Abstract
A self-transferable plasmid of ca. 80 kb, pIP1204, conferred multiple-antibiotic resistance to Klebsiella pneumoniae BM4536, which was isolated from a urinary tract infection. Resistance to β-lactams was due to the blaTEM1 and blaCTX-M genes, resistance to trimethroprim was due to the dhfrXII gene, resistance to sulfonamides was due to the sul1 gene, resistance to streptomycin-spectinomycin was due to the ant3"9 gene, and resistance to nearly all remaining aminoglycosides was due to the aac3-II gene and a new gene designated armA (aminoglycoside resistance methylase). The cloning of armA into a plasmid in Escherichia coli conferred to the new host high-level resistance to 4,6-disubstituted deoxystreptamines and fortimicin. The deduced sequence of ArmA displayed from 37 to 47% similarity to those of 16S rRNA m7G methyltransferases from various actinomycetes, which confer resistance to aminoglycoside-producing strains. However, the low guanine-plus-cytosine content of armA (30%) does not favor an actinomycete origin for the gene. It therefore appears that posttranscriptional modification of 16S rRNA can confer high-level broad-range resistance to aminoglycosides in gram-negative human pathogens.
Despite the development of new β-lactams and fluoroquinolones, aminoglycosides are still used for the treatment of severe infections caused by gram-negative organisms. Bacterial resistance to these drugs has been reported since their introduction into clinical use (37). There are three known mechanisms of resistance to aminoglycosides in bacterial human pathogens: (i) decreased intracellular accumulation of the antibiotic by alteration of outer membrane permeability (15), diminished inner membrane transport (39), or active efflux (22, 25); (ii) modification of the target by mutation in ribosomal proteins or 16S RNA; and (iii) enzymatic modification of the drug (37), which is the most common. Microorganisms that produce aminoglycosides have developed an additional pathway to avoid suicide. This self-defense mechanism involves posttranscriptional methylation of rRNA by use of S-adenosylmethionine as a cofactor (3).
Aminoglycosides act by causing translational errors and by inhibiting translocation (9). Their target sites include ribosomal domains in which the accuracy of the codon-anticodon is assessed (30, 46). In particular, they bind to a highly conserved motif of 16S RNA, which leads to alterations in the ribosome functions (24, 48). Substitution or methylation of bases involved in the binding between 16S rRNA and aminoglycosides can lead to a loss of affinity for the antibiotic and to the resistance of the host (7, 19, 29).
We have studied multiresistant Klebsiella pneumoniae BM4536 and found that a gene which encodes a putative 16S rRNA m7G methyltransferase is responsible for high-level resistance to a large number of aminoglycosides in this clinical isolate.
MATERIALS AND METHODS
Strains and growth conditions.
K. pneumoniae BM4536 was isolated from a urinary tract infection at the Hôpital Saint-Michel in Paris in 2000. Escherichia coli BM694 (20), a spontaneous mutant of E. coli C1a (35) resistant to nalidixic acid, and E. coli INVαF′ (Invitrogen, Paisley, United Kingdom) were used as recipients for conjugation and cloning, respectively. The strains were grown in brain heart infusion broth and agar (Difco Laboratories, Detroit, Mich.) at 37°C.
Susceptibility testing.
Antibiotic susceptibility was determined by disk diffusion on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France). The MICs of the antibiotics were determined by agar dilution with ca. 104 CFU per spot after 18 h of incubation at 37°C.
DNA preparation and transfer.
Isolation of total DNA and small- and large-scale preparation of plasmid DNA were performed as described previously (35). Restriction with endonucleases was done according to the recommendations of the supplier. Amplification of DNA was performed in a 2400 thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) with Taq (Qbiogene, Inc., Carlsbad, Calif.) or Pfu (Stratagene, La Jolla, Calif.) DNA polymerase, as recommended by the manufacturers. PCR elongation times and temperatures were adjusted according to the expected size of the PCR product and to the nucleotide sequences of the primers, respectively. The primers used for detection and identification of various genes from pIP1204 are listed in Table 1. The aminoglycoside resistance gene was amplified with the specific oligodeoxynucleotides metF and metR (Table 1) by using plasmid pIP1204 DNA as a template. The amplification products were purified by using the QIAquick PCR purification kit (QIAGEN, Inc., Chatworth, Calif.).
TABLE 1.
Sequences of primers used for detection of various genes from pIP1204
| Gene | Primer name | Sequence (5′ → 3′) | Position | GenBank accession no. or reference |
|---|---|---|---|---|
| blaTEM | tem1F | TCGGGGAAATGTGCGCG | 4458-4433 | X67018 |
| tem1R | TGCTTAATCAGTGAGGCACC | 3477-3496 | ||
| ISEcp1 | ecpF | AAAAATGATTGAAAGGTGGT | 603-622 | AF454633 |
| blaCTX-M | P2D | CAGCGCTTTTGCCGTCTAAG | NAa | 13 |
| intI1 | intI1F | ACATGTGATGGCGACGCACGA | 1542-1562 | M95287 |
| intI1R | ATTTCTGTCCTGGCTGGCGA | 2110-2091 | ||
| sul1 | sulF1 | ATGGTGACGGTGTTCGGCAT | 4321-4340 | U12441 |
| sulR | CTAGGCATGATCTAACCCTC | 5160-5141 | ||
| aac3-IIa | aac3F | ATGCATACGCGGAAGGCAAT | 105-124 | L22613 |
| aac3R | CTAACCTGAAGGCTCGCAA | 965-947 | ||
| ant3"9 | aad2F | ATGAGGGTAGCGGTGACCAT | 1918-1937 | AF227505 |
| aad2R | TCATTTACCAACTGACTTGA | 2709-2690 | ||
| dhfrXII | dhfr12F | GCCAATCGGGTTATTGGCAA | 151-170 | AF335108 |
| dhfr12R | TGGGAAGAAGGCGTCACCCTC | 507-487 | ||
| armA | metF | CAAATGGATAAGAATGATGTT | 1975-1995 | This work and AY220558 |
| metR | TTATTTCTGAAATCCACT | 2751-2734 |
NA, not applicable.
Conjugation from K. pneumoniae BM4536 to E. coli BM694 was performed with selection on 40 μg of nalidixic acid per ml and 20 μg of amikacin per ml. Transformation of E. coli INVαF′ was done by the protocol provided by Invitrogen on brain heart infusion agar containing 20 μg of amikacin per ml.
DNA sequence determination and analysis.
Both strands of the DNA were sequenced with synthetic oligodeoxynucleotides and a CEQ 2000 DNA analysis system automatic sequencer (Beckman Coulter, Fullerton, Calif.). The nucleotide and deduced amino acid sequences were analyzed with the GCG sequence analysis software package (version 10.1; Genetics Computer Group, Madison, Wis.). Searches of sequences were performed with the BLAST program, available the National Center for Biotechnology Information website (http://www.ncbi.nim.nih.gov/). Multiple-sequence alignment was performed with the ClustalX program, available at the Institut National de la Recherche Agronomique website (http://genome.jouy.inra.fr/); TreeView software; and the PROSITE database, available at ExPASy (http://www.expasy.org).
Enzymes and chemicals.
T4 DNA ligase was from Biolabs (Beverly, Mass.), and RNaseA (bovine pancreas) was from Calbiochem-Behring (La Jolla, Calif.). The following antibiotics were from the indicated providers: amikacin, ampicillin, and kanamycin B, Bristol-Myers Squibb (Princeton, N.J.); tobramycin, Eli Lilly & Co. (Indianapolis, Ind.); gentamicin, isepamicin, and netilmicin, Schering-Plough Research Institute (Kenilworth, N.J.); and apramycin, spectinomycin, and streptomycin, Sigma-Aldrich (Saint Quentin Fallavier, France).
Nucleotide sequence accession number.
The nucleotide sequence of the armA gene and its flanking regions has been deposited in the GenBank data library under accession number AY220558.
RESULTS
Antibiotic resistance of K. pneumoniae BM4536.
Clinical isolate K. pneumoniae BM4536 was resistant to cefotaxime, trimethoprim, and sulfonamides and had an unusual phenotype of broad-spectrum high-level resistance to aminoglycosides, including amikacin, dibekacin, 5-episisomicin, fortimicin, isepamicin, gentamicin, kanamycin, netilmicin, 2′- and 6′-N-ethylnetilmicin, sisomicin, tobramycin, streptomycin, and spectinomycin (Table 2). Resistance to these drugs was transferable en bloc by conjugation to E. coli BM694. Analysis of the plasmid DNA content of one of the transconjugants, BM4537, indicated the presence of plasmid pIP1204, which was ca. 80 kb, as estimated by agarose gel electrophoresis after digestion with PstI or EcoRI (data not shown).
TABLE 2.
MICs of various aminoglycosides for K. pneumoniae BM4536 and E. coli with and without the armA gene
| Strain | MIC (μg/ml)a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AMI | GEN | ISE | NET | TOB | APR | PAR | STR | SPE | |
| K. pneumoniae BM4536 | 512 | 256 | 512 | 256 | 512 | 1 | 2 | 32 | >256 |
| E. coli | |||||||||
| BM694 | 2 | 0.5 | 0.5 | 0.5 | 0.5 | 2 | 4 | 4 | 8 |
| BM694(pIP1204) | 1,024 | 256 | 1,024 | 256 | 256 | 2 | 4 | 8 | 256 |
| INVαF′ | 1 | 0.25 | 0.5 | 0.25 | 0.25 | 0.5 | 0.5 | 1 | 4 |
| INVαF′(pAT780) | 512 | 256 | 512 | 256 | 128 | 0.5 | 0.5 | 1 | 8 |
| INVαF′(pAT781) | 512 | 256 | 256 | 256 | 128 | 0.5 | 0.5 | 1 | 8 |
| INVαF′(pAT782) | 256 | 128 | 256 | 128 | 128 | 0.5 | 0.5 | 1 | 8 |
Abbreviations: AMI, amikacin; GEN, gentamicin; ISE, isepamicin; NET, netilmicin; TOB, tobramycin; APR, apramycin; PAR, paromomycin; STR, streptomycin; SPE, spectinomycin.
Determination of the sequences of the PCR products obtained with the oligonucleotides listed in Table 1 and pIP1204 DNA as a template indicated that resistance to β-lactams was due to the presence of the blaTEM-1 and blaCTX-M3 genes and also revealed the presence of ISEcp1, which is often located upstream from the latter gene (34). Resistance to streptomycin-spectinomycin was due to the ant3"9 gene, resistance to certain aminoglycosides was due to aac3-IIa, and resistance to trimethroprim was due to a dhfrXII gene. Resistance to sulfonamides was mediated by sul1, and detection of the intI1 structural gene suggested the presence of a type I integron.
Characterization of aminoglycoside resistance.
Plasmid DNA from E. coli BM4537 and pUC18 DNA were digested with HindIII, mixed, ligated, and introduced by transformation into E. coli INVαF′. Transformants selected on medium containing amikacin were screened for their plasmid contents by agarose gel electrophoresis of crude bacterial lysates. The smallest plasmid, pAT780, contained a ca. 4-kb insert that was sequenced. The plasmid conferred to the new host high-level resistance to amikacin, 5-episisomicin, isepamicin, fortimicin, gentamicin, netilmicin, 2′- and 6′-N-ethylnetilmicin, sisomicin, and tobramycin but not to apramycin, lividomycin, neomycin B, paromomycin, or ribostamycin (Table 2 and data not shown).
Sequence analysis of the aminoglycoside resistance gene and the deduced protein.
Comparison of the sequence of the insert in pAT780 with sequences in the GenBank data library revealed identity to a portion of plasmid pCTX-M3 from Citrobacter freundii (positions 70,408 to 74,449; GenBank accession number AF550415). The insert contained four open reading frames (ORFs). The sequence of ORF1 (positions 1 to 579), truncated by the HindIII cloning, was found to be identical to that of the 3′ end of ORF513, which specifies a putative recombinase in E. coli (100% identity over 192 amino acids; GenBank accession number AF174129). The sequence of ORF2 (positions 761 to 1652) was homologous to that of a putative transposase of Providencia rettgeri (76% identity over 245 amino acids; GenBank accession number L06418). The sequence of ORF3 (positions 1826 to 2751) did not share homology with any of the sequences in GenBank, but the deduced sequence was found to be similar to that of the 16S rRNA methyltransferase from Streptoalloteichus hindustanus (27% identity over 257 amino acids; GenBank accession number AF038408). The sequence of the last ORF, truncated by the HindIII cloning (positions 3440 to 4042), was homologous to that of the putative transposase of Tn1330 from Yersinia enterocolitica (40% identity over 192 amino acids; GenBank accession number AJ344215).
Within ORF3, a ribosome binding site, GTAGG (positions 1964 to 1968), was found 9 nucleotides upstream from an ATG initiation codon (position 1978), leading to a 771-bp putative coding sequence. Analysis of the upstream sequence indicated a putative promoter consisting of the −35 (TTGACG) (positions 1792 to 1797) and −10 (TACACT) motifs separated by 18 nucleotides (the underlined bases indicate identity with the −35 and −10 consensus promoter elements recognized by the E. coli σ70 factor). The putative aminoglycoside resistance gene, named armA for aminoglycoside resistance methyltransferase, specifies a protein with a calculated mass of 28,811 Da. The overall guanine-plus-cytosine content of the armA gene (30%) was significantly lower than that of the sequence from the flanking DNA regions (ca. 50%).
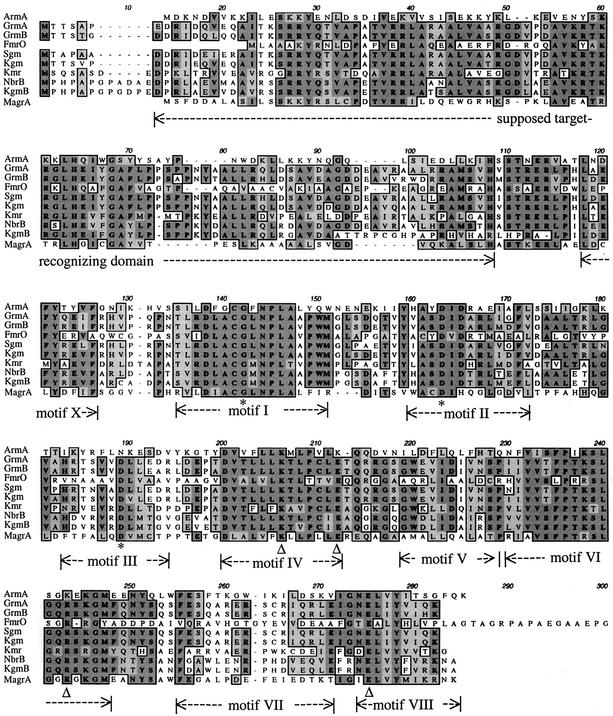
The deduced protein, ArmA, showed homology with a closely related group of 16S rRNA methyltransferases, the Agr (aminoglycoside resistance) family (Table 3 and Fig. 1) (4). These include Grm from Micromonospora purpurea (41) and Micromonospora rosea (17), KgmB from Streptomyces tenebrarius (16), Kmr from Streptomyces kanamyceticus (10), NbrB from Streptoalloteichus hindustanus (accession number AAB95477), Sgm from Micromonospora zionensis (18), Kgm from Streptomyces lividans (40), FmrO from Micromonospora olivasterospora (28), and MagrA from Pseudomonas aeruginosa (accession number AB083212). The levels of identity of ArmA with these proteins were between 21 and 30%, and the levels of similarity were between 37 and 47% (Table 3).
TABLE 3.
Amino acid identities and similarities between 16S rRNA methyltransferases
| Methyltransferasea | % Amino acid similarity or identityb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ArmA | GrmA | GrmB | FmrO | Sgm | Kgm | Kmr | NbrB | KgmB | MagrA | |
| ArmA | 46.9 | 46.0 | 37.3 | 46.2 | 46.2 | 39.4 | 43.3 | 41.2 | 45.0 | |
| GrmA | 25.3 | 95.6 | 41.5 | 96.0 | 98.5 | 70.5 | 74.5 | 70.2 | 49.3 | |
| GrmB | 24.9 | 89.4 | 42.0 | 92.7 | 94.9 | 69.8 | 72.7 | 69.9 | 50.0 | |
| FmrO | 20.8 | 25.3 | 25.9 | 40.2 | 41.1 | 42.9 | 40.0 | 39.7 | 43.6 | |
| Sgm | 26.0 | 89.8 | 86.5 | 24.0 | 94.5 | 69.4 | 72.3 | 68.1 | 48.2 | |
| Kgm | 26.3 | 95.2 | 89.4 | 25.3 | 88.3 | 70.1 | 73.4 | 69.5 | 49.6 | |
| Kmr | 21.5 | 51.4 | 51.1 | 27.4 | 51.8 | 50.7 | 70.6 | 68.7 | 45.0 | |
| NbrB | 25.9 | 59.9 | 55.7 | 28.7 | 57.8 | 58.5 | 54.3 | 89.0 | 49.1 | |
| KgmB | 24.9 | 55.0 | 52.8 | 31.2 | 57.8 | 54.2 | 52.1 | 85.9 | 45.0 | |
| MagrA | 29.8 | 30.8 | 31.5 | 24.7 | 30.4 | 30.8 | 26.6 | 29.9 | 28.2 | |
The methyltransferases were ArmA from K. pneumoniae BM4536, GrmA from M. purpurea (GenBank accession number P24618), GrmB from M. rosea (GenBank accession number P24619), FmrO from M. olivasterospora (GenBank accession number Q08325), Sgm from M. zionensis (GenBank accession number A45282), Kgm from S. lividans (GenBank accession number CAC93944), Kmr from S. kanamyceticus (GenBank accession number CAA75800), NbrB from S. hindustanus (GenBank accession number AAB95477), KgmB from S. tenebrarius (GenBank accession number S17717), and MagrA from P. aeruginosa (GenBank accession number AB083212).
Percent amino acid similarity values are presented in the upper half (above the diagonal space), and percent identity values are given in the lower half (below the diagonal space).
FIG. 1.
Alignment of the deduced sequences of the Agr family. Identical amino acids in all proteins are highlighted with a dark grey background. Homologous amino acids are highlighted with a light grey background. Sequences are from K. pneumoniae (ArmA), M. purpurea (GrmA; GenBank accession number P24618), M. rosea (GrmB; GenBank accession number P24619), M. olivasterospora (FmrO; GenBank accession number Q08325), M. zionensis (Sgm; GenBank accession number A45282), S. lividans (Kgm; GenBank accession number CAC93944), S. kanamyceticus (Kmr; GenBank accession number CAA75800), S. hindustanus (NbrB; GenBank accession number AAB95477), S. tenebrarius (KgmB; GenBank accession number S17717), and P. aeruginosa (MagrA; GenBank accession number AB083212). Due to spatial constraints, the C-terminal extension of FmrO (ATRPVVDVPATARPDADRVDPTG) was omitted. Conserved residues postulated to form the active site in both m7G methyltransferase families are indicated with open triangles. Conserved residues predicted to form the common AdoMet-binding site are indicated with asterisks, and common motifs are delineated by double-head dashed arrows.
Resistance conferred by the armA gene.
The sequence encoding the putative ArmA 16S rRNA methyltransferase was amplified by PCR with primers metF and metR (Table 1); cloned into pCR2, a PCR product cloning vector; and resequenced. The 771-bp EcoRI fragment was ligated into the high-copy-number vector pUC18 to generate pAT781 and into the low-copy-number plasmid pACYC184 to generate pAT782. The recombinant plasmids conferred to E. coli INVαF′ high-level resistance to the 4,6-disubstituted deoxystreptamines and, in addition, to the structurally unrelated compound fortimicin. Susceptibilities to the 4-5-disubstituted deoxystreptamines lividomycin, neomycin B, paromomycin, and ribostamycin were not affected, nor were susceptibilities to apramycin, streptomycin, and spectinomycin (Table 2). Plasmids pIP781 and pIP782 conferred a phenotype of very high-level resistance to the host, indicating a lack of a gene dosage effect for this mechanism. This observation accounts for the fact that plasmid pIP1204, most likely present at a low copy number, is associated with high-level resistance to aminoglycosides that are not substrates for the 3-N-aminoglycoside acetyltransferase type IIa.
DISCUSSION
Aminoglycosides are polycationic molecules, and their positive charges interact with the negatively charged nucleic acids. More specifically, they display high affinities with certain domains of prokaryotic rRNA (31, 32, 33, 44). The flexibility of the aminoglycosides allows their adaptation to the shape of the binding pocket in the internal loops of RNA helix cores to make specific contacts (12, 45, 47). The majority of these antibiotics are composed of amino sugars linked to a 2-deoxystreptamine ring (19). The conserved elements among aminoglycosides are rings I and II and, within ring II, the amino groups at positions 1 and 3. These elements are crucial for the binding to the decoding site of 16S rRNA of prokaryotic ribosomes. The 2-deoxystreptamine ring is substituted, most commonly, at positions 4 and 5, as in the neomycin class, or at positions 4 and 6, as in the kanamycin and gentamicin classes (19).
A highly conserved sequence in the A site of 16S rRNA governs the interaction between the codon and the aminoacyl-tRNA anticodon (8, 27, 48). Most aminoglycosides bind to this region in a pocket created by asymmetry in the internal loop due to the A1408 · A1493 base pair and the single bulged adenine at position 1492 and interfere with several steps of translation, in particular, by inducing codon misreading (9, 36). The link between 16S rRNA and the aminoglycosides is crucial for antimicrobial activity, since mutations in the A site or modification of these antibiotics by bacterial enzymes affects this interaction, leading to resistance. The major difference in the binding sites of these antibiotics between prokaryotic and eukaryotic ribosomes is the nucleotide at position 1408 (E. coli numbering), which is an adenosine in prokaryotes and a guanosine in eukaryotes (31). Study of the interaction of aminoglycosides with small RNA molecules (14, 21, 30) and aptamers (42, 44) combined with the knowledge of the structure of the 30S ribosomal subunit (5) allowed identification of the bases involved in the binding. The nucleotides involved in the activities of the aminoglycosides are confined to a very limited number of bases mainly located in the A site. For example, critical positions for paramomycin binding to the A site include the C1407 · G1494 base pair, A1408, A1493, and U1495 (12). The 16S rRNA alterations which confer resistance to 4,6-linked aminoglycosides could be due to mutations (29) or RNA-modifying enzymes (3, 16, 38), with methylation being the simplest and most common RNA modification.
In prokaryotes, methylation of the bases is usual, whereas in eukaryotes, methylation of the ribose moiety (2′-O-methylation) is far more common. The majority of RNA methylation is carried out by S-adenosyl-l-methionine (AdoMet)-dependent methyltransferases (1, 2, 6). Methyltransferases constitute a large family of ubiquitous enzymes involved in various key biochemical reactions (23). Despite differences in their primary sequences, nine conserved blocks of amino acids have been recognized among these enzymes; and most DNA methyltransferases share a catalytic domain constituted of a β sheet flanked by α helices, related to the nucleotide-binding Rossman fold (23). RNA methylation can have diverse effects on the binding of the aminoglycosides. The naturally occurring m5C1407 located within the aminoglycoside-binding domain of the A site does not adversely affect affinity for the drugs (45). Curiously, the absence of posttranscriptional modification at A1518 and at the adjacent A1519 in strains lacking a functional KsgA methyltransferase leads to kasugamycin resistance (43). By contrast, two methyltransferases with activities involving 16S rRNA that result in resistance to aminoglycosides have been identified. The first one, KamA, which corresponds to the conversion of A1408 to N-1 methyladenosine and which leads to kanamycin and apramycin resistance, has been reported in the istamycin producer Streptomyces tenjimariensis (28). The second, mediated by GmrA, converts G1405 N-7 to 7-methylguanosine and confers resistance to the gentamicin-producing species M. purpurea (3). GmrA belongs to the closely related family of methyltransferases designated Agr, which includes 10 members, all but 1 (MagrA from P. aeruginosa) of which originate in actinomycetes. It has been established for at least two members that modification converts G1405 within 16S rRNA to 7-methylguanosine (3), resulting in high-level resistance to certain aminoglycosides; G1405 is located in an extremity of the A site of the decoding region. Interestingly, although the Agr family of methyltransferases differs from other methyltransferases, structure predictions confirm the presence of a putative AdoMet-binding site, albeit at a location distinct from that in the other proteins (4).
The sequence of 257-amino-acid ArmA protein of K. pneumoniae BM4536 exhibited significant similarity with the sequences of the Agr family (Table 3; Fig. 1). The alternative motifs proposed by Bujnicki and Rychlewski (4) for Agr proteins, which are equivalent to the conserved 9-amino-acid blocks in the nucleic acid methyltransferases, were present in ArmA (Fig. 1). Two of the three amino acids predicted to form the AdoMet-binding site (Gly142 and Asp163) were conserved, and amino acids suspected to interact at the active site were identical or similar to those in the other members of the Agr family (Fig. 1). The resistance phenotype conferred by ArmA (Table 1) strongly suggests modification of G1405, since methylation of this base confers resistance to 4,6-linked 2-deoxystreptamine aminoglycosides but does not affect binding of 4,5-linked derivatives (3, 45).
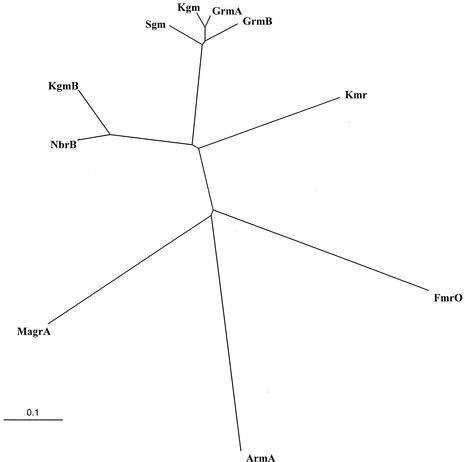
The phylogenetic tree drawn from the primary sequences of the enzymes (Fig. 2) indicated that Sgm, Kgm, GrmA, and GrmB constitute a cluster and that Kgm and NbrB form a second group equally distant from the first cluster and from Kmr. The three remaining proteins, FmrO, MagrA, and ArmA, were similarly distant from each other and from the other Agr enzymes.
FIG. 2.
Phylogenetic relationship among 16S rRNA methyltransferases. The tree was constructed from an analysis of the sequences presented in Fig. 1 by using the bootstrapping method (11). The scale bar represents a 10% difference in amino acid sequence.
The base composition of armA was significantly different from that of the flanking DNA, suggesting acquisition of an exogenous resistance determinant by plasmid pIP1204. However, its low guanine-plus-cytosine content of 30%, very distinct from those of kgmB (71%) and grmA (65%), does not favor a direct and recent origin of the resistance gene in the actinomycetes. There are few nonactinomycete aminoglycoside producers, such as gram-positive bacilli (26), and because of the absence of sequences homologous to armA in the data banks, the identity of the progenitor of the gene remains open.
Posttranscriptional methylation of rRNA, previously confined to aminoglycoside producers, is a new mechanism of resistance to these drugs in human pathogens. This mechanism is particularly efficient since it modifies all 16S rRNA copies and thus leads to high-level resistance to all the aminoglycosides, except streptomycin, used in human therapy, even when the corresponding gene was present at a low copy number (Table 2). This is in contrast to rRNA gene mutations, which must occur in a minimum number of gene copies to confer clinical resistance (19, 29). The armA gene was borne by conjugative plasmid pIP1204, in which it is flanked by putative transposable elements and linked to blaCTX-M, which confers resistance to broad-spectrum cephalosporins by synthesis of an extended-spectrum β-lactamase. All these elements favor dissemination of armA, and it is thus not surprising that we found the gene in various members of the family Enterobacteriaceae isolated from several European countries (data not shown).
Acknowledgments
We thank L. Sañudo and S. Simon for technical assistance.
REFERENCES
- 1.Alexandrov, A., M. R. Martzen, and E. M. Phizicky. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8:1253-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anantharamian, V., E. V. Koonin, and L. Aravind. 2002. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 4:71-75. [PubMed] [Google Scholar]
- 3.Beauclerk, A. A., and E. Cundliffe. 1987. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J. Mol. Biol. 193:661-671. [DOI] [PubMed] [Google Scholar]
- 4.Bujnicki, J. M., and L. Rychlewski. 2001. Sequence analysis and structure prediction of aminoglycoside-resistance 16S rRNA:m7G methyltransferases. Acta Microbiol. Pol. 50:7-17. [PubMed] [Google Scholar]
- 5.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, X., and R. J. Roberts. 2001. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 29:3784-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cundliffe, E. 1990. Recognition sites for antibiotics within rRNA, p. 479-490. In W. E. Hill, P. B. Moore, A. Dahlberg, D. Schlessinger, R. A. Garrett, and J. R. Warner (ed.), The ribosome: structure, function, and evolution. American Society for Microbiology, Washington, D.C.
- 8.Cunningham, P. R., K. Nurse, A. Bakin, C. J. Weitzmann, M. Pflumm, and J. Ofengand. 1992. Interaction between the two conserved single-stranded regions at the decoding site of small subunit ribosomal RNA is essential for ribosome function. Biochemistry 31:12012-12022. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J., and B. D. Davis. 1968. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. J. Biol. Chem. 243:3312-3316. [PubMed] [Google Scholar]
- 10.Demydchuk, J., Z. Oliynyk, and V. Fedorenko. 1998. Analysis of a kanamycin resistance gene (kmr) from Streptomyces kanamyceticus and a mutant with increased aminoglycoside resistance. J. Basic Microbiol. 38:231-239. [DOI] [PubMed] [Google Scholar]
- 11.Felsentein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 13.Gniadkowski, M., I. Schneider, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamasaki, K., J. Killian, J. Cho, and R. R. Rando. 1998. Minimal RNA constructs that specifically bind aminoglycoside antibiotics with high affinities. Biochemistry 37:656-663. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E. W. 1981. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. J. Antimicrob. Chemother. 8:249-276. [DOI] [PubMed]
- 16.Holmes, D. J., and E. Cundliffe. 1991. Analysis of a ribosomal RNA methylase gene from Streptomyces tenebrarius which confers resistance to gentamicin. Mol. Gen. Genet. 229:229-237. [DOI] [PubMed] [Google Scholar]
- 17.Kelemen, G. H., E. Cundliffe, and I. Financsek. 1991. Cloning and characterization of gentamicin-resistance genes from Micromonospora purpurea and Micromonospora rosea. Gene 98:53-60. [DOI] [PubMed] [Google Scholar]
- 18.Kojic, M., L. Topisirovic, and B. Vasiljevic. 1992. Cloning and characterization of an aminoglycoside resistance determinant from Micromonospora zionensis. J. Bacteriol. 174:7868-7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labigne-Roussel, A., G. Gerbaud, and P. Courvalin. 1981. Translocation of sequences encoding antibiotic resistance from the chromosome to a receptor plasmid in Salmonella ordonez. Mol. Gen. Genet. 182:390-408. [DOI] [PubMed] [Google Scholar]
- 21.Lynch, S. R., R. L. Gonzalez, and J. D. Puglisi. 2003. Comparison of X-ray crystal structure of the 30S subunit-antibiotic complex with NMR structure of decoding site oligonucleotide-paramomycin complex. Structure 11:43-53. [DOI] [PubMed] [Google Scholar]
- 22.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyl-transferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 24.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 25.Moore, R. A., D. Deshazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nara, T., I. Kawamoto, R. Okachi, and T. Oka. 1977. Source of antibiotics other than Streptomyces. J. Antibiot. 30:S174-S189. [PubMed] [Google Scholar]
- 27.Ogle, J. M., E. Brodersen, W. M. Clemons, M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 28.Ohta, T., and M. Hasegawa. 1993. Analysis of the self-defense gene (fmrO) of a fortimicin A (astromicin) producer, Micromonospora olivasterospora: comparison with other aminoglycoside-resistance-encoding genes. Gene 127:63-69. [DOI] [PubMed] [Google Scholar]
- 29.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. Bottger, and R. J. Wallace, Jr. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573-1581. [DOI] [PubMed] [Google Scholar]
- 30.Purohit, P., and S. Stern. 1994. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature 370:659-662. [DOI] [PubMed] [Google Scholar]
- 31.Recht, M. I., S. Douthwaite, K. D. Dahlquist, and J. D. Puglisi. 1999. Effect of mutations in the A site of 16S rRNA on aminoglycoside antibiotic-ribosome interaction. J. Mol. Biol. 286:33-43. [DOI] [PubMed] [Google Scholar]
- 32.Recht, M. I., S. Douthwaite, and J. D. Puglisi. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Recht, M. I., D. Fourmy, S. C. Blanchard, K. D. Dahlquist, and J. D. Puglisi. 1996. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J. Mol. Biol. 262:421-436. [DOI] [PubMed] [Google Scholar]
- 34.Saladin, M., V. T. B. Cao, T. Lambert, J.-L. Donay, J.-L. Hermann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philipon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schroeder, R., C. Waldsich, and H. Wank. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 19:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skeggs, P. A., D. J. Holmes, and E. Cundliffe. 1987. Cloning of aminoglycoside-resistance determinants from Streptomyces tenebrarius and comparison with related genes from other actinomycetes. J. Gen. Microbiol. 133:915-923. [DOI] [PubMed] [Google Scholar]
- 39.Taber, H. W., J. P. Mueller, P. F. Miller, and A. Arrow. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano, E., J. White, C. J. Thompson, and M. J. Bibb. 1995. Construction of thiostrepton-inducible high-copy-number expression vectors for use in Streptomyces spp. Gene 166:133-137. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, C. J., P. A. Skeggs, and E. Cundliffe. 1985. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer, Micromonospora purpurea. Mol. Gen. Genet. 201:168-173. [DOI] [PubMed] [Google Scholar]
- 42.Tok, J. B. H., J. Cho, and R. R. Rando. 2000. RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct. Nucleic Acids Res. 15:2902-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Buul, C. P., and P. H. Knippenberg. 1985. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in ribosomal RNA. Gene 38:65-72. [DOI] [PubMed] [Google Scholar]
- 44.Wallace, S. T., and R. Schroeder. 1998. In vitro selection and characterization of streptomycin-binding RNAs: recognition discrimination between antibiotics. RNA 4:112-123. [PMC free article] [PubMed] [Google Scholar]
- 45.Wong, C.-H., M. Hendrix, E. S. Priestley, and W. A. Greenberg. 1998. Specificity of aminoglycoside antibiotics for the A-site of the decoding region of ribosomal RNA. Chem. Biol. 5:397-406. [DOI] [PubMed] [Google Scholar]
- 46.Woodcock, J., D. Moazed, M. Cannon, J. Davies, and H. F. Noller. 1991. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J. 10:3099-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 22:6437-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1999. Recognition of the codon-anticodon helix by ribosomal RNA. Science 285:1722-1725. [DOI] [PubMed] [Google Scholar]