Abstract
An in vitro protein selection method, ribosome display, has been applied to comprehensively identify and map the immunologically relevant proteins of the human pathogen Staphylococcus aureus. A library built up from genomic fragments of the virulent S. aureus COL strain (methicillin-resistant S. aureus) allowed us to screen all possible encoded peptides for immunoreactivity. As selective agents, human sera exhibiting a high antibody titer and opsonic activity against S. aureus were used, since these antibodies indicate the in vivo expression and immunoreactivity of the corresponding proteins. Identified clones cluster in distinct regions of 75 genes, most of them classifiable as secreted or surface-localized proteins, including previously identified virulence factors. In addition, 14 putative novel short open reading frames were identified and their immunoreactivity and in vivo mRNA expression were confirmed, underscoring the annotation-independent, true genomic nature of our approach. Evidence is provided that a large fraction of the identified peptides cannot be expressed in an in vivo-based surface display system. Thus, in vitro protein selection, not biased by the context of living entities, allows screening of genomic expression libraries with a large number of different ligands simultaneously. It is a powerful approach for fingerprinting the repertoire of immune reactive proteins serving as target candidates for active and passive vaccination against pathogens.
Staphylococcus aureus is a gram-positive bacterial pathogen that causes diseases ranging from minor skin infections to life-threatening deep infections such as pneumonia, endocarditis, meningitis, osteomyelitis, postoperative wound infections, septicemia, and toxic shock syndrome. Hospitalized patients are at particular risk, and the emergence of drug resistance has made many of the available antibiotics ineffective. Approximately 40 to 60% of staphylococcal isolates are resistant to multiple antibiotics, including methicillin and vancomycin. The need for new drugs in order to treat S. aureus infections is unmatched with the current situation, as only a limited number of antibiotics have received market approval in the last decades. Passive or active immunization might therefore provide a solution to this problem; however, still no effective vaccine against S. aureus is available. Consequently, to produce an effective vaccine it is utterly important to find antigens that raise the production of protective antibodies. Specifically, proteins conserved among different isolates are well-suited for the development of defined vaccines, as nonprotein compounds like polysaccharides often vary among isolates, frequently even specify a serotype, and also fail to induce a long-lasting immune response, except for the more costly conjugate vaccines.
More recently, whole-genome, bioinformatic-based approaches have been reported (21, 27). Proteins from Neisseria meningitidis and Streptococcus pneumoniae predicted to be cell surface localized or secreted have been expressed as recombinant proteins and empirically tested in animal models. This approach is highly dependent on proper gene identification and annotation and is difficult to apply to the roughly 40% of genes, on average, to which no function can be assigned in a given genome (19). Another intrinsic shortcoming is that it is not known how many of these proteins are actually expressed in vivo during an infection and are able to induce an immune response in humans. Both aspects are requirements for effective vaccines. A functional selection to identify proteins with these characteristics would therefore be advantageous.
The human humoral immune system is able to raise a specific antibody profile upon encountering a given pathogenic microorganism. These antibodies can be seen as an inverse, immunological blueprint suitable to identify the corresponding proteins out of proteomic samples or expression libraries. A number of these identified proteins from S. aureus were shown to induce antibodies that possess antistaphylococcal activity (2-4, 25). Thus, the comprehensive identification of the antibody repertoire against a given pathogen is a prerequisite for an effective vaccine and, moreover, could give more insight into the pathogenesis of a particular infection.
In recent years, several in vitro protein selection technologies have emerged (8, 9, 17, 22) that, by circumventing the need to introduce the encoding DNA of interest into a cellular host, provide two very distinct main properties. First, they allow access to library complexities that are basically limited only by the amount of DNA that is added to cell-free protein synthesis systems, thereby exceeding in vivo display and selection systems by several orders of magnitude. Second, virtually any peptide or protein that can be produced by the ribosomal machinery can be displayed and selected for, because the in vitro environment excludes any interference with a host's physiology (e.g., toxic proteins and membrane penetration).
Here we present the application of functional in vitro protein selection technology for identification of immunoreactive peptides on a genomic scale for the first time. Using an S. aureus-derived genomic library, in vitro selection was successfully performed with high-titer sera against S. aureus, thereby expanding in vitro protein selection also for the simultaneous use of a multiple ligand set. Emphasis was set to characterize the selection dynamics and to confirm the immunogenicity of the identified antigens by peptide enzyme-linked immunosorbent assay (ELISA). Finally, an expression comparison with an in vivo based surface display system was performed.
MATERIALS AND METHODS
Template design.
The following oligodeoxyribonucleotides were used for PCR to generate DNA templates for in vitro selection: ICC277 (5′-CGAATAATACGACTCACTATAGGGAGACCACAACGGTTTCCCACTAGTAATAATTTTGTTTAACTTTAAGAAGGAGATATATCCATGCAGACCTTGGCCGGCCTCCC-3′), which introduces T7 RNA polymerase promoter, 5′ stem loop, and a ribosome binding site, and ICC202 (5′-GGCCCACCCGTGAAGGTGAGCCGGCGTAAGATGCTTTTCTGTGAC-3′), which introduces a 3′ stem loop structure. These primers amplify myc9 or S. aureus genomic fragments ligated into vector pMAL4.1 (4) with three glycines as linker and the first 89 amino acids from β-lactamase (bla) as tether. A 390-bp DNA sequence encoding nine copies of the myc epitope was ligated into the SmaI restriction site of pMAL4 (pMAL4 contains an additional guanine between the SmaI and NotI site compared to pMAL4.1, thereby restoring the reading frame of bla). From this vector myc9-bla was constructed by PCR amplification. The construction of the genomic S. aureus library was described previously (4). Plasmid DNA from pMAL4.1 containing frame-selected genomic regions was used as the template for PCR. The Expand High-Fidelity PCR system kit (Roche) was used for all PCRs.
In vitro selection.
One selection cycle was carried out with the following modifications to the described procedure (7): in vitro transcription was performed with RIBOMAX T7 large-scale RNA production systems (Promega) as suggested by the manufacturer, and RNA was purified with YM-100 Microcon columns (Amicon). A 22-μl in vitro translation reaction contained 2 μg of RNA, 4.4 μl of Premix Z, 200 mM potassium glutamate, 13.8 mM magnesium acetate, 8 μl of Escherichia coli S-30 extract (2.6 μg of protein/μl), and 1.9 mM methionine and was carried out for 9 min at 37°C. Anti-ssrA oligonucleotide (8) was omitted, as it did not show any obvious effect in our selection of genomic DNA fragments (data not shown). Immunoprecipitation of the ribosomal complexes was accomplished with MAGmol Protein G Microbeads (Miltenyi Biotec). The translation reaction mixture was transferred to a prechilled tube containing 35.2 μl of 10% milk-WBT buffer [WBT buffer is 50 mM Tris(Ac)2 (pH 7.5), 150 mM NaCl, 50 mM Mg(Ac)2, 0.1% Tween 20] mixed with 52.8 μl of WBTH buffer (WBT buffer and heparin [2.5 mg/ml]), 1 μg of 9E10 myc antibody, or 10 μg of E. coli cell, and extract-preadsorbed high-titer immunoglobulins G (IgGs) were used as indicated. Serum selection and treatment was performed as described previously (4). After incubation on ice for 90 min, 50 μl of protein G Microbeads were added, and incubation was prolonged for another 90 min. The immunoprecipitate was applied to a precooled microcolumn (Miltenyi Biotec), washed five times with ice-cold WBT buffer, and incubated with 20 μl of EB20 elution buffer [50 mM Tris(Ac)2 (pH 7.5), 150 mM NaCl, 20 mM EDTA, Saccharomyces cerevisiae RNA (50 μg/ml; Sigma)] for 5 min at 4°C. Elution was completed by eluting with 100 μl of EB20. Reverse transcription (RT)-PCR was performed with Superscript II (Roche Diagnostics). After four rounds of selection the library fragments were cloned into the transcriptionally silenced vector pMAL4.34 (Hanner et al., unpublished data) with FseI and NotI, selected with ampicillin and kanamycin, and sequenced.
Peptide ELISA and Western blotting.
Peptides were synthesized according to their deduced amino acid sequence by 9-fluoroenylmethoxy carbonyl chemistry. ELISA and Western blotting were performed as described previously (4). Values above double the background plus 1 standard deviation (optical density at 450 nm = 0.2) were calculated as positive in ELISA. Numbers in Table 3 were calculated from ELISA values as follows: <0.2 = −; <0.5 = 1; <0.8 = 2; >0.8 = 3. FhuA expression analysis was performed as described previously (5).
TABLE 3.
Results of ELISA with synthesized peptidesa
| ORF name | Position (amino acids) | Healthy-subject high-titer serum
|
|||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3* | 4* | 9* | 15* | 35* | 2 | 5 | 6 | 7 | 8 | 10 | 11 | 14 | 16 | 17 | 18 | 19 | 20 | 21 | 24 | 26 | 28 | 29 | 31 | 32 | 34 | 36 | 37 | 38 | 39 | 40 | 41 | ||
| SA0006 | 114-130 | 2 | 2 | 2 | — | ND | 1 | — | 1 | 1 | 1 | 1 | 1 | — | 3 | 1 | 2 | 1 | 1 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA0103 | 84-100 | 3 | 2 | 2 | 2 | ND | 2 | 1 | 2 | 2 | 3 | 3 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA0195 | 78-128 | 1 | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 | 1 |
| SA0200 | 323-345 | 1 | — | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | — | — | 1 | 1 | — | 1 | 1 | 1 | — | 1 | 1 | 1 | — | 1 | 1 |
| SA0317 | 147-162 | — | 1 | 1 | — | — | 1 | 1 | — | 1 | 1 | — | 1 | — | — | — | — | — | — | — | — | — | 1 | — | — | — | 1 | — | 1 | — | — | — | — |
| SA0835 | 6-54 | 3 | 1 | 2 | 1 | 1 | 3 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | 3 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | — | 2 | 1 | 1 | 1 | — | 2 | 2 |
| SA1062 | 63-81 | 2 | — | 2 | 2 | ND | 1 | — | 2 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | — | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA1112 | 13-37 | 2 | 3 | 2 | 2 | ND | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA1743 | 127-152 | 2 | 1 | 2 | 2 | ND | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | — | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA1777 | 13-29 | — | — | 1 | — | — | 1 | — | — | 1 | 1 | — | 1 | — | — | — | — | — | — | — | — | — | 1 | — | 1 | — | 1 | — | 1 | — | — | — | — |
| SA1919 | 120-135 | 2 | 3 | 2 | 2 | ND | 2 | 1 | 3 | 2 | 2 | 2 | 3 | 2 | 3 | 2 | 3 | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA2125 | 293-312 | 3 | 3 | 3 | 2 | ND | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 3 | 2 | 2 | 2 | 2 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA2347 | 205-224 | 3 | 2 | 2 | 2 | ND | 2 | 1 | 2 | 2 | 2 | 3 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA2437 | 83-102 | 3 | 3 | 3 | 2 | ND | 2 | 1 | 3 | 2 | 3 | 2 | 3 | 2 | 3 | 2 | 3 | 2 | 3 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA2505 | 824-832 | 2 | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | 1 | 1 | — | — | — | — | — | — | — | — | — | — | — | — | 1 | 1 | 2 |
| SA2511 | 131-148 | 2 | — | 1 | 1 | ND | — | — | 1 | — | 2 | 2 | 1 | — | 1 | 1 | 1 | 1 | 1 | — | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| SA2652 | 601-617 | 1 | 1 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 |
| SA2659 | 90-107 | — | — | — | 3 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| SAN7 | 1-18 | 1 | — | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | — | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | — | 2 |
| SAN10 | 27-35 | 2 | 2 | 1 | 1 | 2 | 1 | — | 1 | 2 | — | 1 | 1 | 1 | — | 1 | 1 | 1 | 1 | 1 | — | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | — | 1 | — | 1 |
Semiquantitative PCR.
Equal amounts of cDNA template from the individual rounds and an appropriate amount of the input library were used in a PCR with primers ICC277 and ICC202. After analysis by gel electrophoresis and photometry, the PCR products were used as templates for semiquantitative PCR with 30 cycles. ICC509 (5′-CACCCAACTGATCTTCAGC-3′) hybridizing 40 bp downstream of the SmaI site in the bla gene was used as the antisense primer in all reactions. The following sense primers were utilized to amplify specific regions of the open reading frames (ORFs): 5′-TATGTTTATAACGATGGGA-3′ for SA0021, 5′-GTTGGTCGTAAATACGGGC-3′ for SA0195, 5′-CGTGCAATTATAGCTATTG-3′ for SA0200, 5′-TTCACAGTCTTAGCAATTG-3′ for SA0334, 5′-GTAGTTAGCATTGCGACGG-3′ for SA0835, 5′-ACAGAAGACCTAAATTTCC-3′ for SA1777, 5′-TTGATGGCGTTCATGCACC-3′ for SA2659, and 5′-AATGGGTTATCGGTCAATG-3′ for SA2702.
RT-PCR.
The following primers were used for RT-PCR: IsaA sense, 5′-GCGCTGAAGTAAACGTTGATCAAGCA-3′; IsaA antisense, 5′-TAGAATCCCCAAGCACCTAAACCTTG-3′; PSAN1 sense, 5′-TGTGCTATTATTCCCCCTATTG-3′; PSAN1 antisense, 5′-TGGTAGGTTAGTACAAGGAG-3′; SAN1 sense, 5′-GTCCATGGAAGTTACCACC-3′; SAN1 antisense, 5′-GTGGCAGCGCGTATGC-3′; SAN2 sense, 5′-CAGTGTTTCAATAATCAGCCAAC-3′; SAN2 antisense, 5′-AGCTGATGCATTTTGCTTTACC-3′; SAN3 sense, 5′-ATCAGGTGTGTGATGATAATCC-3′; SAN3 antisense, 5′-GAAAAAATTAACAGCAGCAGCG-3′; SAN4 sense, 5′-GCTTTTGCGACATTAGCGC-3′; SAN4 antisense, 5′-AAAACTTAAACAGTTGAGAGATGC-3′; SAN5 sense, 5′-AATCTTTTGATGTTTCATTTTCAACTG-3′; SAN5 antisense, 5′-GCCTAAATTAGTTATTCTAATGGG-3′; SAN6 sense, 5′-GCCAGTCACTTCTCGTTC-3′; SAN6 antisense, 5′-CGGTTGAATGCCATGTAAAAGC-3′; SAN7 sense, 5′-TATAGCATTTACCGCTTGATTTG-3′; SAN7 antisense, 5′-CAGACAACAAGCTATTGAAAG-3′; SAN8 sense, 5′-TAATCAAAACTGTCACCTTTATCAG-3′; SAN8 antisense, 5′-CGGAGGTCGAATAACATGC-3′; SAN9 sense, 5′-TTGAGTTTGTGCGTTTGCTG-3′; SAN9 antisense, 5′-ATGCCAATACACAGCCTAAC-3′; SAN10 sense, 5′-ATGCTTTTGAGTTACTTCATAATAC-3′; SAN10 antisense, 5′-AGAACATCGGTTATCATTTTTTAAAC-3′; SAN11 sense, 5′-TGTGTACCTATGAATTGTTGCG-3′; SAN11 antisense, 5′-CCTGATGATATTTATGTCAACAATG-3′; SAN12 sense, 5′-GATTGACTGCCACCACCG-3′; SAN12 antisense, 5′-CATTCTACATCATCAAGTAATAACC-3′; SAN13 sense, 5′-CATCTGGGTGAACTTGAACAG-3′; and SAN13 antisense, 5′-GATATCCCTAATGAAACAACTGG-3′. RNA was extracted from an S. aureus COL strain (optical density at 600 nm = 1.7 [BHI medium]) as described previously (13) and used for RT (Superscript II; Roche Diagnostics). Two microliters of cDNA was used for PCR with 25 to 30 cycles (94°C for 30 s, 42°C for 30 s, 72°C for 2 min). As a negative control, an appropriate amount of RNA was directly used for PCR. The amplified products were cloned and confirmed by sequencing.
Bioinformatic analysis.
The localization of the selected clones was determined by BLAST analysis against the S. aureus COL genome (http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl) (20) as well as ORF set on a local BLAST server and the localization data stored in a Sybase relational database. TM/S prediction of transmembrane domains or signal peptides was done with the following programs: TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) (10) and SignalP V1.1 (http://www.cbs.dtu.dk/services/SignalP/index.html) (18).
RESULTS
In vitro selection of antigens by ribosome display.
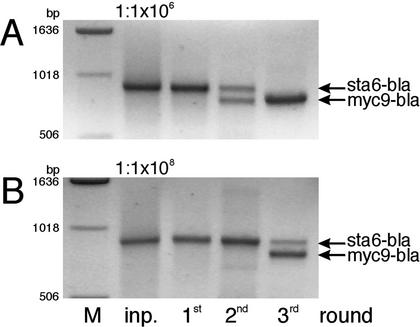
In order to adapt the ribosome display method (8) for selection of immunoreactive peptides, we first employed a well-characterized antibody-epitope system. Nine consecutive repeats of the myc epitope were cloned in frame with a fragment of the β-lactamase gene serving as a tether, interspersed by a flexible glycine linker to facilitate independent folding of the polypeptide of interest. After a number of modifications and optimization steps (see Materials and Methods), we were able to specifically select and enrich the myc-containing sequence very efficiently and dependent on myc antibody presence. A sample containing a 106-fold molar excess of an unrelated RNA over the myc-containing RNA was subjected to three rounds of selection and amplification in the presence of myc antibody. Figure 1A shows that only the myc-containing DNA fragment was present after the third round of selection. At a dilution of 1 to 1 × 108, the myc DNA fragment is dominant over the unrelated one after only three rounds of selection (Fig. 1B). Note that this in vitro expression and selection system has the intrinsic capability to screen libraries with complexities exceeding 1012 members; here it was sufficient to demonstrate the sensitivity of our system to screen rather low complexity libraries required for covering microbial genomes.
FIG. 1.
Ribosome display with antibodies as selective agents. RNA encoding the myc epitope sequence was mixed with an unrelated fragment (sta6-bla) at a dilution of 1:106 (A) or 1:108 (B). Three selection cycles were performed with antibodies against the myc epitope. Shown are PCR products of the input mix (inp.) and the cDNA of the three selection cycles as indicated. On the left the length of the DNA marker (lane M) is depicted.
For the actual selection of antigenic regions from S. aureus we transferred a previously described genomic library derived from the methicillin-resistant S. aureus strain COL (4) into the ribosome display context by PCR. The IgG fraction from precharacterized sera from healthy individuals exhibiting a high titer and opsonizing activity against S. aureus as previously described was used for selection (4). After four rounds of selection, the resulting pool was cloned into a transcriptionally silenced vector and sequenced. As a first confirmation on the immunoreactivity of the selected pool, immune precipitation experiments with six different clones that were frequently found revealed that they all precipitated with sera used for selection but not with infant serum as a negative control (data not shown).
Bioinformatic analysis.
Based on sequence determination from 1,576 selected clones, bioinformatic analysis revealed the following picture.
(i) Selected clones cluster in distinct regions.
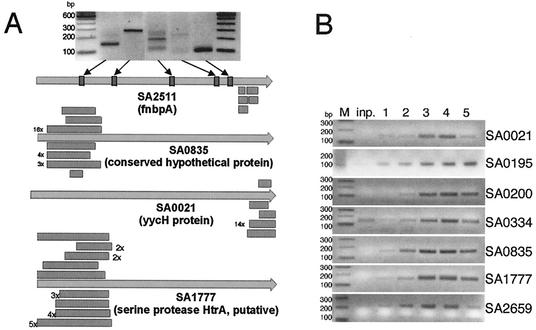
Selected clones cluster in distinct regions of 75 different ORFs in an overlapping fashion (Fig. 2A). A number of control PCRs with primers matching ORF regions other than the identified ones revealed that these regions were indeed present in the input library but evidently were not selected and enriched (Fig. 2A and data not shown). A representative subset of identified genes can be seen in Table 1.
FIG. 2.
Evaluation of antigenic epitopes. (A) Alignment of recovered epitopes to individual ORFs from S. aureus. SA2511 (coding for fibronectin-binding protein, FnbpA), SA0835, SA0021, and SA1777 are shown as examples. For fnbpA, PCR products amplified from the input library are shown. The sense primer binds to the indicated regions of fnbpA, whereas the antisense oligonucleotide hybridizes to the constant region of the β-lactamase spacer. (B) Enrichment of frequently found S. aureus library fragments during the selection process. Enrichment was determined by semiquantitative PCR with clone-specific primers. The sense primer hybridizes to the indicated S. aureus library fragment (compare Table 1). The antisense primer binds to the constant β-lactamase spacer. Each lane shows the PCR product of a different template: inp., DNA of input library; 1 to 5, cDNA of the first through fifth round, respectively, of ribosome display screened with normal high titer serum. Marker (M) length is shown on the left.
TABLE 1.
Immunoreactive S. aureus genes identified in this studya
| TIGR locus | Gene name or function | No. of clonesb | Reactivityc
|
IPd | % Conservatione | TM/Sf | Region(s)g (amino acid) | |
|---|---|---|---|---|---|---|---|---|
| P sera | N sera | |||||||
| SA0006 | DNA gyrase, A subunit (gyrA) | 5 | 9/14 | 15/19 | ND | 81.08 | ND | 89-130 |
| SA0021 | yycH protein (yycH) | 53 | ND | 100.00 | ND | 8-24, 394-494 | ||
| SA0050 | Methicillin-resistant surface protein (pls) | 6 | + | 54.05 | ND | 1191-1255, 1389-1496 | ||
| SA0103 | Drug transporter, putative | 8 | 14/14 | 18/19 | ND | 100.00 | ND | 84-100 |
| SA0195 | Maltose ABC transporter, permease protein | 100 | 14/42 | 4/32 | + | 100.00 | ND | 78-129 |
| SA0200 | Phosphoglycerate transporter family protein | 379 | 42/42 | 26/32 | + | 94.59 | ND | 323-345 |
| SA0317 | Lipase precursor, interruption | 33 | 2/42 | 10/32 | ND | 75.00 | ND | 54-92 |
| SA0334 | Conserved hypothetical protein | 168 | + | 100.00 | TM/S | 1-43 | ||
| SA0754 | Multidrug resistance protein | 3 | 5/5 | ND | 70.27 | ND | 155-175 | |
| SA0835 | Conserved hypothetical protein | 65 | 40/42 | 30/32 | + | 100.00 | TM/S | 6-54 |
| SA0856 | Clumping factor A (clfA) | 6 | 2/5 | ND | 21.62 | ND | 87-98 | |
| SA0946 | Conserved hypothetical protein | 5 | 2/5 | ND | 75.68 | TM/S | 105-122 | |
| SA0950 | Na+/H+ antiporter, MnhF component (mnhF) | 66 | 2/5 | ND | 66.67 | ND | 1-21 | |
| SA1045 | Iron compound ABC transporter | 10 | 5/5 | ND | 100.00 | ND | 1-18 | |
| SA1062 | Bifunctional autolysin (atl) | 19 | 8/14 | 13/18 | ND | 100.00 | ND | 64-81 |
| SA1112 | Conserved hypothetical protein | 349 | 14/14 | 18/18 | ND | ND | −/− | 13-37 |
| SA1138 | Cell wall surface anchor family protein | 4 | ND | ND | ND | 53-73 | ||
| SA1168 | Fibrinogen-binding protein (fib) | 3 | ND | 100.00 | ND | 127-141 | ||
| SA1259 | Conserved hypothetical protein | 13 | 2/5 | ND | ND | TM/S | 196-218 | |
| SA1743 | Amino acid permease | 9 | 14/14 | 17/18 | ND | 94.59 | ND | 127-152 |
| SA1777 | Serine protease HtrA, putative | 73 | 17/42 | 9/32 | ND | 100.00 | ND | 1-30 |
| SA1806 | Cell wall surface anchor family protein | 9 | 7/14 | 16/18 | ND | 100.00 | ND | 508-523 |
| SA1919 | Transcriptional regulator | 3 | 13/14 | 18/18 | ND | 100.00 | ND | 120-135 |
| SA2125 | Peptidase, M20/M25/M40 | 3 | 13/14 | 18/18 | ND | 97.30 | ND | 293-312 |
| SA2347 | Drug resistance transporter, EmrB/QacA subfarr | 5 | 14/14 | 18/18 | ND | 100.00 | ND | 205-224 |
| SA2437 | Bicyclomycin resistance protein (bcr) | 8 | 13/14 | 18/18 | ND | 100.00 | ND | 54-104 |
| SA2505 | Cell wall surface anchor family protein | 5 | 19/42 | 7/32 | + | ND | ND | 829-849 |
| SA2511 | Fibronectin-binding protein A (fnbA) | 8 | 13/14 | 12/18 | ND | 100.00 | ND | 129-196, 876-913 |
| SA2652 | Clumping factor B (clfB) | 17 | 41/42 | 32/32 | ND | 100.00 | ND | 711-751 |
| SA2659 | Zinc metalloproteinase aureolysin | 12 | 4/42 | 1/32 | ND | 97.30 | ND | 90-107 |
Listed is a representative subset of 75 genes for which three or more independent clones were identified as mapping to the same region. The Institute for Genomic Research (TIGR) locus and gene name are according to the CMR database (20).
Number of clones located in the respective gene.
Reactivity in peptide ELISA with patient (P) sera or normal-subject high-titer (N) sera (as described in reference 4). Data are presented as number of serum samples positive for reactivity/total number of serum samples tested.
Immunoprecipitation reaction: +, positive; ND, not determined.
Conservation of respective genes in 36 S. aureus isolates as determined by DNA chip hybridization (6). ND, not determined.
Prediction of transmembrane domain or signal peptide (TM/S) (ND, not determined).
Selected region(s) in relative amino acid position.
(ii) Specific enrichment of selected genes.
Shown in Table 2 is the specific enrichment of selected genes allocated to functional classification (TIGRFAM) (20) in comparison to its percentage present in the unselected whole genome. The majority of the identified ORFs can be assigned to cell envelope function (25%), transporter proteins (24%), or pathogenesis- or toxin-involved functions (9%), among them those described to provide protective activity in challenge models (coding for clumping factor A [clfA], fibronectin-binding protein A [fnbpA] and fibrinogen-binding protein [fib]) (12, 14, 23) (Table 1). This result underscores the above-mentioned notion that a functional screen using human sera as selective agent should identify proteins accessible to the immune system and expressed during an infection in vivo. In addition, most selected genes with no assigned function (encoding hypothetical proteins) were found to contain a predicted signal sequence required for export and/or a trans-membrane domain typical for membrane associated proteins, thereby extending the above notion also into this group (Table 1).
TABLE 2.
Grouping of genes identified in this study into functional categories according to the CMR databasea
| Main cellular role | Cellular subrole | In vitro selection
|
Whole genome
|
||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Cell envelope | Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | 3 | 4.00 | 43 | 1.44 |
| Other | 16 | 21.33 | 74 | 2.47 | |
| Total | 19 | 25.33 | 170 | 5.69 | |
| Cellular processes | Cell division | 1 | 1.33 | 18 | 0.60 |
| Pathogenesis | 4 | 5.33 | 58 | 1.94 | |
| Toxin production and resistance | 3 | 4.00 | 87 | 2.91 | |
| Total | 8 | 10.67 | 206 | 6.89 | |
| Hypothetical proteins | Conserved | 10 | 13.33 | 594 | 19.87 |
| Other | 3 | 4.00 | 403 | 13.48 | |
| Total | 13 | 17.33 | 997 | 33.34 | |
| Protein fate | Degradation of proteins, peptides, and glycopeptides | 3 | 4.00 | 46 | 1.54 |
| Protein and peptide secretion and trafficking | 1 | 1.33 | 18 | 0.60 | |
| Total | 4 | 5.33 | 88 | 2.94 | |
| Transport and binding proteins | Amino acids, peptides, and amines | 4 | 5.33 | 61 | 2.04 |
| Anions and cations | 4 | 5.33 | 86 | 2.88 | |
| Carbohydrates, organic alcohols, and acids | 3 | 4.00 | 38 | 1.27 | |
| Other | 7 | 9.33 | 36 | 1.20 | |
| Total | 18 | 24.00 | 290 | 9.70 | |
| Other Total | 13 | 17.33 | 1,239 | 41.44 | |
| Grand Total | 75 | 100.00 | 2,990 | 100.00 | |
Genes were grouped according to the CMR database (TIGRFAM) (20) in comparison to the number of genes and respective percentages in the genome. A small number of genes fit into more than one classification group (compare CMR [http://www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl]).
(iii) Variation in number of clones matching a particular ORF.
The number of clones matching a particular ORF varies considerably, most probably reflecting corresponding antibody abundance and/or affinity (4). The relative identification frequency of the antigens was dependent on the selection round (see below).
(iv) Orientation of clone clusters.
Several clone clusters fall into the orientation opposite that of annotated genes. The transcriptional activity from these clusters and a brief characterization are described below and in Table 4. However, it emphasizes the annotation-independent true genomic nature of our approach and reveals completely new putative targets for vaccine development.
TABLE 4.
Selected novel putative ORFs
| ORFa | No.b | Result ofc:
|
Genome positiond (nucleotides) | Orientatione | ORF length (amino acids) | Selected clone region (nucleotides) | Opposite ORFf | Name of opposite ORF productf | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Peptide ELISA | RT-PCR | NB | ||||||||
| PSAN1 | 6 | ND | + | + | 1553-1443 | − | 37 | 1535-1464 | SAA0002 | Tetracycline resistance protein |
| SAN1 | 9 | N: 5/5 | + | ND | 11772-11671 | − | 34 | 11751-11708 | SA0008 | Histidine ammonia lyase |
| SAN2 | 8 | N: 4/5 | + | ND | 135574-135699 | + | 42 | 135601-135660 | SA0120 | Bacterial regulatory proteins, gntR |
| SAN3 | 9 | ND | + | ND | 301822-301718 | − | 35 | 301816-301743 | SA0263 | Peptidoglycan hydrolase |
| SAN4 | 69 | N: 4/5 | + | ND | 382929-382792 | − | 46 | 382878-382808 | SA0379 | phi PVL ORF 15 and 16 homologue |
| SAN5 | 7 | ND | − | ND | 1038052-1037975 | − | 26 | 1038085-1038018 | SA1028 | Hypothetical protein |
| SAN6 | 21 | ND | + | + | 1616061-1616270 | + | 70 | 1616109-1616161 | SA1579 | Conserved hypothetical protein |
| SAN7 | 86 | P: 36/42 N: 29/32 | + | + | 1858035-1858460 | + | 142 | 1858134-1858159 | SA1806 | LPXTG-motif cell wall protein |
| SAN8 | 5 | ND | + | ND | 2122207-2122449 | + | 81 | 2122291-2122377 | SA2055 | rsbW protein |
| SAN9 | 5 | ND | + | + | 2221258-2221758 | + | 167 | 2221486-2221680 | SA2150 | mrp protein |
| SAN10 | 256 | P: 34/42 | + | + | 2276258-2276142 | − | 39 | 2276261-2494441 | SA2194 | Hyaluronate lyase |
| N: 26/32 | ||||||||||
| SAN11 | 8 | N: 3/5 | − | ND | 2290161-2290319 | + | 53 | 2290173-2290241 | SA2208 | tRNA pseudouridine synthase A |
| SAN12 | 4 | ND | + | ND | 2723814-2723680 | − | 45 | 2723822-2723753 | SA2661 | Hypothetical protein |
| SAN13 | 3 | N: 3/5 | − | ND | 2728321-2728259 | − | 21 | 2728315-2728266 | SA2664 | Mannose-6-phosphate isomerase |
Consecutive numbering of novel ORFs, SAN, S. aureus New; prefix P, localization on plasmid of S. aureus COL strain.
Number of selected clones located in the respective putative ORF.
Peptide ELISA results show reactivity with patient sera (P) or healthy-subject high-titer sera (N) (as described in reference 4). Expression analysis by RT-PCR and Northern blotting (NB) were also carried out. ND, not determined.
Position of the putative ORFs on the genome according to CMR database (20).
Orientation of the putative ORFs on the plus strand (+) or negative strand (−) of the genome.
The opposite ORF and its name are the ORF number and name of the gene that is located in antisense direction to the putative novel ORF.
(v) Gene conservation among 36 S. aureus isolates.
A comprehensive study on gene conservation among 36 S. aureus isolates was recently performed by DNA chip hybridization technology (6). Of the immunodominant ORFs identified in our study, 66% were present in all of the 36 different strains tested and 82% of the ORFs were conserved in more than 90% of the strains (Table 1).
Repetition of the selection with a similar serum set revealed a largely overlapping set of ORFs, indicating the reproducibility of the screen (data not shown).
Selection dynamics of the identified clones.
In order to visualize the enrichment of frequently identified clones during the selection cycles, semiquantitative PCRs using DNA fragment-specific primers were performed. Input DNA library as well as DNA pools from the consecutive rounds of selection was used as a template. Examples of the gradual enrichment of individual fragments are shown in Fig. 2B. Interestingly, the dynamics and competitive nature of selection for individual fragments is visible. The peak amount of DNA from a number of clones can be seen in rounds three and four, leveling off in the fifth round (ORFs SA0021 and SA2659). Sequencing a number of clones from the third round of selection revealed a wider variety of different clones with low frequency. In the fifth round, however, the pool is dominated by very few clones derived from ORF SA0195 and SA0200 (data not shown). This indicates that at levels where the selection has advanced to the stage that every synthesized peptide—at limiting ribosomes—is able to interact with its cognate antibody, these peptides outcompete others dependent on antibody abundance and/or affinity. This selection dynamic is also observed in in vitro selection of nucleic acids (26).
Serological confirmation of selected peptides.
Next, peptides corresponding to the identified regions of 25 more or less frequently selected clones were synthesized and tested by peptide ELISA. A large fraction of sera from healthy individuals exhibiting a high titer against S. aureus as well as from patients suffering from an S. aureus infection showed reactivity towards the selected peptides (Tables 1 and 3). This demonstrates that the identified peptides interact with the selective agent independently of the ribosome display context. However, the identification of immunodominant peptides is highly dependent on the choice of serum used. This is best exemplified with the aureolysin-derived peptide (Table 3 [SA2659]). It reacts strongly only with one of the sera used in the serum pool for selection but not with any of the other normal high-titer sera tested.
The presence of specific antibodies is molecular proof for the expression of the corresponding antigens in vivo during exposure, colonization, or disease. Many of the epitopes identified have corresponding antibodies in both healthy-subject and patient sera. However, since these different encounters with the human host require different virulence factors (adhesins and toxins) and other bacterial components, it is not surprising that the antigen pattern revealed by sera from healthy individuals with high titers of antibodies (nasally colonized and noncolonized) and from patients with different S. aureus infections varies. Indeed, we have identified epitope-specific antibodies, which seemed to be more prevalent—meaning higher antibody levels and/or more-reactive sera were observed—in one of the two donor populations. For example, SA0317 mainly reacted with the sera with high titers of antibody, whereas SA0195 and SA2505 reacted more frequently with individual patient sera.
Characterization of newly identified putative short ORFs.
Fourteen clone groups are located in the opposite orientation to annotated genes, indicating the presence of putative antisense transcripts, which could encode immunodominant peptides. Analysis of the surrounding areas revealed possible ORFs with sizes varying between 21 and 167 amino acids in length taking into account the closest start (including TTG codons) and stop codon in frame with the selected peptide (Table 4). BLAST analysis of these putative ORFs against the nonredundant database did not reveal any considerable similarity to any annotated sequence (data not shown).
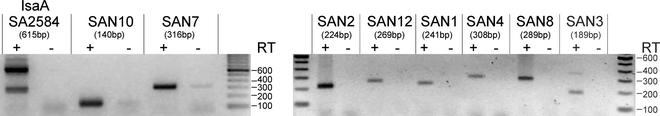
All peptides derived from these clones that were tested in peptide ELISA reacted strongly with a variety of sera (Tables 3 and 4), to an extent similar to that observed with peptides derived from annotated genes. However, we cannot exclude the possibility of cross-reactive antibodies in the sera. To confirm the actual in vivo transcription of these putative short ORFs, total mRNA samples from S. aureus grown in BHI medium were used for mRNA expression analysis. RT-PCR analyses with primers specific for corresponding short ORFs were performed and confirmed the mRNA presence for most putative ORFs that were tested (Fig. 3 and Table 4). Northern analysis with five strand-specific single-stranded probes revealed the existence of mRNA transcripts (Table 4 and data not shown).
FIG. 3.
Transcriptional analysis of putative short ORFs. In vivo transcription of putative novel ORFs was determined by RT-PCR. RNA was extracted from S. aureus COL. RT-PCR with specific primers for the indicated novel ORFs and the IsaA gene as a control was carried out. The expected length of the RT-PCR products is shown in parentheses. Lanes: −, controls without RT, followed by PCR; +, lanes subjected to RT-PCR.
In vivo expression analysis of in vitro-selected peptides.
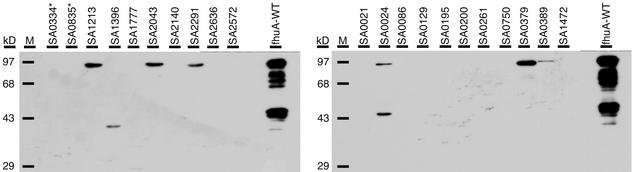
Finally, we wanted to experimentally verify the theoretical advantage of this in vitro expression and selection system (ribosome display) for displaying a variety of genomically encoded peptides. Certain peptides might interfere with the physiology in general or cannot be presented in an in vivo-based display system. Clones corresponding to 19 different ORFs that were identified in the in vitro selection were transferred to an FhuA-based surface display system in E. coli (5). The resulting clones were grown and protein extracts were taken. The expression and stability of the FhuA fusion proteins were tested with an FhuA-specific antibody and Western analysis. As can be seen in Fig. 4, only 6 out of 19 clones tested show full-length FhuA fusion protein, probably indicating that the majority of fusion proteins cannot be displayed on the surface of E. coli and may therefore be rapidly degraded in the cytoplasm or periplasm. In additional experiments, more clones were tested, and approximately 55% of clones that were identified by ribosome display did not show reactivity in Western blots when expressed in the FhuA platform. A similar percentage of reactivity was observed upon testing genomically encoded peptides from Streptococcus pyogenes (data not shown), indicating the general nature of this phenomenon.
FIG. 4.
In vivo expression analysis of in vitro-selected peptides. Assessment of in vivo expression by inserting 19 clones identified by in vitro selection into loop 5 of the fhuA gene. Recombinant E. coli was induced for fhuA expression, and extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with antibodies against FhuA. As controls, wild-type fhuA (positive control) and two clones (SA0835 and SA0334, indicated by an asterisk) containing a stop codon in the library fragment (negative controls), respectively, are shown.
DISCUSSION
In this study we describe the identification of novel antigens from the pathogen S. aureus by in vitro protein selection. For the first time we show that ribosome display allows the simultaneous selection for a set of interacting proteins from genomic expression libraries using a polyclonal IgG preparation and not a single ligand as a selective agent. The display of all genomically encoded peptides provides an effective means of selection independent of gene annotation and reading frame, as shown here with the identification of 14 immunoreactive peptides encoded in putative novel short ORFs.
The use of human antibodies exhibiting a high titer of antibody against S. aureus allowed the selection of proteins, which are expressed in vivo during an infection and are able to induce an immune response, thereby revealing new possible candidate vaccine peptides against S. aureus. However, note that some protective proteins might be nonimmunogenic or immunorecessive in S. aureus infections and may be only immunogenic upon massive encounter as recombinant proteins and therefore will be missed by our approach.
Although most of the identified peptides reacted with high-titer and healthy-patient sera to various extents, some alterations for individual peptides could be seen. Antibodies missing from sera of acute-phase patients can be considered especially important, since it can be speculated that those provide protection in the healthy individuals. It is an important task to identify these missing antibodies and their corresponding epitopes for the design of protective or preventive vaccination. However, it is beyond the scope of the present work. Notably, for some genes a cytoplasmic rather than a surface localization was expected from the function they were assigned to. However, even peptides derived from these genes react very strongly with a number of different sera. This could indicate that the localization of the respective proteins is not restricted to the cytoplasm but is also accessible to an immune attack (compare, e.g., reference 16), or they act highly immunogenic after destruction of the bacterial cell, leading to the release of cytoplasmic proteins and subsequent raising of specific antibodies. However, no highly abundant cytoplasmic proteins (e.g., ribosomal proteins or EF-Tu) were identified, making this interpretation less likely.
Surprisingly, a number of identified clones mapped to orientations opposite from those of known genes. Reactivity with serum and transcriptional activity in vivo was confirmed, therefore raising the possibility for the existence of novel putative short ORFs. Final proof would be the isolation of the corresponding proteins; these proteins are potential novel targets for vaccine development. However, it should be noted that positive serum reactivity could also arise from cross-reacting antibodies. On the other hand, it was recently observed that antisense transcription of bacterial and even mammalian genomes is a general phenomenon (24, 28), allowing the assumption that some of these antisense transcripts are translated in vivo. The concept of nontraditional or “hidden” antigens is known for T-cell epitopes (15) but has to our knowledge never been demonstrated to this extent for B-cell epitopes. Moreover, the evidence provided here for the existence of antisense transcripts to known genes could also indicate a regulatory function on the RNA level, as it was shown that antisense transcripts very efficiently turn down the expression of the corresponding genes (11). Interestingly, some of the putative short ORFs lie opposite to genes involved in virulence, like tet (which codes for tetracycline resistance protein), mrp (which codes for methicillin resistance protein), or hysA (which codes for hyaluronate lyase) (Table 4).
Finally we wanted to directly compare the expressibility of the identified clones in an in vivo-based system. About 55% of the identified clones could not be expressed in an FhuA-based surface display system in E. coli. Besides the possibility that toxic fragments interfere with growth, it is known that membrane permeation of certain protein domains is a challenge due to the high selectivity of the outer membrane translocase (1). Besides for bacterial surface display, membrane permeation is also required for (nonlytic) phage display (e.g., M13 based) as well as yeast display; our observation can therefore probably be generalized also to these systems, although the extent to which the generalization applies might vary. Interestingly, a major class of peptides that cannot be displayed belongs to membrane-associated transporter proteins (compare Fig. 4 and Table 1).
Taken together, our report provides evidence that in vitro protein selection allows the rapid and comprehensive identification of immunodominant peptides from genomic expression libraries and therefore to extensively map the human antibody response against a particular pathogen. Moreover, individual humoral responses between healthy individuals or patients can be compared on a large scale when performing screens with only one serum sample.
Furthermore, the identified peptides are excellent vaccine candidates. Due to the specific mapping of immunoreactive regions within a protein, it spares the often-difficult expression of the corresponding full-length recombinant protein by providing the possibility of using defined synthetic peptides for testing and formulation of vaccine preparations. These advantages suggest in vitro protein selection as a generally applicable approach for characterization of the humoral immune response and therefore aiding the identification of active or passive vaccine candidates.
TABLE 3.
—Continued
| Patient serum | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 15 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 42 | 44 | 45 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 |
| — | 2 | 1 | 1 | — | 1 | 1 | 1 | — | — | 2 | 1 | — | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 1 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 1 | 2 | 3 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| — | — | — | 1 | 2 | — | — | — | — | — | 1 | 2 | 2 | 1 | — | 1 | — | — | 2 | 1 | — | — | — | 2 | — | — | — | 2 | — | — | — | — | 1 | — | — | — | — | 1 | — | — | 1 | — |
| 1 | 3 | 1 | 3 | 3 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 3 | 2 | 1 | 2 | 1 | 2 | 3 | 2 | 1 | 1 | 1 | 3 | 1 | 3 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 2 |
| — | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 2 | 3 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | — | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | — | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 3 | 2 | 2 | 1 | 2 | 2 | 3 | 3 | 1 | 1 | 1 |
| 1 | 2 | — | 2 | — | 2 | 1 | 2 | 1 | — | 1 | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 2 | 3 | 1 | 3 | 2 | 3 | 2 | 3 | 2 | 2 | 2 | 3 | 3 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 2 | 3 | 1 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| — | — | — | 1 | 1 | 1 | — | 1 | — | — | — | 3 | 1 | 2 | — | 1 | — | — | 1 | 3 | — | — | — | 1 | — | 3 | — | — | — | — | — | 1 | 1 | — | — | — | — | 3 | 1 | 1 | — | — |
| 2 | 3 | — | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 3 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 2 | 3 | — | 2 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 2 | 3 | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 1 | 2 | 3 | 2 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 2 | 3 | 1 | 3 | 2 | 3 | 2 | 3 | 2 | 1 | 2 | 3 | 2 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| — | — | — | — | — | — | — | — | — | — | 1 | 1 | — | — | 1 | — | — | — | 1 | — | — | — | — | — | 2 | 1 | — | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | — | 1 | 2 | 2 | 2 | — |
| 1 | 3 | — | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 1 | 2 | 1 | 3 | 3 | 2 | 1 | 1 | 3 | 2 | 3 | 3 | 3 | 2 | 2 | 3 | 1 | 3 | 3 | 2 | 1 | 2 | 2 | 3 | 1 | 2 | 1 | 3 | 2 | 1 | — | 1 | 3 | 3 | 2 | 2 | 2 | 2 | 1 | 3 | 2 | 2 |
| — | — | — | — | — | — | — | — | — | — | — | — | 1 | — | — | — | — | — | 1 | — | — | — | — | 1 | — | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 1 | 1 | — | 2 | 2 | 1 | — | 1 | 1 | 1 | — | 1 | 2 | 1 | 1 | 3 | — | 2 | 2 | 1 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 2 | 2 | 1 | — | — | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 2 | 1 |
| 1 | — | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 3 | 2 | 2 | 1 | 1 | 1 | — | 2 | 2 | 2 | — | — | 1 | 2 | 1 | 3 | — | 2 | 1 | 1 | — | — | 1 | 2 | 1 | 1 | 1 | 1 | 1 | — | 2 | 1 |
Acknowledgments
We thank Wolfgang Zauner for peptide synthesis and stimulating discussions, Markus Hanner for plasmid pMAL4.34, and Christine Triska and Ika Baumgartner for technical assistance.
Editor: D. L. Burns
REFERENCES
- 1.Braun, P., G. Gerritse, J. M. van Dijl, and W. J. Quax. 1999. Improving protein secretion by engineering components of the bacterial translocation machinery. Curr. Opin. Biotechnol. 10:376-381. [DOI] [PubMed] [Google Scholar]
- 2.Burnie, J. P., R. C. Matthews, T. Carter, E. Beaulieu, M. Donohoe, C. Chapman, P. Williamson, and S. J. Hodgetts. 2000. Identification of an immunodominant ABC transporter in methicillin-resistant Staphylococcus aureus infections. Infect. Immun. 68:3200-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colque-Navarro, P., M. Palma, B. Soderquist, J. I. Flock, and R. Mollby. 2000. Antibody responses in patients with staphylococcal septicemia against two Staphylococcus aureus fibrinogen binding proteins: clumping factor and an extracellular fibrinogen binding protein. Clin. Diagn. Lab. Immunol. 7:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etz, H., D. B. Minh, T. Henics, A. Dryla, B. Winkler, C. Triska, A. P. Boyd, J. Sollner, W. Schmidt, U. Von Ahsen, M. Buschle, S. R. Gill, J. Kolonay, H. Khalak, C. M. Fraser, A. Von Gabain, E. Nagy, and A. Meinke. 2002. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 99:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etz, H., D. B. Minh, C. Schellack, E. Nagy, and A. Meinke. 2001. Bacterial phage receptors, versatile tools for display of polypeptides on the cell surface. J. Bacteriol. 183:6924-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanes, J., L. Jermutus, S. Weber-Bornhauser, H. R. Bosshard, and A. Plückthun. 1998. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc. Natl. Acad. Sci. USA 95:14130-14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanes, J., and A. Plückthun. 1997. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94:4937-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, M., and M. J. Taussig. 1997. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 25:5132-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann, K., and W. Stoffel. 1993. TMBASE: a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 11.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josefsson, E., O. Hartford, L. O'Brien, J. M. Patti, and T. Foster. 2001. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 184:1572-1580. [DOI] [PubMed] [Google Scholar]
- 13.Kornblum, J. S., S. J. Projan, S. L. Moghazeh, and R. P. Novick. 1988. A rapid method to quantitate non-labeled RNA species in bacterial cells. Gene 63:75-85. [DOI] [PubMed] [Google Scholar]
- 14.Mamo, W., M. Boden, and J. I. Flock. 1994. Vaccination with Staphylococcus aureus fibrinogen binding proteins (FgBPs) reduces colonisation of S. aureus in a mouse mastitis model. FEMS Immunol. Med. Microbiol. 10:47-53. [DOI] [PubMed] [Google Scholar]
- 15.Mayrand, S. M., and W. R. Green. 1998. Non-traditionally derived CTL epitopes: exceptions that prove the rules? Immunol. Today 19:551-556. [DOI] [PubMed] [Google Scholar]
- 16.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemoto, N., E. Miyamoto-Sato, Y. Husimi, and H. Yanagawa. 1997. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 414:405-408. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Nierman, W., J. A. Eisen, and C. M. Fraser. 2000. Microbial genome sequencing 2000: new insights into physiology, evolution and expression analysis. Res. Microbiol. 151:79-84. [PubMed] [Google Scholar]
- 20.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 22.Roberts, R. W., and J. W. Szostak. 1997. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 94:12297-12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schennings, T., A. Heimdahl, K. Coster, and J. I. Flock. 1993. Immunization with fibronectin binding protein from Staphylococcus aureus protects against experimental endocarditis in rats. Microb. Pathog. 15:227-236. [DOI] [PubMed] [Google Scholar]
- 24.Selinger, D. W., K. J. Cheung, R. Mei, E. M. Johansson, C. S. Richmond, F. R. Blattner, D. J. Lockhart, and G. M. Church. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262-1268. [DOI] [PubMed] [Google Scholar]
- 25.Vytvytska, O., E. Nagy, M. Bluggel, H. E. Meyer, R. Kurzbauer, L. A. Huber, and C. S. Klade. 2002. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics 2:580-590. [DOI] [PubMed] [Google Scholar]
- 26.Wilson, D. S., and J. W. Szostak. 1999. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68:611-647. [DOI] [PubMed] [Google Scholar]
- 27.Wizemann, T. M., J. H. Heinrichs, J. E. Adamou, A. L. Erwin, C. Kunsch, G. H. Choi, S. C. Barash, C. A. Rosen, H. R. Masure, E. Tuomanen, A. Gayle, Y. A. Brewah, W. Walsh, P. Barren, R. Lathigra, M. Hanson, S. Langermann, S. Johnson, and S. Koenig. 2001. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yelin, R., D. Dahary, R. Sorek, E. Y. Levanon, O. Goldstein, A. Shoshan, A. Diber, S. Biton, Y. Tamir, R. Khosravi, S. Nemzer, E. Pinner, S. Walach, J. Bernstein, K. Savitsky, and G. Rotman. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21:379-386. [DOI] [PubMed] [Google Scholar]