Abstract
The Escherichia coli O26 serogroup includes important food-borne pathogens associated with human and animal diarrheal disease. Current typing methods have revealed great genetic heterogeneity within the O26 group; the data are often inconsistent and focus only on verotoxin (VT)-positive O26 isolates. To improve current understanding of diversity within this serogroup, the genomic relatedness of VT-positive and -negative O26 strains was assessed by comparative genomic indexing. Our results clearly demonstrate that irrespective of virulence characteristics and pathotype designation, the O26 strains show greater genomic similarity to each other than to any other strain included in this study. Our data suggest that enteropathogenic and VT-expressing E. coli O26 strains represent the same clonal lineage and that VT-expressing E. coli O26 strains have gained additional virulence characteristics. Using this approach, we established the core genes which are central to the E. coli species and identified regions of variation from the E. coli K-12 chromosomal backbone.
Escherichia coli is a normal part of the microflora of the gastrointestinal tract that can be beneficial to the host. However, certain E. coli types have been associated with disease in humans and animals. E. coli strains associated with diarrheal disease have been subdivided into six different categories or pathotypes based on clinical features, virulence, and adherence properties (24). E. coli strains of serogroup O26 have been associated with major outbreaks of infant diarrhea since 1951 and give rise to watery diarrhea, vomiting, and fever in infants (24). Strains from this serotype have also been shown to harbor the Shiga toxin- or verotoxin (VT)-producing lambda prophage (37) which causes hemorrhagic colitis and hemolytic uremic syndrome infection: such strains are called enterohaemorrhagic E. coli (EHEC). Verotoxin-producing E. coli (VTEC) of the O26 serotype are the most common non-O157 cause of hemolytic-uremic syndrome infection in Germany and other European countries (15, 28, 38), but cases have also been reported worldwide (16, 19, 28). Interestingly, in Brazil, O26 strains have only been implicated in diarrhea in children but not associated with hemolytic-uremic syndrome (29).
Strains of this serogroup are also of veterinary importance, as they have been isolated from a variety of animals, including healthy cattle and pigs (5, 20) as well as diarrheic calves (17), diarrheic lamb and goats (8), and mastitic cattle (9). Therefore, animals not only act as an important reservoir for O26 strains but may also be infected by these pathogens. In contrast, E. coli O157 does not cause disease in animals.
The clonal diversity of isolates from different food products as well as from human and animal subjects has been studied to determine the reservoirs and routes of transmission of O26 strains through the food chain. However, most research has concentrated only on VT-positive O26 strains, reflecting the seriousness of hemolytic-uremic syndrome infection and the rising numbers of these isolates. These studies imply considerable genetic heterogeneity within the O26 serogroup, but the data from different typing methods (multilocus enzyme electrophoresis, random amplification of polymorphic DNA, and pulsed-field gel electrophoresis) do not allow direct comparison (29, 33, 42, 44, 45). Our aim was to assess the clonal diversity of the O26 serogroup, including both VT-positive and -negative strains, using an approach we call comparative genomic indexing (CGI). In this study we used an E. coli K-12 microarray as the baseline for determining the genomic variation between O26 isolates.
Comparisons of the genome sequences of the pathogenic O157 strain EDL933 with the laboratory E. coli K-12 strain MG1655 revealed that they have a common K-12 chromosomal backbone punctuated by unique genomic regions reflecting deletion and insertion events (30). It was expected that CGI would allow the definition of the core genes common to pathogenic strains and the commensal E. coli K-12 and also identify regions of differences between these strains.
Strains chosen in this study were mainly of veterinary origin and were isolated in the United Kingdom, where these zoonotic pathogens are found in the food chain and associated with human and animal disease. It was known at the outset that half of the O26 strains were verotoxin positive, while the other half were not. Additional strains whose toxin status was known were randomly chosen from serotypes associated with EPEC and enterohemorrhagic E. coli (EHEC) infection (O157, O86, O55, O111, O126, and untypeable); a control commensal strain (O29) was also examined.
MATERIALS AND METHODS
Bacterial strains, identification, and growth condition.
The bacterial strains used in this study are shown in Table 1. These include clinical and field isolates of E. coli from diseased and healthy animals, typed strains from the Veterinary Laboratories Agency reference laboratory and genetically characterized E. coli K-12 derivatives. Verocytotoxin and cytolethal distending toxins were detected by standard protocols (18). Strain EC720/98 was designated untypeable after failing to agglutinate any of 164 different serogroup-specific antisera (VLA Diagnostics Unit).
TABLE 1.
E. coli strains used in this study and analysis of their virulence characteristicsa
| Strain no. | Serotype | Detailsb (reference) | Animal species | Clinical symptom(s) | Toxin produced | Virulence determinant(s) | Designationc |
|---|---|---|---|---|---|---|---|
| E33167 | O26:K60 | PHLS, London | Human | Not known | VT | hlyA, eae β | Atypical EHEC |
| EC537/01 | O26:K60 | Winswick | Bovine | Not known | VT | hlyA, eae β | Atypical EHEC |
| EC1840/00 | O26:K60 | Barnow Gurney (Langford) | Bovine | Diarrhea | VT | hlyA, eae β | Atypical EHEC |
| EC172/00 | O26:K60 | Langford | Bovine | Collapsed, dehydrated | VT | hlyA, eae β | Atypical EHEC |
| EC1722/00 | O26:K60 | Langford | Bovine | Diarrhea | VT | hlyA, eae β | Atypical EHEC |
| EC183/99 | O26:K60 | Preston | Bovine | Diarrhea | VT | hlyA, eae β | Atypical EPEC |
| EC681/99 | O26:K60 | Penrith | Ovine | Routine survey | VT | hlyA, eae β | Atypical EPEC |
| EC335/98 | O26:K60 | Sweden | Ovine | Diarrhea | None | hlyA, eae β | Atypical EPEC |
| EC38/99 | O26:K60 | Penrith | Bovine | Diarrhea | None | eae β | Atypical EPEC |
| EC622/99 | O26:K60 | Winchester | Porcine | Routine survey | None | eae β | Atypical EPEC |
| EC225/00 | O26:K60 | Powys (Wales) | Bovine | Dysentry | None | hlyA, eae β | Atypical EPEC |
| EC459/01 | O26:K60 | Haverford West (Carmarthen) | Bovine | Mastitis | None | eae β | Atypical EPEC |
| EC1725/00 | O26:K60 | Kirby Moorside (Thirsk) | Bovine | Diarrhea, runny nose | None | eae β | Atypical EPEC |
| EC1388/00 | O26:K60 | Aberyswyth | Ovine | Nephrosis | None | eae β | Atypical EPEC |
| EC720/98 | Untypeable | Sutton Bonington | Bovine | Healthy | VT | hlyA, eae β | Atypical EHEC |
| EC370/98 | O86:K61 | Inverness | Avian (migrating) | Dead | CLDT | eae γ | Atypical EPEC |
| EC558/00 | O86:K61 | Inverness | Avian (migrating) | Dead | CLDT | eae γ, eae δ | Atypical EPEC |
| EC157/99 | O157:H7 | Thirsk | Bovine | 0157 tracebackd | VT | hlyA, eae γ | Typical EHEC |
| EDL933 | O157:H7 | Sequenced strain (29) | VT | hlyA, eae γ | Typical EHEC | ||
| E140065 | O157:H7 | PHLS, London | Human | Not known | VT | hlyA, eae γ | Typical EHEC |
| E139229 | O157:H7 | PHLS, London | Human | Not known | VT | hlyA, eae γ | Typical EHEC |
| EC1727/00 | O55:K+ | Langford | Bovine | Diarrhoea | None | None | Neither |
| SO55 | O55:K+ | O antigen strain collection, Weybridge | None | hlyA, eae β | Atypical EPEC | ||
| SO111 | O111:K+ | O antigen strain collection, Weybridge | None | bfp, eae β, eae δ | Typical EPEC | ||
| SO126 | O126:K+ | O antigen strain collection, Weybridge | None | eae β | Atypical EPEC | ||
| SO29 | O29:K+ | O antigen strain collection, Weybridge | None | None | Neither | ||
| GC4468 | K-12 Δ(argF-lac)169 IN(rrnD-rrnE)1 rpsL179 | Lab strain (31) | |||||
| KL773 | K-12 Δ(gpt-lac)5 Δ(galK-bioD)76 rpsL171 metE70 | Lab strain (6) | |||||
| MG1655 | K-12 | Sequenced strain (4) |
The serotype, geographic location, animal origin (where applicable), and disease (where applicable) are shown. All strains with the EC or S prefix were obtained from the Veterinary Laboratories Agency-Weybridge diagnostic laboratory. For EC strains, the last two numbers denote the year the isolate was collected; the S strains were from the Veternary Laboratories Agency-Weybridge reference laboratory collection. Virulence determinants which were successfully amplified by PCR are shown for each strain.
PHLS, Public Health Laboratory Service, Colindale, London, United Kingdom.
Atypical EHECs do not possess either the γ-intimin or hemolysin A or are not VT positive. Atypical EPECs do not possess either the β, δ, or α intimin or bundle forming pili genes but may possess VT or hemolysin genes.
isolate from a cattle farm which was the source of an O157 human outbreak.
API 20E miniaturized biochemical test strips (Biomerieux) were used for the differential identification of Enterobacteriaceae. They were inoculated with bacterial suspensions, incubated at 37οC for 24 h, and read according to the manufacturer's instructions.
DNA isolation and PCR amplification of protein coding sequences.
For preparation of genomic DNA, cells were grown overnight in Luria-Bertani (LB) broth at 37°C, and DNA was isolated with the Qiagen DNeasy Tissue kit (no. 69504; Qiagen). Oligonucleotide primers and PCR conditions used for amplification of the eae γ, eae β, eae α, and eae δ genes were essentially as described by McGraw et al. (26) and Adu-Bobie et al. (1). PCR amplification of the bfp gene was performed with primers (with restriction enzyme sites underlined) bfpAF (CGGCGGATTCTGGTTTCTAAAATCATGAATAAG) and bfpAR (CGGCAAGCTTCTTCATAAAATATGTAACTTTAT). PCR amplification of the hlyA gene was performed with primers hlyCF (GCTATGGGCCTGTTCTCCTCTG) and hlyAR (TGTCTTGCGTCATATCCATTCTCA).
E. coli microarray construction.
The microarrays used in this study featured 4,262 of the 4,279 protein-coding sequence (CDS) identified in E. coli K-12 strain MG1655 (http://www. ncbi.nlm.nih.gov). Entire CDS were amplified with specific primer pairs (Sigma-Genosys) with some minor modifications. PCRs were performed in a total volume of 100 μl with 40 ng of E. coli MG1655 chromosomal DNA, 60 pmol of each primer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate mix, and HotStart Taq DNA polymerase (Qiagen). PCR amplifications were performed with an MWG RoboAmp 4200 liquid handling robot for 30 cycles for 1 min at 94°C, 0.5 min at 60°C, and 3 min at 72°C, following an initial enzyme activation step at 95°C for 15 min.
Agarose gel electrophoresis was used to perform quality control on all PCR products Oligonucleotides were removed from the PCR mix by isopropanol precipitation. DNA was resuspended in 40 μl of spotting solution containing 50% dimethyl sulfoxide and 0.3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). PCR products were spotted onto to gamma amino propylsilane-coated (GAP) slides (Corning) with a Stanford arrayer (40). The DNA was UV cross-linked to slides with a Stratalinker at 300 mJ (Stratagene). Subsequently, the slides were washed in a 95°C water bath for 2 min and in 95% ethanol for 1 min and dried by centrifugation at 185 × g prior to storage at room temperature.
Probe preparation and hybridization.
For each microarray hybridization reaction, genomic DNAs from a reference strain (E. coli K-12 MG1655) and a test strain were fluorescently labeled with indodicarbocyanine and indocarbocyanine, respectively, with the protocol of DeRisi (http://www.microarrays.org/pdfs/GenomicDNALabel_A.pdf). The genomic DNA was not sheared or digested with restriction enzymes prior to labeling. Labeled reference and test DNAs were combined in a 15-μl hybridization solution (3× SSC, 25 mM HEPES [pH 7.0], 1.87 μg of E. coli tRNA per μl, 0.2% sodium dodecyl sulfate, and 5× Denhardt's solution) and added to a microarray slide. Hybridizations were performed for 14 to 18 h at 63°C. The slides were then washed in 2× SSC-0.1% sodium dodecyl sulfate at 65°C for 5 min, followed by 1× SSC at room temperature for 5 min, and finally in 0.2× SSC at room temperature for 5 min. They were dried by centrifugation at 185 × g for 5 min. At least two hybridization reactions were performed for each test strain.
Microarray data analysis.
The processed slides were scanned with a GenePix 4000A scanner (Axon Instruments, Inc.). Fluorescent spots and the local background intensities were quantified with Genepix Pro software (Axon Instruments, Inc.). The data were filtered so that spots with a reference signal lower than the background plus 2 standard deviations of the background were discarded. Signal intensities were corrected by subtracting the local background, then the red/green (indodicarbocyanine/indocarbocyanine) ratios were calculated. To compensate for unequal dye incorporation, data centering was performed by bringing the median Ln(red/green) for each block to 0 (one block being defined as the group of spots printed by the same pin) with the following equation: ln(Ti) = ln(Ri/Gi) − c, where T is the centered ratio, i is the gene index, R and G are the red and green intensities, respectively, and c is the 50th percentile of all red/green ratios. Centered data (i.e., normalized) from all strains were used for all subsequent analyses and subjected to average-linkage hierarchical clustering with the Pearson correlation coefficient (13), in the GeneSpring microarray analysis software version 5.0 (Silicon Genetics). Only those CDS or genes with a good reference signal (or reading) in at least 22 of the 26 test strains upon hybridization to the microarray were considered for clustering. Approximately 70% of the CDS passed this test, leaving a total of 3,039 CDS in the data set. The CDS data set is available as supplemental data at http://www.defra.gov.uk/corporate/vla/aboutus/publicat.htm.
RESULTS
Validation of E. coli microarrays.
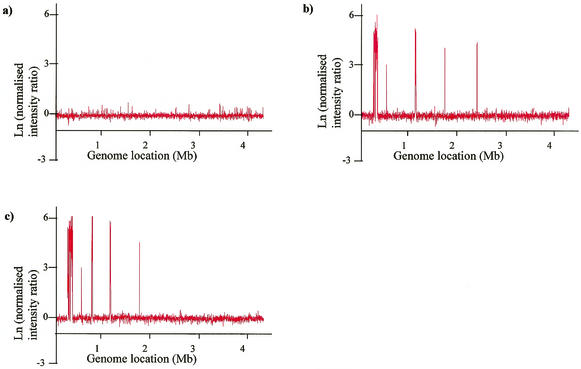
A whole-genome E. coli K-12 microarray based on the sequence of MG1655 was constructed and validated with two well-characterized deletion derivatives of E. coli K-12, GC4468 and KL773 (Table 1). Figure 1 shows the hybridization intensity ratio for each CDS in MG1655, GC4468, and KL773 following hybridization with genomic DNA and normalization of data (see Materials and Methods). The intensity ratio for all CDS in MG1655 was around 1 (i.e., natural log [ln] value around 0; Fig. 1a), confirming that test and control DNA had hybridized equally well to all CDS, regardless of which dye had been used for labeling.
FIG. 1.
Validation of CGI approach by identification of known deletions. The ln(normalized intensity ratio) at 635 nm versus 532 nm from microarray hybridization is shown for each CDS of three E. coli K-12 control strains against MG1655. The strains were (a) MG1655, (b) GC4468, and (c) KL773.
The intensity ratios for strains GC4468 and KL773 clearly identified regions of deleted CDS. These regions were seen as peaks, with an ln(red/green) value equal to or greater than 2, indicating the presence of a particular CDS in the control strain but its absence in the test strain (Fig. 1b and c). This cutoff enabled 100% of the deleted CDS to be detected in these strains with no false-positives. Based on these results, a cutoff of ln(red/green) = 2 was chosen to define the absence of a CDS. In this study it is recognized that with this cutoff value, certain divergent genes which may be present but did not hybridize under these conditions will be defined as absent. However, this criterion enabled us to identify 95.1% of the CDS which were known to be absent from EDL933 (30) following hybridization of genomic DNA from E. coli O157 EDL933 to the K-12 microarray, with no false-positives.
Categorization of strains with conventional virulence markers.
Our collection of E. coli strains whose toxin status was already known were further characterized with respect to selected virulence determinants: bfpA, hlyA, eae γ, eae β, eae α, and eae δ (Table 1). Toxin testing had already shown that half of the O26 strains and all of the O157s were VT positive, a diagnostic feature of EHEC strains. However, typical EHECs also possess the plasmid-encoded hlyA gene (27) and the gamma intimin (γ eae) gene (1). All four O157 strains fitted this criterion, but the VT-positive O26 strains possessed the β eae and not the γ eae gene and were designated atypical EHECs. The VT-negative O26 strains also possessed the β eae gene, a typical EPEC feature, but did not possess the plasmid-encoded bfp gene typical of EPEC strains (18).
Two VT-negative strains also possessed the EHEC-associated hlyA gene. Therefore, these strains were also designated atypical EPECs. In fact, only one strain in this study showed typical EPEC features and belonged to the O111 serogroup. All other strains were characterized as either atypical EPECs (O55, O126, and O86) or neither EPEC or EHEC (O55 and O29) based on their virulence characteristics. Two O86 strains were designated atypical EPECs because they were positive for the cytolethal distending toxin, a feature common to many EPEC strains, but showed the presence of the γ eae gene associated with EHECs.
In a separate study, this variant intimin was further characterized (23). An untypeable VT-positive strain (EC720/98) of bovine origin also included in the study, showed virulence characteristics similar to those of other VT-positive O26 strains and was designated an atypical EHEC. In summary, our pathogenic E. coli strains were a heterogeneous group of organisms with respect to their virulence characteristics. This heterogeneity has been observed previously in clinical isolates (27, 28), although virulence characteristics are still routinely used for pathotype determination.
Comparative genomic indexing of E. coli strains.
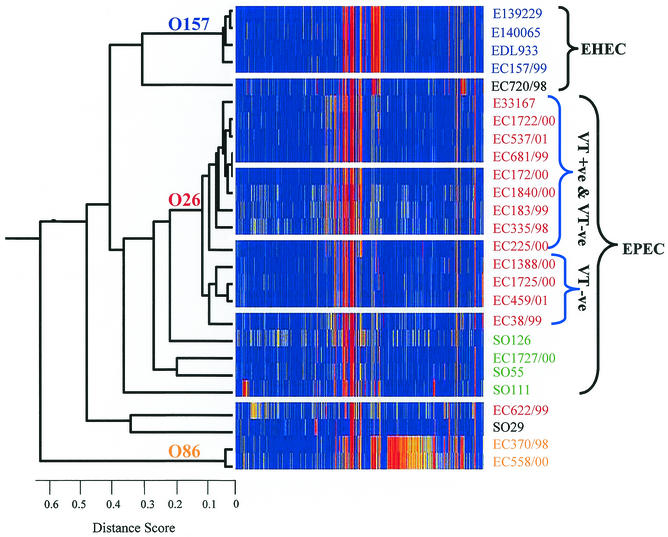
Microarrays were used to compare the relatedness of the 26 E. coli strains by CGI (Table 1). Following microarray hybridization, the scanned data was centered (i.e., normalized; see Materials and Methods), and the presence or absence of genes was determined (see supplementary data). In the first dimension or vertical axis of the hierarchical clustering, the relationship among genomes of each strain was assessed with the Pearson coeffecient correlation pairwise similarity function, where the linkage distance between strains is represented by branch lengths or distance score in the resulting hierarchical cluster (13). The higher the correlation between strains, the smaller the distance score (Fig. 2). The second dimension of the hierarchical clustering was used to group genes with a similar profile for each strain along the horizontal axis, also with the Pearson correlation coefficient. This enabled the clear identification of groups of genes absent in the majority of E. coli field strains as a central cluster (shown in red in Fig. 2; branches for this clustering have not been shown).
FIG. 2.
Genomic index of 26 pathogenic E. coli strains. An average linkage hierarchical clustering of the E. coli strains was compiled in GeneSpring version 5.0 from CGI data with the Pearson coefficient correlation. CDS present (ln[red/green] = 1) are shown in blue, absent CDS (ln [red/green] ≥ 2) are shown in red, and CDS with ln(red/green) between 1.5 and 2.0 are shown in yellow. Strains of the O26 serotype are labeled in red; other strains in the EPEC cluster are labeled in green, O157 strains are labeled in blue, and O86 strains are labeled in orange. The scale for distance score (ds) is also shown.
Hierarchical clustering showed that 13 of the 14 O26 strains included in this study formed a discrete cluster within a major group, which was designated the EPEC cluster (Fig. 2). Other strains within this cluster included strains of serotypes O111, O126, and O55. The distance score of the O26 cluster (0.114) indicates that strains of this serogroup have a greater genomic relatedness to each other than to any other strain included in the study, despite differences in their virulence and verotoxin characteristics (Table 1). Within the O26 cluster, a smaller subcluster of nine strains (distance score = 0.086) was also present. It included all the VT-positive O26 strains and two VT-negative strains (EC335/98 and EC225/00) which possessed the EHEC-associated hlyA gene (Table 1). Of the remaining VT-negative O26 strains, four formed a subcluster (distance score = 0.092). One VT-negative strain (EC622/99) fell outside the O26 cluster, showing greater genomic similarity to an O29 strain; we subsequently reconfirmed this strain to be of the O26 serotype.
In general, VT-negative and VT-positive O26 E. coli strains (with the exception of EC622/99) were found to be 92.7% similar, and on average 7.3% of the total number of CDS present in MG1655 were absent from this group (Table 2). More than 94% of the absent CDS were conserved within each VT group and between 87% and 94% were conserved between the VT-negative and VT-positive strains. In fact, most large regions of genes absent from the K-12 chromosomal backbone were commonly missing from the majority of E. coli strains included in this study, and not only strains of the O26 serogroup (Table 3). One region was found to differentiate the VT-positive from the VT-negative strains; all VT-positive strains lacked a 5.2-kb region (yagP-yagT) which was present in all VT-negative strains (see supplemental data at http://www.defra.gov.uk/corporate/vla/aboutus/publicat.htm). This could prove to be a useful diagnostic feature.
TABLE 2.
Summary of E. coli genomic indexing dataa
| Serotype | Toxinb | CDS absent (%) |
|---|---|---|
| O26:K60 | VT | 7.2 (0.9) |
| O26:K60 | None | 7.3 (0.6) |
| O86:K61 | CLDT | 19.5 (0.7) |
| O157:H7 | VT | 9.8 (0.2) |
| O55:K+ | None | 5.9 (0.8) |
| Untypeable | VT | 11 |
| O111:K+ | None | 10 |
| O126:K+ | None | 7.4 |
| O29:K+ | None | 5.1 |
| K-12 ΔgalK-bioD | N/A | 2.6 |
| K-12 Δgpt-lac | N/A | 3.2 |
| K-12 | N/A | 0 |
The percentage of CDS absent in the test strains, with respect to the control MG1655 strain, was assessed by microarray analyses. The average value of the percentage of CDS missing for each serogroup (where n > 1) was calculated, and the standard deviation is given in parentheses.
CLDT, cytolethal distending toxin; N/A, not applicable.
TABLE 3.
Regions of the K-12 chromosomal backbone with more than 10 consecutive genes absent from at least two strainsa
| Start | End | Strains which lack this region |
|---|---|---|
| b0245 | b0254 (perR) | E139229; E140065; E33167; EC1388; EC172; EC1722; EC1725; EC1727; EC1840; EC225; EC335; EC370; EC459; EC622; EDL933; SO29; SO55 |
| b0245 | b0255 (yi91a) | EC38; EC720; SO111; EC157; EC681∗; EC558∗ |
| b0264 (insB_2) | b0280 (yagN) | E140065; EC720; EC157∗; EC370∗ |
| b0274 (insB_3) | b0286 (yagT) | EC370; EC558 |
| b0504 (ybbS) | b0521 (arcC) | EC370; EC558 |
| b0557 (ybcU) | b0566 (envY) | EC1388; EC622 |
| b0933 (ycbE) | b0942 | EC370; EC558 |
| b1021 (ycdP) | b1031 (ycdV) | EC370; EC558 |
| b1136 (icdA) | b1146 | EC720; EC172*; SO111; EC370∗; EC38∗ |
| b1137 (ymfD) | b1157 | E139229; E140065∗; EC157; EC1727∗; EC558∗; EC622*; EDL933∗ |
| b1149 (ymfN) | b1161 (ycgX) | EC183; EC681; EC172∗; EC1722∗; EC720∗; SO126∗; SO29 |
| b1159 (mcrA) | b1172 | E139229; E140065; EC370∗; EC558∗; EC157∗ |
| b1305 (pspB) | b1321 (ycjX) | EC558; EC370∗ |
| b1345 | b1360 | EC720; O126; SO29∗; EC622∗ |
| b1382 (ynbE) | b1401 (ydbA_1) | E139229; E140065; EC558; EC720; EC157; EC370∗; EDL933∗; SO111∗ |
| b2030 (trs5_7) | b2039 (rfbA) | EC38; EC622 |
| b2030 (trs5_7) | b2041 (rfbB) | E139229; E140065; EDL933; EC157; SO55; SO126; SO29∗; EC720∗; SO111∗; EC1727∗ |
| b2043 (wcaM) | b2062 (wza) | EC370; EC558; SO111∗ |
| b2347 (yfdC) | b2363 | EC370; EC558; EC1727∗; EC622∗; EC720∗ |
| b2355 (yfdL) | b2365 (dsdX) | E139229; EDL933 |
| b2622 (intA) | b2644 (yfjY) | EC1388; EC1727; EC183; EC1840; EC370; EC537; EC558; EC157; EC720∗; EC622∗; EC459∗ |
| b2622 (intA) | b2646 (ypjF) | E139229; E140065; E33167; EC172; EC1722; EC1725; EC225; EC335; EC38; EC681; EDL933; SO111; SO126; SO29; SO55 |
| b2648 | b2659 | EC558; SO29; EC335∗ |
| b2968 (yghD) | b2986 (yghT) | E139229; E140065; EDL933; EC157 |
| b3318 (rplW) | b3338 (yheB) | EC335; EC558; EC681; SO126 |
| b3320 (rplC) | b3338 (yheB) | SO111; EC157 |
| b3321 (rpsJ) | b3338 (yheB) | EC172; EC183 |
| b3322 (pinO) | b3338 (yheB) | E139229; E140065; E33167; EC1388; EC1722; EC1727; EC1840; EC225; EC370; EC38; EC459; EDL933; EC537∗; EC1725∗ |
| b3574 (yiaJ) | b3583 (sgbE) | E139229; E140065; EDL933; EC157 |
| b3622 (rfaL) | b3632 (rfaQ) | E139229; E140065; E33167; EC1388; EC172; EC1722; EC1725; EC1727; EC38; EC183; EC1840; EC225; EC335; EC459; EC537; EC681; EC720; EDL933; SO111; SO126; EC157; SO55 |
| b3894 (fdoG) | b3907 (rhaT) | EC370; EC558 |
| b4273 (yi22_6) | b4282 (yjhE) | EC183; EC681; EC1722∗; EC622∗ |
| b4274 (yjgW) | b4283 (yi91b) | EC1388; EC38; SO111; SO126; EC157; EC370∗; EC558 |
| b4285 | b4308 (yjhR) | EC157; EC720∗; EC558∗; EC370∗ |
| b4294 (insA_7) | b4308 (yjhR) | EC183; EC335; EC622; SO111; SO126 |
| b4295 (yjhU) | b4308 (yjhR) | E139229; E140065; E33167; EC1388; EC172; EC1722; EC1725; EC1727; EC1840; EC225; EC459; EC537; EC681; EDL933; SO29; SO55; EC38∗ |
| b4338 (yjiP) | b4351 (mrr) | SO111; SO126; SO29 |
| b4341 (yjiS) | b4351 (mrr) | EC1388; EC1725; EC459 |
| b4342 (yjiT) | b4351 (mrr) | EC1727; EC38; SO55 |
The start and end positions of regions that are absent from the K-12 chromosomal backbone are given by the gene identifier and gene name (in parentheses), where applicable. Strains belonging to the O26 serotype are shown in bold. The names of some clinical isolates have been shortened by omitting the suffix. ∗, strain in which the missing region has a significant overlap but starts before or extends beyond or is not as large as the positions given but is still missing at least 10 consecutive genes.
Twelve other non-O26 strains were also included in this study, of which the majority of EPEC strains (both typical and atypical EPECs) clustered within the EPEC cluster. This included strains of serotypes O111, O126, and O55 but not O86. Although both strains from the last serotype were highly correlated to each other (distance score = 0.042), the O86 group showed low correlation to other strains in this study (distance score = 0.61). These strains also showed the greatest differences in comparison to the K-12 chromosomal backbone, with approximately 20% of the CDS being absent (Table 2) and missing many regions of the K-12 chromosomal backbone which were present in other strains (Table 3). These O86 strains had the largest number of genes with an ln(red/green) score of between 1.5 and 2.0 (Fig. 2), indicating that many of the genes are present but have low sequence homology with the respective K-12 genes. Biochemical testing (with API 20E strips) was used to confirm the species characteristics and showed that these O86 strains possessed typical E. coli attributes such as being indole positive, hydrogen sulfide negative, and citrate negative. Interestingly, the two O55 strains, although having different pathotype designations based on their virulence characteristics (Table 1), also clustered together, showing a closer relationship to each other than to any other strain used in this study (distance score = 0.19).
All four O157 strains formed a discrete cluster reflecting their close relationship (distance score = 0.046), in agreement with PCR and toxin data (Fig. 2). It was noted that the O157 strains had several unique regions missing from the K-12 chromosomal backbone. These included a 9-kb fragment from yiaJ to sgbE and a much larger 24-kb fragment from yghD to yghT (Table 3). An untypeable strain, EC720/98, which showed virulence characteristics similar to those of the VT-positive O26 strains (Table 1), was positioned close to the O157 cluster; the number of absent CDS was slightly higher in the EC720/98 strain (11%) than the O157 strains included in this study (9.8%; Table 2).
The CGI approach allowed detection of all K-12 genes missing from the chromosomal backbone, for each strain included in this study (see supplemental data). The absent genes are represented in red in Fig. 2. Further analysis of the missing genes showed that many could be grouped into regions that were absent from several strains. Table 3 shows regions of the K-12 chromosome where 10 or more consecutive genes were absent in at least two strains. Five regions of the K-12 chromosome had genes missing in 22 or more strains. These included b0245 to perR; intA to yfjY/yfjP; rplW/rplC/rpsJ/pinO to yheB; waaL (rfaL) to waaQ (rfaQ); and insA7/yjhU to yjhR. These regions were mainly composed of genes expressing hypothetical proteins, with the exception of the waa locus, which is involved in lipopolysaccharide biosynthesis. Three of the regions (bO245, intA, and yjhU) were also flanked by (or contained) transposases, insertion sequence elements, and/or a tRNA-like genes at the 5′ end. Such genes are commonly found at the sites of integration of foreign DNA, such as pathogenicity islands. The rfb locus, which is involved in synthesis of the structurally diverse O antigen polymer, had at least eight consecutive genes of the locus missing in all strains (see supplemental data), and 10 or more genes missing in the four O157 strains as well as SO55, EC38/99, and EC622/00.
Therefore, our data clearly demonstrate that comparative genomic indexing is a valuable tool for studying the phylogenomics of E. coli pathogens and for defining the core genes present in all strains included in this study.
Functional analysis of core genes.
To gain more information concerning the absent genes and also to define the core genes present in all strains, the E. coli CDS were grouped into functional groups defined by the clusters of orthologous groups of proteins (COGs; http://www.ncbi.nlm.nih.gov/COG). Each COG includes orthologous proteins, which are proteins connected through vertical evolutionary descent, and serves as a platform for functional annotation. The COG information is based on 30 genomes and breaks down into 17 broad functional categories, which include function unknown (39). Genes in each COG category for the E. coli genome were analyzed against our data set with GeneSpring software. The results show that every gene in our data set assigned to the following COG functional categories were present for all strains (Table 4): cell division and chromosome partitioning; coenzyme metabolism; energy production and conversion; nucleotide transport and metabolism; posttranslational modification; protein turnover and chaperones; and translation, ribosome structure, and biogenesis. For groups involved with information storage and processing, only the DNA replication, recombination, and repair functional category showed a high number of genes to be absent. Many genes in the general function prediction, function unknown, and not in COGs categories were missing from our data set, as they did not pass our filtering criteria (see Materials and Methods), so their status was not determined (data not shown).
TABLE 4.
Assignment of absent genes to functional categories with the E. coli COG databasea
| Functional category | Total no. of genes in data set | Genes absent in at least 22 of 26 strains of E. coli (%) |
|---|---|---|
| Metabolism | ||
| Amino acid transport and metabolism | 307 | 1.6 |
| Carbohydrate transport and metabolism | 301 | 3.7 |
| Secondary metabolite biosynthesis, transport, and catabolism | 43 | 4.7 |
| Coenzyme metabolism | 102 | 0 |
| Energy production and conversion | 217 | 0 |
| Lipid metabolism | 64 | 6.3 |
| Nucleotide transport and metabolism | 70 | 0 |
| Cellular processes | ||
| Cell division and chromosome partitioning | 30 | 0 |
| Posttranslational modification, protein turnover, and chaperones | 97 | 0 |
| Signal transduction mechanisms | 94 | 1.1 |
| Inorganic ion transport and metabolism | 150 | 1.3 |
| Cell envelope biogenesis, outer membrane | 199 | 6 |
| Cell motility and secretion | 87 | 14 |
| Information storage and processing | ||
| DNA replication, recombination, and repair | 174 | 10 |
| Translation, ribosome structure, biogenesis | 125 | 0 |
| Transcription | 211 | 5 |
| Poorly characterized | ||
| General function prediction | 271 | 4 |
| Function unknown | 207 | 2 |
| Not in COGs | 340 | 19 |
The 3,039-gene data set was used to derive this table (Materials and Methods).
Approximately 19% of the genes assigned to the not in COGs category were absent in a majority of the E. coli field strains analyzed (at least 22 of 26 strains). Therefore, the largest number of “absent” genes belonged to the not in COGs category. Most of the absent genes were hypothetical proteins with putative or unknown function and included a majority of the genes from the bO245, intA, pinO, and yjhU region (Table 3). Approximately 14% of genes assigned to the cell motility and secretion category were also missing, and these included putative outer membrane and fimbrial proteins such as ychD and smfD. Approximately 10% of genes assigned to the DNA replication, recombination, and repair category was also absent. The CDS for the majority of these absent genes were transposases, although genes whose product may be involved with DNA repair (e.g., yfjY) or frameshift suppression (e.g., yjhR) were also absent.
The data summarized in Table 4 show that a core pool of genes involved with metabolism, various cellular processes (excluding cell motility and secretion), and information storage and processing (excluding DNA replication, recombination, and repair) have been conserved and maintained in all strains. Conversely, many genes of unknown function, external origin, or facultative function were absent from the chromosomal backbone of a majority of strains and could be assigned as expendable. Such patterns of gene stability within E. coli populations are consistent with suggestions by Dobrindt et al. (10), Lan and Reeves (22), and Woodward and Charles (43). Preliminary analysis of the expendable regions suggest that gene insertions with no similarity to the K-12 genome are harbored in many of these regions (data not shown).
DISCUSSION
E. coli strains of serogroup O26 have long been established as etiological agents of human and animal disease. They have been classified into two different pathotypes, EPECs and EHECs, mainly due to differences in disease symptoms and virulence characteristics (24, 27). It is vitally important to determine the clonal relationship within strains of this serogroup to understand the mechanisms of transfer of these pathogens through the food chain. Different typing methods used to assess the clonal lineage within the O26 serogroup have mostly focused on EHEC O26 (VT-positive) strains and have suggested that genetic heterogeneity exists, although the results from different studies are often contradictory. To improve our understanding of the clonal diversity within the O26 serogroup (i.e., both VT-positive and -negative O26 strains), we used a more broad-based genetic approach. The CGI technique allowed comparison of the genome of field isolates of the O26 serogroup with each other and with strains from other EPEC and EHEC serotypes. Several studies have used DNA microarrays to compare the genomes from different subgroups or strains within other bacterial species (2, 11, 12, 14, 21, 32, 34).
Our results (Fig. 2) clearly divided the strains into EHECs (O157), EPECs (O111, O126, O26, and O55), and others (O86 and O29). Furthermore, all O26 strains (excluding EC622/99) clustered within the EPEC cluster irrespective of their pathotype designation (most strains in this study had been shown to possess a mixture of EHEC and EPEC virulence features; Table 1). The CGI results showed a similar percentage of the K-12 chromosomal backbone to be absent in the O26 serogroup and the majority of absent genes were missing from both VT-positive and VT-negative O26 strains (Table 2); only one region on the K-12 chromosome was found to differentiate them (yagP-yagT). These results indicate greater genetic homogeneity within this serogroup than previously proposed and suggest a common clonal lineage of both EPEC and EHEC O26 strains.
Also included in the study were two VT-negative O26 strains (EC335/98 and EC225/00) which possessed the EHEC-associated hlyA gene. Interestingly, both of these strains clustered with the VT-positive O26 strains, showing higher correlation to these strains than to the remaining VT-negative strains. Several studies (29, 45) have shown that O26 strains classed as atypical EPECs (stx eae+) belonged to the classical EHEC O26 serotypes (O26:H11 and O26:H−). This suggests that classification of O26:H11 and O26:H− strains as EHECs may be misleading. Based both on our findings and those of others (29, 45), the proposition that stx eae+ O26 strains could be EHECs which have lost their stx genes or the progenitors of EHEC O26 strains (45) is more likely. This suggestion is further substantiated by a demonstration by Schmidt et al. (35) that an Stx2-converting phage isolated from E. coli O157 was able to infect and lysogenize various E. coli strains, including both EPEC and EHEC O26 strains. Therefore it is likely that stx hlyA O26 strains are O26 strains which have yet to acquire (or have lost) the verotoxin-producing prophage and hlyA plasmid, while stx hlyA+ strains are at an intermediate stage, i.e., have already acquired the virulence plasmid but not the VT-expressing prophage. Therefore, division of this serotype into the pathotypes EPEC and EHEC may be misleading, as VT-positive O26 strains are likely to arise from VT-negative strains and vice versa. Such an environment of genomic exchange would also result in coevolution of VT-positive and VT-negative O26 strains.
Conversely, it could be postulated that in the absence of VTEC genes in the environment, the O26 subpopulation would accumulate different niche-adaptive genes. Studies involving VTEC and O157 isolation from animals (3, 7) have shown the prevalence of these bacteria to be much lower in pigs (7.5% VTEC and 4% O157) than in sheep (66.6% VTEC and 22% O157) and cattle (21.1% VTEC and 15.7% O157). As a result, the O26 subpopulation in pigs (which are less likely to encounter VTECs) have probably accumulated different gene transfer and recombination events than found in the O26 bacteria from ruminants. The resulting difference in genotype could explain our CGI results, which showed that EC622/99, the only strain of porcine origin included in this study, clustered outside the O26 cluster; the O26 cluster included strains of mostly bovine and ovine origin (Fig. 2).
With CGI and the COG database, we identified the core gene pool involved with essential cellular functions, which was maintained in all 26 E. coli field strains used in this study (Table 4 and supplemental data). This collection of genes, which were common to all the pathogenic and nonpathogenic field strains examined as well as the laboratory-adapted MG1655, is the minimal requirement for these bacteria to be classed as the same species. The expendable genes, which were regions of the K-12-chromosomal backbone missing in the field strains, were regions representing adaptation and evolution of these organisms to a different host environment and/or an ecological niche. Six regions of expendable genes missing from the K-12 chromosomal backbone for the majority of strains were identified (Table 3 and supplemental data); this included genes in the O-antigen locus (rfb) and the core oligosaccharide domain (waa locus). Other variable regions of the E. coli chromosome were also identified, e.g., intA, pinO, and yjhU (Table 3).
In our study, most of the E. coli strains were clustered by their serotype, showing serotyping to be an useful indicator of genetic diversity in a clonally structured population such as E. coli, where associations between loci are nonrandom (41). The localized horizontal gene transfer in E. coli populations would not destroy linkage disequilibrium due to its low frequency but would maintain useful variation within a subpopulation (25), as evidenced by our CGI data. Therefore, these variable regions help make the bacterial genome a dynamic structure and contribute to intraspecies variation. Future analyses of these regions of variability will provide further insight into the clonal diversity within the O26 serogroup and will give a better understanding of the genomic transition between VT-positive and VT-negative O26 strains. It will also identify the genomic differences between serogroups which enable them to adapt to distinct host environments.
Therefore, the CGI approach has provided a valuable tool for understanding the clonality of pathogenic E. coli, by defining the core genome and identifying regions of variation. CGI overcomes the limitations inherent in focusing on a particular set of related proteins or group of genes, which may reflect a partial phylogeny. However, because this method relies on CDS microarrays, it cannot detect the single nucleotide changes which cause protein polymorphism and allelic variation.
In summary, we have clearly demonstrated that strains of serogroup O26, both VT positive and negative, have a common clonal lineage and that VT-negative strains are likely to have lost their stx genes or to be progenitors of EHEC O26 strains. Further insight into clonality may be gained by focusing on the hypervariable regions that we have identified on the E. coli chromosome. Future epidemiological studies of outbreaks of this important food-borne pathogen should consider genetic analysis of both VT-positive and VT-negative O26 strains, as the latter may well be an important environmental reservoir that can give rise to EHEC O26 infections.
Acknowledgments
M.F.A. and M.J.W. are grateful for funding from the Veterinary Laboratories Agency (VLA) seedcorn fund. J.C.D.H., A.T., and S.L. acknowledge funding from the BBSRC.
We thank Mary Berlyn for E. coli K-12 strains and VLA, Weybridge Diagnostics Unit, for veterinary strains. We are also very grateful to S. Gordon at VLA for many helpful discussions and suggestions.
Editor: F. C. Fang
REFERENCES
- 1.Adu-Bobie, J., G. Frankel., C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson., W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., D. Geier, H. Steinruck, S. Zimmermann, and F. Scheutz. 1993. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J. Clin. Microbiol. 31:2483-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Caprioli, A., A. Nigrelli, R. Gatti, M. Zavanella, A. M. Blando, F. Minelli, and G. Donelli. 1993. Characterisation of verocytotoxin-producing Escherichia coli isolated from pigs and cattle in northern Italy. Vet. Rec. 133:323-324. [DOI] [PubMed] [Google Scholar]
- 6.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, P. A., C. A. Siddons, A. T. Gerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119: 245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cid, D., J. A. Ruiz-Santa-Quiteria, I. Marin, R. Sanz, J. A. Orden, R. Amils, and R. de la Fuente. 2001. Association between intimin (eae) and espB gene subtypes in attaching and effacing Escherichia coli strains isolated from diarrhoeic lambs and goat kids. Microbiology 147:2341-2353. [DOI] [PubMed] [Google Scholar]
- 9.Correa, M. G., and J. M. Marin. 2002. O-serogroups, eae gene and EAF plasmid in Escherichia coli isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 85:125-132. [DOI] [PubMed] [Google Scholar]
- 10.Dobrindt, U., U. Hentschel, J. B. Kaper, and J. Hacker. 2002. Genome plasticity in pathogenic and non-pathogenic enterobacteria. Curr. Top. Microbiol. Immunol. 264:157-175. [PubMed] [Google Scholar]
- 11.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates with a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geue, L., M. Segura-Alvarez, F. J. Conraths, T. Kuczius, J. Bockemuhl, H. Karch, and P. Gallien. 2002. A long-term study on the prevalence of shiga toxin-producing Escherichia coli (STEC) on four German cattle farms. Epidemiol. Infect. 129:173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gioffre, A., L. Meichtri, E. Miliwebsky, A. Baschkier, G. Chillemi, M. I. Romano, S. Sosa Estani, A. Cataldi, R. Rodriguez, and M. Rivas. 2002. Detection of Shiga toxin-producing Escherichia coli by PCR in cattle in Argentina. Evaluation of two procedures. Vet. Microbiol. 87:301-313. [DOI] [PubMed] [Google Scholar]
- 17.Gunning, R. F., A. D. Wales, G. R. Pearson, E. Done, A. L. Cookson, and M. J. Woodward. 2001. Attaching and effacing lesions in the intestines of two calves associated with natural infection with Escherichia coli O26:H11. Vet. Rec. 148:780-782. [DOI] [PubMed] [Google Scholar]
- 18.Guth, B. E., R. Giraldi, T. A. Gomes, and L. R. Marques. 1994. Survey of cytotoxin production among Escherichia coli strains characterised enteropathogenic (EPEC) by serotyping and presence of EPEC adherence factor (EAF) sequences. Can. J. Microbiol. 40:341-344. [DOI] [PubMed] [Google Scholar]
- 19.Hiramatsu, R., M. Matsumoto, Y. Miwa, Y. Suzuki, M. Saito, and Y. Miyazaki. 2002. Characterization of Shiga toxin-producing Escherichia coli O26 strains and establishment of selective isolation media for these strains. J. Clin. Microbiol. 40:922-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, R. E., R. A. Wilson, M. S. Holland, V. Yuzbasiyan-Gurkan, T. P. Mullaney, and D. G. White. 1999. Characterization of eae+ Escherichia coli isolated from healthy and diarrheic calves. Vet. Microbiol. 66:251-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacteriumtuberculosis. Genome Res. 11:547-554. (Erratum, Genome Res. 11:1796.) [DOI] [PMC free article] [PubMed]
- 22.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 23.La Ragione, R. M., I. M. McLaren, G. Foster, W. A. Cooley, and M. J. Woodward. 2002. Phenotypic and genotypic characterisation of avian Escherichia coli O86:K61 isolates possessing a gamma-like intimin. Appl. Environ. Microbiol. 68:4932-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohaemorrhagic and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 25.Maynard-Smith, J., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 27.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paciorek, J. 2002. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J. Med. Microbiol. 51:548-556. [DOI] [PubMed] [Google Scholar]
- 29.Peixoto, J. C., S. Y. Bando, J. A. Ordonez, B. A. Botelho, L. R. Trabulsi, and C. A. Moreira-Filho. 2001. Genetic differences between Escherichia coli O26 strains isolated in Brazil and in other countries. FEMS Microbiol. Lett. 196:239-244. [DOI] [PubMed] [Google Scholar]
- 30.Perna, N. T., G. Plunkett 3rd, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157: H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 31.Porter, R. D., M. W. Lark, and K. B. Low. 1981. Specialized transduction with lambda plac5: dependence on recA and on configuration of lac and att lambda. J. Virol. 38:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rios, M., V. Prado, M. Trucksis, C. Arellano, C. Borie, M. Alexandre, A. Fica, and M. M. Levine. 1999. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J. Clin. Microbiol. 37:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. A. Falkow. 2000. Whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, V. K. 2002. Detection and quantitation of enterohemorrhagic Escherichia coli O157, O111, and O26 in beef and bovine feces by real-time polymerase chain reaction. J. Food. Prot. 65:1371-1380. [DOI] [PubMed] [Google Scholar]
- 37.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 38.Sramkova, L., M. Bielaszewska, J. Janda, K. Blahova, and O. Hausner. 1990. Vero cytotoxin-producing strains of Escherichia coli in children with haemolytic uraemic syndrome and diarrhoea in Czechoslovakia. Infection 18:204-209. [DOI] [PubMed] [Google Scholar]
- 39.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, A., S. Lucchini, and J. C. D. Hinton. 2001. It's easy to build your own Microarrayer! Trends Microbiol. 9:154-156. [DOI] [PubMed] [Google Scholar]
- 41.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodward, M. J., and H. P. Charles. 1982. Genes for l-sorbose utilisation in Escherichia coli. J. Gen. Microbiol. 128:1969-1980. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, W. L., M. Bielaszewska, A. Liesegang, H. Tschape, H. Schmidt, H. M. Bitzan, and H. Karch. 2000. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J. Clin. Microbiol. 38:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, W. L., M. Bielaszewska, J. Bockemuhl, H. Schmidt, F. Scheutz, and H. Karch. 2000. Molecular analysis of H antigens reveals that human diarrheagenic Escherichia coli O26 strains that carry the eae gene belong to the H11 clonal complex. J. Clin. Microbiol. 38:2989-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]