transduction of the hfq1::Ω allele. We are indebted to C. Condon for careful reading of the manuscript and M. Perez who contributed to this work as an undergraduate student. This work was supported by Université Paris VII (Plan Quadriennal), the Centre National de la Recherche Scientifique (UPR 9073), the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires of the Ministère de l’Education Nationale de la Recherche et de la Technologie. M.F. is the recipent of a grant from the M.N.R.T. F.B. is the recipient of fellowships from the Consiglio Nazionale delle Ricerche, Rome and Università degli Studi di Milano, Milan, Italy.
REFERENCES
- 1.Kramer R.A., Rosenberg,M. and Steitz,J.A. (1974) Nucleotide sequences of the 5′ and 3′ termini of bacteriophage T7 early messenger RNAs synthesized in vivo: evidence for sequence specificity in RNA processing. J. Mol. Biol., 89, 767–776. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar N. (1996) Polyadenylation of mRNA in bacteria. Microbiology, 142, 3125–3133. [DOI] [PubMed] [Google Scholar]
- 3.August J.T., Ortiz,P.J. and Hurwitz,J. (1962) Ribonucleic acid-dependent ribonucleotide incorporation. I: Purification and properties of the enzyme. J. Biol. Chem., 237, 3786–3793. [PubMed] [Google Scholar]
- 4.Edmonds M. (1982) Poly(A) adding enzymes. In Boyer,P. (ed.), The Enzymes, Vol. XV, Part B. Academic Press, New York, NY, pp. 217–244. [Google Scholar]
- 5.Cao G.-J. and Sarkar,N. (1992) Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc. Natl Acad. Sci. USA, 89, 7546–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu F., Lin-Chao,S. and Cohen,S.N. (1993) The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc. Natl Acad. Sci. USA, 90, 6756–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Pandit,S. and Deutscher,M.P. (1998) Polyadenylation of stable RNAs in vivo. Proc. Natl Acad. Sci. USA, 95, 12158–12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., Reimers,S., Pandit,S. and Deutscher,M.P. (2002) RNA quality control: degradation of defective transfer RNA. EMBO J., 21, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Hara E.B., Chekanova,J.A., Ingle,C.A., Kushner,Z.R., Peters,E. and Kushner,S.R. (1995) Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 1807–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argaman L., Hershberg,R., Vogel,J., Bejerano,G., Wagner,E.G.H., Margalit,H. and Altuvia,S. (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol., 11, 941–950. [DOI] [PubMed] [Google Scholar]
- 11.Dreyfus M. and Régnier,P. (2002) The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell, 111, 611–613. [DOI] [PubMed] [Google Scholar]
- 12.Briani F., Del Vecchio,E., Migliorini,D., Hajnsdorf,E., Régnier,P., Ghisotti,D. and Dehò,G. (2002) RNase E and polyadenyl polymerase I are involved in maturation of CI RNA, the P4 phage immunity factor. J. Mol. Biol., 318, 321–331. [DOI] [PubMed] [Google Scholar]
- 13.van Meerten D., Zelwer,M., Régnier,P. and van Duin,J. (1999) In vivo oligo(A) insertions in phage MS2: role of Escherichia coli poly(A) polymerase. Nucleic Acids Res., 27, 3891–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu F. and Cohen,S.N. (1995) RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature, 374, 180–183. [DOI] [PubMed] [Google Scholar]
- 15.Coburn G.A. and Mackie,G.A. (1998) Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol., 279, 1061–1074. [DOI] [PubMed] [Google Scholar]
- 16.Hajnsdorf E., Steier,O., Coscoy,L., Teysset,L. and Régnier,P. (1994) Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams-rnb-pnp mutant. EMBO J., 13, 3368–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marujo P.E., Hajnsdorf,E., Le Derout,J., Andrade,R., Arraiano,C.M. and Régnier,P. (2000) RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA, 6, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coburn G.A. and Mackie,G.A. (1996) Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′ exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem., 271, 15776–15781. [DOI] [PubMed] [Google Scholar]
- 19.Blum E., Carpousis,A.J. and Higgins,C.F. (1999) Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem., 274, 4009–4016. [DOI] [PubMed] [Google Scholar]
- 20.Haugel-Nielsen J., Hajnsdorf,E. and Régnier,P. (1996) The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J., 15, 3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 21.Yehudai-Resheff S. and Schuster,G. (2000) Characterization of the E.coli poly(A) polymerase: nucleotide specificity, RNA binding affinities and RNA structure dependence. Nucleic Acids Res., 28, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y. and Cohen,S.N. (2000) Unpaired terminal nucleotides and 5′ monophosphorylation govern 3′ polyadenylation by Escherichia coli poly(A) polymerase I. Proc. Natl Acad. Sci. USA, 97, 6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajnsdorf E. and Régnier,P. (1999) E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol., 286, 1033–1043. [DOI] [PubMed] [Google Scholar]
- 24.He L., Söderbom,F., Wagner,E.G.H., Binnie,U., Binns,N. and Masters,M. (1993) PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol., 9, 1131–1142. [DOI] [PubMed] [Google Scholar]
- 25.Mikkelsen N.D. and Gerdes,K. (1997) Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol. Microbiol., 26, 311–320. [DOI] [PubMed] [Google Scholar]
- 26.Söderbom F., Binnie,U., Masters,M. and Wagner,E.G.H. (1997) Regulation of plasmid R1 replication: pcnB and RNase E expedite the decay of the antisense RNA, copA. Mol. Microbiol., 26, 493–504. [DOI] [PubMed] [Google Scholar]
- 27.Hajnsdorf E. and Régnier,P. (2000) Host factor HFq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl Acad. Sci. USA, 97, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moll I., Leitsch,D., Steinhauser,T. and Bläsi,U. (2003) RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep., 4, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuppli D., Miranda,G., Tsui,H.-C.T., Winckler,M.E., Sogo,J.M. and Weber,H. (1997) Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 10239–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Régnier P. and Hajnsdorf,E. (1991) Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J. Mol. Biol., 217, 283–292. [DOI] [PubMed] [Google Scholar]
- 31.Walsh A.P., Tock,M.H., Mallen,M.H., Kaberdin,V.R., von Gabain,A. and McDowall,K.J. (2001) Cleavage of poly(A) tails on the 3′-end of RNA by ribonuclease E of Escherichia coli. Nucleic Acids Res., 29, 1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajnsdorf E., Braun,F., Haugel-Nielsen,J. and Régnier,P. (1995) Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc. Natl Acad. Sci. USA, 92, 3973–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Pandit,S. and Deutscher,M.P. (1998) 3′ Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sippel A.E. (1973) Purification and characterization of adenosine triphosphate:ribonucleic acid adenyltransferase from Escherichia coli. Eur. J. Biochem., 37, 31–40. [DOI] [PubMed] [Google Scholar]
- 35.Mohanty B.K. and Kushner,S.R. (2000) Polynucleotide phosphorylase functions both as a 3′→5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arluison V., Derreumaux,P., Allemand,F., Folichon,M., Hajnsdorf,E. and Régnier,P. (2002) Structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol., 320, 705–712. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher M.A., Rearson,R.F., Moller,T., Valentin-Hansen,P. and Brennan,R.G. (2002) Structures of the pleiotropic translational regulator HFq and an HFq-RNA complex: a bacterial Sm-like protein. EMBO J., 21, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonnleitner E., Moll,I. and Bläsi,U. (2002) Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology, 148, 883–891. [DOI] [PubMed] [Google Scholar]
- 39.Binns N. and Masters,M. (2002) Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol. Microbiol., 44, 1287–1298. [DOI] [PubMed] [Google Scholar]
- 40.Régnier P. and Portier,C. (1986) Initiation, attenuation and RNase III processing of transcripts from Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol., 187, 23–352. [DOI] [PubMed] [Google Scholar]