Abstract
Bloom syndrome is a disorder of profound and early cancer predisposition in which cells become hypermutable, exhibit high frequency of sister chromatid exchanges, and show increased micronuclei. BLM, the gene mutated in Bloom syndrome, has been cloned previously, and the BLM protein is a member of the RecQ family of DNA helicases. Many lines of evidence suggest that BLM is involved either directly in DNA replication or in surveillance during DNA replication, but its specific roles remain unknown. Here we show that hBLM can suppress both the temperature-sensitive growth defect and the DNA damage sensitivity of the yeast DNA replication mutant dna2-1. The dna2-1 mutant is defective in a helicase-nuclease that is required either to coordinate with the crucial Saccharomyces cerevisiae (sc) FEN1 nuclease in Okazaki fragment maturation or to compensate for scFEN1 when its activity is impaired. We show that human BLM interacts with both scDna2 and scFEN1 by using coimmunoprecipitation from yeast extracts, suggesting that human BLM participates in the same steps of DNA replication or repair as scFEN1 and scDna2.
Eukaryotic genomes encode hundreds of genes with homology to DNA helicases. Many of these show functional overlap, making it difficult to assign the precise DNA structures that are unwound by the respective gene products during DNA replication, recombination, and repair. One class of human helicase, the RecQ family, has received special attention in the past few years, because mutations in three different RecQ helicase family members give rise to three distinct human disorders leading to cancer predisposition and/or segmental premature aging, Bloom syndrome, Werner syndrome, and Rothmund–Thomson syndrome (1–3). The genes affected in the diseases, BLM, WRN, and RECQ4, have a single, sequence-specific homolog in yeast called SGS1 (4, 5). Both hBLM and hWRN have been shown to be biologically active in yeast by demonstrating the effects of their expression on sgs1Δ mutants. mWRN suppresses the slow-growth phenotype and the inhibition of type II recombination at telomeres observed in est2Δsgs1Δ survivors (6). Both hBLM and hWRN suppress the excessive illegitimate and homologous recombination phenotypes of sgs1Δ mutants (7). However, hBLM and hWRN are not completely interchangeable in yeast. hBLM, but not hWRN, restores slow growth in the sgs1Δtop3 mutants (7–10). Similarly, hBLM, but not hWRN, suppresses the premature aging phenotype of yeast sgs1Δ mutants (8). Differential function in yeast is consistent with structural and biochemical differences between BLM and WRN. BLM and WRN are homologous mainly in the helicase domain but are divergent in N-terminal and C-terminal domains. WRN, but not BLM, contains an intrinsic nuclease activity in addition to helicase activity (see ref. 2 for review). There are also clear differences in physiological function, because the symptoms of Werner and Bloom syndromes and types of cellular damage in humans differ. Taken together, the sgs1 suppression studies clarify the function of BLM and WRN by suggesting that they suppress recombination. Nevertheless, the suppression studies leave many questions unanswered, because there are many mechanisms by which a gene product might suppress recombination.
SGS1 interacts genetically with several other yeast helicase-encoding genes, including the DNA2 helicase-nuclease identified in our laboratory (11–13). Although Dna2 is not a member of the RecQ helicase subfamily, and encodes both a nuclease and helicase activity, sgs1Δdna2-2 mutants are synthetically lethal, suggesting that their defects are synergistic (12, 14). DNA2 is essential and is required for the elongation stage of DNA replication in vivo and in vitro (11, 15–18). High-copy-number suppression of the temperature-sensitive growth defect of a yeast rad27Δ strain by DNA2 (and vice versa), synthetic lethality of the dna2-1 allele with a rad27Δ mutation and with a DNA ligase mutant, and biochemical evidence for an interaction between Dna2 and the RAD27 gene product FEN1 suggest that Dna2 may be involved in Okazaki fragment maturation (19–22). Interaction of DNA2 with lagging strand-replication functions has also been shown in Schizosaccharomyces pombe (23). A strand-displacement model for removal of RNA primers during Okazaki fragment processing was proposed that incorporated roles for both FEN1 and Dna2 (24). In this model, pol δ displaces the RNA primer of the previously synthesized Okazaki fragment, providing a flap structure that is endonucleolytically processed by sequential action of Dna2 and FEN1 in preparation for ligation (24). In vitro reconstitution of the processing reaction showed that, if the flap was >30 nt, then Dna2 was required to stimulate the endonucleolytic activity of FEN1; however, Dna2 had no effect if the flap was <30 nt (24, 25). Thus, Dna2 is likely to be essential for Okazaki fragment maturation only in cases where FEN1 activity is somehow impaired (25, 26).
Dna2 is conserved in primary sequence from yeast to human (23, 27, 28). In Xenopus, depletion of Xenopus laevis DNA2 (xDNA2) from egg extracts leads to inhibition of DNA replication, and Caenorhabditis elegans dna2 mutants appear to be replication-deficient (29), so the function of Dna2 in DNA replication is also conserved (27). The human genome contains three genes related to DNA2 in the helicase domain, although only one of them also shows conservation throughout the length of the protein, including the nuclease domain (27, 28). Yeast dna2 mutants are sensitive to x-rays, methyl methanesulfonate (MMS), bleomycin, and hydroxyurea (HU) (15, 16). The mechanism by which Dna2 participates in base excision repair (MMS sensitivity) may be related to its role in DNA replication, because reconstitution of the repair reaction requires the same proteins as those involved in Okazaki fragment maturation (26). Some hypomorphic alleles of dna2 induce high levels of recombination in the yeast rDNA, and yeast dna2 mutants have short lifespans (12). Subnuclear localization of Saccharomyces cerevisiae (sc) Dna2 in yeast is notable. The bulk of scDna2 is localized to telomeres in the G1 and G2 phases of the cell cycle (16). The scDna2 protein is released in S phase and is then found throughout the replicating chromatin. The scDna2 protein is also released from telomeres by treatment of cells with agents that cause DNA damage (16). The scDna2 relocalization from telomeres in response to DNA damage requires the checkpoint gene MEC1 (unpublished data). All of these phenomena suggest that scDna2 is a major suppressor of genome instability.
In this study, we show that both the human DNA2 and BLM genes can suppress the temperature-sensitive growth and DNA repair defects of dna2 mutants, suggesting that hBLM is involved in DNA replication.
Materials and Methods
Strains. All of the strains used in this study were derived from W303 (MATa ade2-1 can1-100 trp1-1 leu2-3,112 his3-11,15 ura3). The dna2-1 derivative has been described (12). The rad27Δ and BJ5459 strains have been described (15). Yeast cultures, transformations, and other techniques were as described (30). All media containing galactose also contained raffinose.
Plasmids and Constructs. hBLM was excised from pBluescript II KS(+)-human (h) BLM (gift of Y. Furuichi, AGENE Research Institute, Kanagawa, Japan) with NotI/XhoI and inserted into the yeast expression vector pRS316 behind the GAL promoter. The helicase defective derivative was made by subcloning the K695T mutation from pC4 BLM K695T (gift of N. Neff, New York Blood Center, New York) on the EcoRI/SalI fragment into the corresponding sites of the recipient plasmid. The HindIII/XhoI fragment of pcDNA3-FLAG-mWRN and the NotI/XhoI fragment of pcDNA3-FLAG-hRecQ4 were cloned into the corresponding sites of plasmid pRS316/GAL, respectively. (mWRN and hRECQ4 were obtained from Y. Furuichi.) A hDNA2 (GenBank accession no. P51530) fragment containing the complete ORF was cloned by PCR by using pBluescript II SK(+)-hDNA2 (gift of N. Nobuo, Kazusa DNA Research Institute, Chiba, Japan) as a template, and constructed as follows. Based on the nucleotide sequence of the hDNA2 cDNA, a sense primer, 5′-CGGGATCCCGTCCAGGATGGAGCAGCTGAAC-3′ (BamHI site underlined) and an antisense primer, 5′-CCGCTCGAGCGGTTACTTGTCATCGTCATCCTTGTAGTCTTCTCTTTGAAAGTCACCCAA-3′ (XhoI site underlined) were used to amplify the complete ORF. The antisense primer contains a stretch of 24 nt (boldface) that encodes a FLAG tag in-frame to Glu-1060 of hDna2. The PCR fragment was digested with BamHI and XhoI and inserted into the yeast expression vector pRS316 behind the GAL promoter. pYEp24–3a, containing the RAD27 gene on a YEp24-based vector, pJDG-Rad27-myc, encoding scFEN1-myc, and pJDG-DNA2-hemagglutinin (HA) have been described (15).
Measurement of MMS and HU Sensitivity. Overnight cultures were diluted to an OD of 0.2 and grown for an additional 4–5 h. Tenfold serial dilutions from each strain were spotted onto synthetic complete plates containing galactose with or without MMS (0.005%) or HU (150 mM). The plates were then incubated for the time indicated. For each genotype tested, at least two independent strains were assayed.
Protein Blotting. The dna2-1 cells transformed with pRS316, pGAL-hBLM, or pGAL-hDNA2-FLAG were grown in SC raffinose minus uracil (Ura) to an OD of 0.5, and the expression was induced by adding 2% galactose. The cells were then cultured for the time indicated, and extracts were prepared. The cells, 0.5 ml of culture, were pelleted, washed, and resuspended in 100 μl of 50 mM NaOH (pH 10.5) containing 2 mM EDTA, 1 mM PMSF, 2% SDS, 10% glycerol, 5% mercaptoethanol, and a mixture of protease inhibitors (Roche Molecular Biochemicals), and boiled for 5 min. Debris was removed by centrifugation and the supernatant was adjusted to pH 7.0 with 1 M HCl and supplemented with bromophenol blue. Equal amounts of crude lysates were electrophoresed in SDS/7.5% polyacrylamide gels, blotted to nitrocellulose membranes, and analyzed with monoclonal anti-FLAG M2 (Sigma) or polyclonal anti-BLM antibodies (gift of N. Neff). scFen1-myc was detected by anti-myc (9E10, Babco, Richmond, CA) antibodies. scDNA2-HA was detected by anti-HA (12CA5, Roche Molecular Biochemicals) antibodies. Secondary antibodies were purchased from Bio-Rad and detected by using an ECL kit (Amersham Pharmacia) following the manufacturer's instructions.
Immunoprecipitation. Yeast cells were grown in yeast extract/peptone/raffinose (YPR) at 30°C until they reached an OD of 0.5, at which point 2% galactose was added. The cells were then cultured for a further 4 h and pelleted by centrifugation. The pellets were washed with sterile water twice, then resuspended in 200 μl of lysis buffer (50 mM Tris·HCl, pH 7.6/150 mM NaCl/0.2% Nonidet P-40/10% glycerol/15 mM DTT/2 mM EDTA/1 mM PMSF, and a mixture of protease inhibitors). Glass beads (500 μl) were added, and the cells were lysed by vortexing for 5 min. The extracts were clarified by 15 min of centrifugation at 4°C. The supernatant was diluted with lysis buffer and incubated at 4°C for 2 h with protein G-Sepharose beads bound to anti-BLM or anti-myc antibodies or with protein A-Sepharose beads bound to anti-HA antibodies. The beads were washed four times with lysis buffer and boiled in sample buffer, and the immunoprecipitates were resolved by SDS/7.5% PAGE.
Results
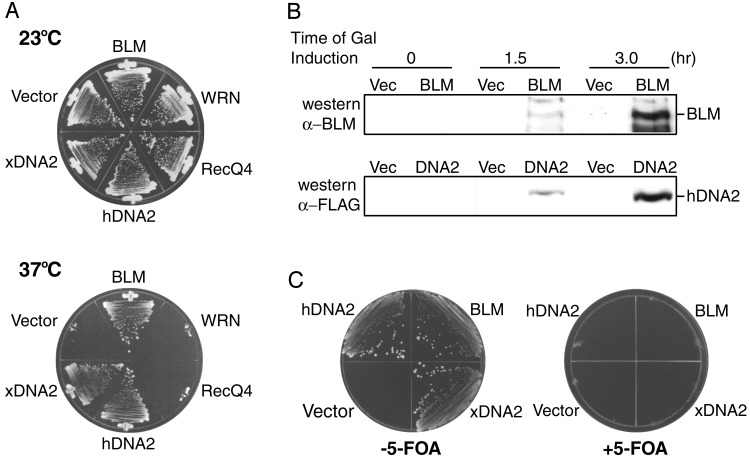
Suppression of Strain dna2-1. Because of the genetic interaction between dna2 and sgs1, we tested whether any of the human RecQ helicase disease genes could suppress any of the phenotypes of dna2 mutants. The putative human DNA2 ORF, hDNA2 (28), the human BLM gene, hBLM (31), the murine FLAG-WRN gene, mWRN (32), and the human FLAG-RECQ4 gene, hRECQ4 (33), were each subcloned into the yeast single-copy, centromeric vector pRS316 under the control of the GAL1,10 yeast promoter. As we had previously shown for the yeast DNA2 gene and the Xenopus DNA2 gene (xDNA2) (11, 27, 34), human DNA2 (hDNA2) suppressed dna2-1 (Fig. 1A), suggesting that this is the functional human Dna2 homolog. Suppression by xDNA2 is shown as a control. As shown in Fig. 1 A, hBLM also suppressed the temperature-sensitive growth defect of a dna2-1 mutant. Surprisingly, the complementation of yeast dna2-1 by hBLM is as efficient as complementation by hDNA2 (Fig. 1 A). Suppression by hBLM required galactose induction. Western blotting revealed that the hBLM protein was not expressed at detectable levels on glucose medium (data not shown), but was present at significant levels after 3 h of galactose induction (Fig. 1B). We conclude that high levels of hBLM can function in yeast to compensate for loss of dna2 function in DNA replication.
Fig. 1.
(A) hBLM and hDNA2 complement the dna2-1 temperature-sensitive phenotype. Strain dna2-1 was transformed with the following plasmids: empty vector (pRS316), pGAL-hBLM, pGAL-FLAG-mWRN, pGAL-FLAG-hRecQ4, pGAL-hDNA2-FLAG, and pGAL-xDNA2. Transformants were streaked onto SC galactose minus Ura and incubated at 23°C(Upper) and 37°C(Lower) for 10 days. (B) Analysis of expression levels for the hBLM and hDNA2 proteins in dna2-1 cells. Strains dna2-1 transformed with empty vector (pRS316), pGAL-hBLM, and pGAL-hDNA2-FLAG were grown in SC galactose minus Ura and harvested at the indicated times to prepare protein extracts. Extracts were prepared by the alkaline lysis method. Equal amounts of protein were loaded onto SDS/7.5% PAGE and immunoblots were probed with either anti-BLM (Upper) or anti-FLAG (Lower) antibodies. The positions of the hBLM and hDNA2 bands are indicated on the right. (C) Strain dna2-1 carrying empty vector (pRS316), pGAL-hBLM, pGAL-hDNA2-FLAG, or pGAL-xDNA2 was streaked onto SC galactose minus Ura plates with (Right) or without (Left) 5-FOA, and grown at 37°C for 7 days.
To ensure that the complementation was caused by the gene introduced on the plasmid and not by chromosomal reversion or suppression, the transformants were also tested on 5-fluoroorotic acid (5-FOA) plates, on which only cells that have lost the URA3 vector plasmid and have become Ura- can grow. There was no growth at 37°C on 5-FOA, indicating that suppression requires the presence of the hBLM, hDNA2, and xDNA2 plasmid-encoded genes (Fig. 1C).
Neither mWRN nor hRECQ4 suppressed the temperature-sensitive growth defect of a dna2-1 mutant (Figs. 1 A and 2C). mWRN and hRECQ4 were expressed under these conditions of induction, but at much lower levels than hBLM (data not shown). Because mWRN and hRECQ4 were not expressed at high levels, and because several attempts to improve expression were unsuccessful, we can draw no conclusions at this time about their ability or inability to suppress dna2-1 replication defects. Thus, the remainder of our studies address primarily hBLM.
Fig. 2.
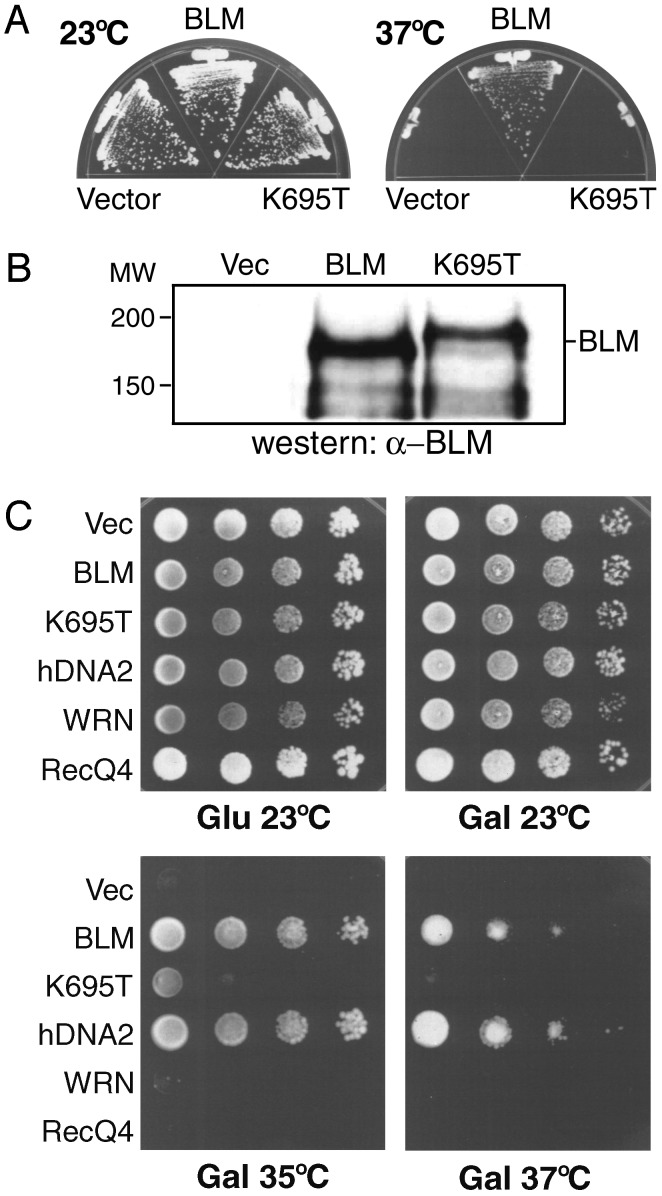
The helicase activity of hBLM is required for complementation of dna2-1. (A) hBLM helicase activity is required for complementation. The dna2-1 cells carrying empty vector (pRS316), pGAL-hBLM, or pGAL-hBLM(K695T) were streaked onto SC galactose minus Ura plates and grown at 23°C or 37°C for 7 days. The K695T mutation changes lysine to arginine at amino acid position 695 within the helicase domain of hBLM. (B) Expression of hBLM(K695T). Strain dna2-1 transformed with various vectors were analyzed by Western blotting with anti-hBLM antibody as described in Fig. 1B. Lane Vec, dna2-1 transformed with empty vector (pRS316); lane BLM, pGAL-hBLM; lane K695T, pGAL-hBLM(K695T). The position of hBLM is indicated. The helicase mutant protein always migrates slightly more slowly than the wild-type protein, as noted (9). (C) Complementation assay at different temperatures. Exponentially growing yeast cultures of dna2-1 cells carrying empty vector (pRS316), pGAL-hBLM, pGAL-hBLM(K695T), pGAL-FLAG-mWRN, pGAL-FLAG-hRecQ4, and pGAL-hDNA2-FLAG were spotted in 10-fold serial dilutions onto SC glucose minus Ura or SC galactose minus Ura plates and grown at 23°C, 35°C, and 37°C for 5–7 days.
Failure of hBLM(K695T) to Suppress Strain dna2-1. To explore the mechanism of suppression by hBLM, a mutation that inactivates the hBLM helicase, K695T (9), was expressed in the same way in the dna2-1 strain. No complementation occurred at 37°C (Fig. 2 A). Because the mutant hBLM and wild-type proteins were expressed at similar levels (Fig. 2B), the helicase activity of hBLM is essential to suppress the dna2-1 defect. We next examined the complementation by a more quantitative dilution assay. The suppression of yeast dna2-1 by hBLM is as efficient as suppression by hDNA2, and the hBLM(K695T) mutant was unable to suppress the dna2-1 defect at 37°C (Fig. 2C). A very low level of viability (10-3) was restored to dna2-1 by hBLM(K695T) at 35°C, which might be caused by stabilization of partially active Dna2 protein by the helicase-dead hBLM protein at the lower temperature (see below).
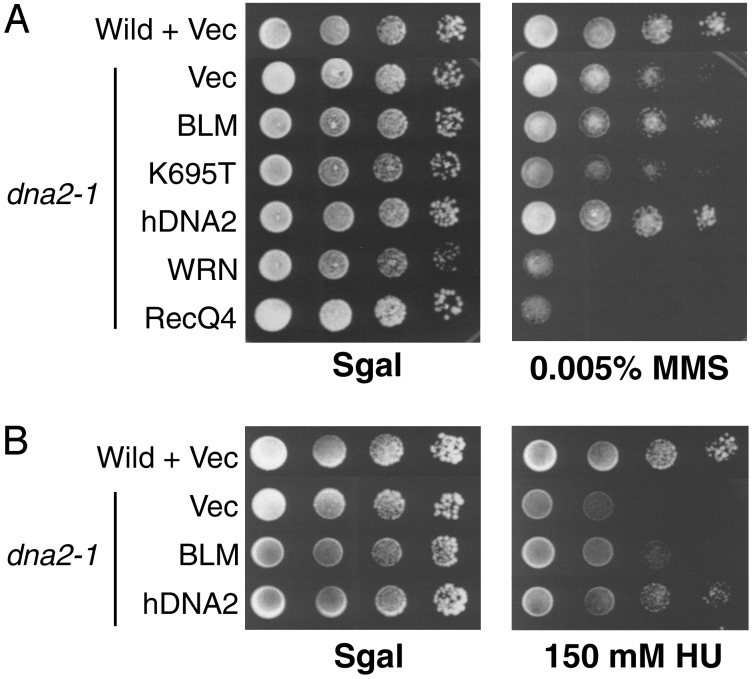
Suppression of HU and MMS Sensitivity of dna2-1 Mutants by hBLM. dna2-1 mutants, like other dna2 mutants (15, 16), are sensitive to MMS, a DNA alkylating agent, and HU, an inhibitor of ribonucleotide reductase, even at the temperature permissive for growth in the absence of drugs (Fig. 3). hDNA2 suppresses both the MMS and HU sensitivity of dna2-1 at the permissive temperature (Fig. 3). Wild-type hBLM also suppresses sensitivity to 0.005% MMS (Fig. 3A) and 150 mM HU (Fig. 3B), but the hBLM helicase mutant (K695T) does not. This finding demonstrates that the ability of hBLM to act as a suppressor of dna2-1 is not uniquely specific for DNA replication, but also includes the ability to suppress sensitivity to DNA-damaging agents. The low level of MMS used here is thought to cause lethal damage during S phase, because it induces the S-phase checkpoint (35). At higher levels of MMS, even hDNA2 fails to complement the damage sensitivity of dna2-1 mutants (data not shown). Both mWRN and hRECQ4 expression caused increased sensitivity of dna2-1 to MMS.
Fig. 3.
Complementation of sensitivity of dna2-1 to MMS and HU. Exponentially growing yeast cultures of wild-type cells carrying empty vector (pRS316) and dna2-1 cells carrying empty vector, pGAL-hBLM, pGAL-hBLM(K695T), pGAL-FLAG-mWRN, pGAL-FLAG-RecQ4, and pGAL-hDNA2-FLAG were spotted in 10-fold serial dilutions onto SC galactose minus Ura plates with or without MMS or HU at the concentrations indicated and grown at the permissive temperature for 6 days. This experiment was performed in duplicate with identical results.
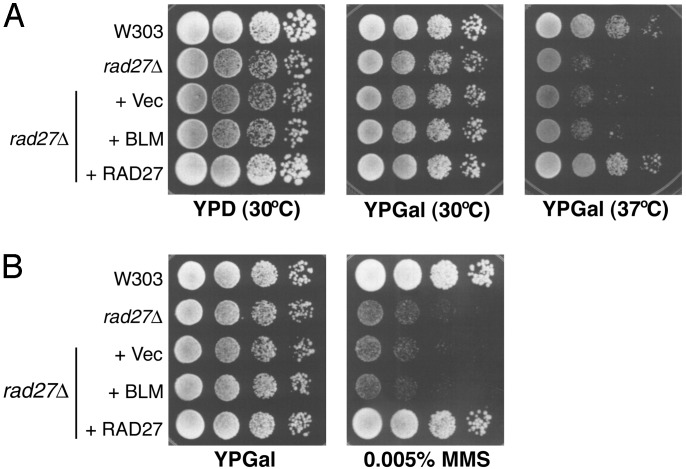
Lack of Suppression of Yeast rad27Δ by hBLM. hBLM might function at either of two phases in the replication process to suppress the growth defect of dna2-1 mutants. hBLM might reduce the need for Dna2 during its otherwise essential role in fork propagation, or hBLM might efficiently mediate repair of toxic structures arising because of dna2 insufficiency. For instance, BLM has been proposed to function in the resolution of regressed DNA replication forks (see ref. 2 for review). If hBLM were involved in repair of toxic intermediates, then hBLM might suppress defects in mutants affecting proteins that participate in Okazaki fragment processing along with scDNA2. As described in the introduction, the essential function of DNA2 can be provided by overproduction of scFEN1, and mutants lacking both FEN1 and Dna2 are inviable, suggesting that they interact or perform redundant functions in the same step of DNA replication. scFEN1 is encoded by the RAD27 gene. rad27Δ mutants are viable at 30°C, presumably because another protein can perform a redundant function at this temperature, but rad27Δ mutants are not viable at 37°C. We next asked if hBLM might also suppress the temperature-sensitive growth of a rad27Δ strain. As shown in Fig. 4A, hBLM does not suppress temperature-sensitive growth of the rad27Δ mutant. rad27Δ mutants are also sensitive to MMS. hBLM fails to suppress the MMS sensitivity of the rad27Δ mutant at 30°C (Fig. 4B). We conclude that either the damage in rad27Δ mutants is repaired by a different mechanism than damage in dna2-1 mutants, or hBLM is specifically compensating for Dna2 in propagating the yeast-replication fork.
Fig. 4.
Lack of complementation of temperature-sensitive growth and MMS sensitivity of strain rad27Δ. Exponentially growing yeast cultures of W303 wild type, rad27Δ, rad27Δ/vector, rad27Δ/pGAL-hBLM, and rad27Δ/Yep24-Rad27 were spotted in 10-fold serial dilution onto plates. (A) Growth at 30°Cor37°C. (B) Growth in the presence and absence of 0.005% MMS. Cells were grown for 3 days.
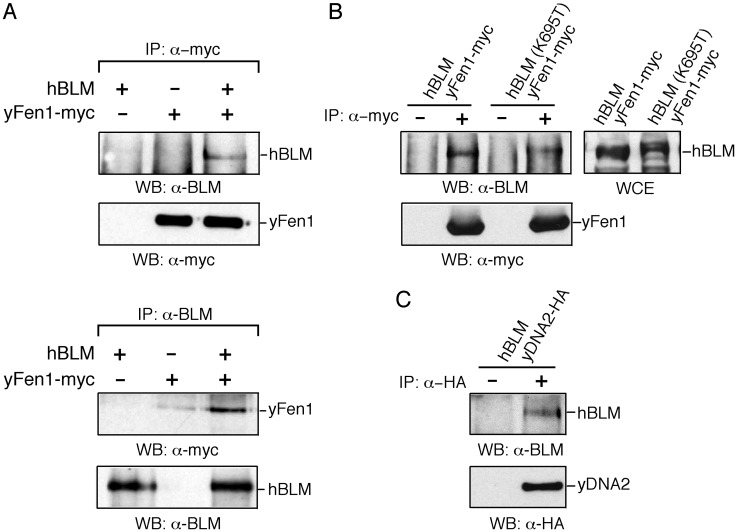
Physical Interaction Between hBLM and scFEN1 and Between hBLM and scDna2. To obtain further evidence that hBLM is associated with the replication fork, we wanted to show coimmunoprecipitation of hBLM with a yeast-replication fork protein. We chose scFEN1, because the human WRN helicase/nuclease has been shown to interact with FEN1 (36, 37), and the same might also be true of hBLM, although it has not been reported. As shown in Fig. 5A, we did find convincing coimmunoprecipitation of hBLM and scFEN1-myc, which suggests that both scFEN1 and hBLM reside in the same complex, although we cannot determine whether the interaction is direct or indirect. We also examined the interaction of scFEN1-myc with the hBLM K695T mutant protein, which did not complement the dna2-1 phenotypes (Figs. 2 A and 3A). Similar to the wild-type hBLM, the K695T mutant protein coprecipitated with scFEN1 (Fig. 5B). Thus, we speculate that the K695T protein is present at the replication fork but does not suppress dna2-1 phenotypes because it is catalytically inactive.
Fig. 5.
Interaction between hBLM and replication fork proteins. (A) Interaction between hBLM and scFEN1. Extracts from BJ5459 transformed with pGAL-hBLM, pJDG-Rad27-myc encoding scFEN1-myc, or both were prepared, and immunoprecipitations were performed with either anti-myc (Upper) or anti-BLM (Lower) antibodies. Washed immunoprecipitates were loaded onto SDS/7.5% PAGE and immunoblotted with either anti-BLM or anti-myc antibodies, as indicated. (B) Interaction of the hBLM(K695T) mutant protein with scFEN1. Extracts were prepared from BJ5459 cells carrying pJDG-Rad27-myc and pGAL-hBLM or pGAL-BLM(K695T) and subjected to immunoprecipitation with anti-myc antibodies. The immunoprecipitates were immunoblotted with either anti-BLM or anti-myc antibodies, as indicated. BLM(K695T) always migrates slightly more slowly than wild-type BLM protein (see also Fig. 2B). (C) Interaction of hBLM with scDNA2. Extracts were prepared from BJ5459 cells carrying pGAL-hBLM and pJDG-DNA2-HA and subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitates were immunoblotted with either anti-BLM or anti-HA antibodies, as indicated. WCE, whole-cell extract before immunoprecipitation; IP, immunoprecipitation/immunoprecipitate; WB, Western blot of immunoprecipitate; -, control with no antibody in IP mix; +, presence of indicated antibody in IP mix.
scFEN1 associates with the scDna2 helicase/nuclease when they are overexpressed (19). We therefore examined whether hBLM interacts with scDna2. As shown in Fig. 5C, hBLM also coimmunoprecipitates with scDna2-HA in extracts. These results suggest that hBLM participates in the same steps of DNA replication or repair as scFEN1 and scDna2.
Discussion
In summary, expression of hBLM in dna2 mutants suppresses both their replication and DNA repair defects, and with a similar efficiency to hDNA2. hBLM does not suppress rad27Δ mutants, however, suggesting that hBLM is specifically suppressing dna2 defects. This finding suggests that hBLM may be at the replication fork, and, supporting this, hBLM coimmunoprecipitates with scFEN1 and scDna2. This result forges a direct link between hBLM and the replication apparatus. Similarly, depletion of xBLM from Xenopus in vitro replication extracts suggests that xBLM is involved in DNA replication (38).
Suppression of the dna2-1 temperature-sensitive growth and repair defect by hBLM helicase is intriguing. The question arises whether hBLM can functionally substitute for scDna2 protein or if the role of the hBLM protein is indirect, involving additional molecular transactions leading to genomic stability of the dna2 strain. The Dna2 and BLM helicases have opposite directionalities on simple duplex substrate DNAs (39, 40). This finding alone may suggest that the suppression is not caused by direct replacement of Dna2 activities. Because scDna2 and hBLM interact, one form of indirect suppression might be stimulation or stabilization of scDna2 activities. Several lines of evidence involving BLM, WRN, and FEN1 proteins may provide additional clues as to the function of BLM in suppression of the dna2 defect. (i) We have demonstrated that overproduction of scFEN1 nuclease suppresses the dna2-1 defect (19). (ii) We have shown that hBLM and scFEN1 interact physically. (iii) WRN and FEN1 interact and the interaction enhances FEN1 cleavage activity (36). (iv) hBLM is thought to recruit several repair/ checkpoint proteins to arrested replication forks (41, 42). Therefore, it is conceivable that the hBLM/scFEN1 interaction at the replication fork leads to either enhanced cleavage by scFEN1 or enhanced recruitment of scFEN1 to the flap, effectively raising the local concentration of the protein and reducing the need for scDna2. Suggesting that some component of the maturation system, such as a helicase, may be missing in vitro, pol δ cannot strand displace rapidly enough in a reconstituted model system containing purified pol δ, proliferating cell nuclear antigen, RFC, FEN1, Dna2, and ligase to account for rates of Okazaki fragment maturation in vivo (25, 26). Thus, hBLM might stimulate flap formation. By stimulating scFEN1 in these or in other ways, hBLM might offset the need for full scDna2 activity. Yeast rad27Δsgs1 double mutants are inviable, suggesting a similar possible role for the yeast hBLM homolog Sgs1 helicase (43). Fibroblasts from patients with Bloom syndrome show a retarded rate of DNA chain growth that might be explained by a model in which BLM is essential for efficient Dna2/FEN1-mediated maturation of replication intermediates (44, 45).
It has recently been suggested that an important function of hBLM and its yeast ortholog, Sgs1, may be to aid the replisome in copying DNA sequences that form alternative structures (46, 47). Both hBLM and Sgs1 proteins were shown to have significantly greater affinity for G quartet DNA and more rapid kinetics of unwinding of G DNA than of Holliday junctions (47). Dna2 helicase might perform a similar function, in addition to or instead of its proposed role in Okazaki fragment processing, which might account for the suppression of dna2-1 by hBLM and the synthetic lethality of sgs1 and dna2-2. dna2-2 mutants show increased pausing in the ribosomal DNA (48), and Dna2 is localized to telomeres and participates in their stable maintenance (13). Both of these DNA regions have G-rich repeats. hBLM has also recently been linked to telomere function in several studies (49, 50). Taken together, we conclude that BLM plays an important role in DNA replication and repair, possibly directly, or, alternatively, as a molecular matchmaker at a crossroad between DNA replication and repair.
Acknowledgments
We thank Dr. Norma Neff for hBLM gene alleles and anti-hBLM antibody and for critical reading of the manuscript; Alexander Varshawsky for critical reading of the manuscript; Dr. Y. Furuichi for hBLM, mWRN, and hRECQ4 genes; and N. Nobuo for the hDNA2 EST clone. This work was supported by U.S. Public Health Service Grant GM25508.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: sc, Saccharomyces cerevisiae; xDNA2, Xenopus laevis DNA2; HU, hydroxyurea; MMS, methyl methanesulfonate; Ura, uracil; 5-FOA, 5-fluoroorotic acid.
References
- 1.Ellis, N. A. (1997) Curr. Opin. Genet. Dev. 7 354-363. [DOI] [PubMed] [Google Scholar]
- 2.Oakley, T. J. & Hickson, I. D. (2002) DNA Repair 1 175-207. [DOI] [PubMed] [Google Scholar]
- 3.Hickson, I. (2003) Nat. Rev. Cancer 3 169-178. [DOI] [PubMed] [Google Scholar]
- 4.Gangloff, S., McDonald, J. P., Bendixen, C., Arthur, L. & Rothstein, R. (1994) Mol. Cell. Biol. 14 8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watt, P. M., Louis, E. J., Borts, R. H. & Hickson, I. D. (1995) Cell 81 253-260. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, H. & Sinclair, D. A. (2001) Proc. Natl. Acad. Sci. USA 98 3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamagata, K., Kato, J., Shimamoto, A., Goto, M., Furuichi, Y. & Ikeda, H. (1998) Proc. Natl. Acad. Sci. USA 95 8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heo, S.-J., Tatebayashi, K., Ohsugi, I., Shimamoto, A., Furuichi, Y. & Ikeda, H. (1999) Genes Cells 4 619-625. [DOI] [PubMed] [Google Scholar]
- 9.Neff, N. F., Ellis, N. A., Ye, T. Z., Noonan, J., Huang, K., Sanz, M. & Proytcheva, M. (1999) Mol. Biol. Cell 10 665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu, L., Davies, S., North, P. S., Goulaouic, H., Riou, J. F., Turley, H., Gatter, K. C. & Hickson, I. D. (2000) J. Biol. Chem. 275 9636-9644. [DOI] [PubMed] [Google Scholar]
- 11.Budd, M. E. & Campbell, J. L. (1995) Proc. Natl. Acad. Sci. USA 92 7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mays Hoopes, L. L., Budd, M., Choe, W., Weitao, T. & Campbell, J. L. (2002) Mol. Cell. Biol. 22 4136-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabre, F., Chan, A., Heyer, W.-D. & Gangloff, S. (2002) Proc. Natl. Acad. Sci. USA 99 16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frei, C. & Gasser, S. M. (2000) Genes Dev. 14 81-96. [PMC free article] [PubMed] [Google Scholar]
- 15.Budd, M. E. & Campbell, J. L. (2000) Mutat. Res. 459 173-186. [DOI] [PubMed] [Google Scholar]
- 16.Choe, W., Budd, M., Imamura, O., Hoopes, L. & Campbell, J. L. (2002) Mol. Cell. Biol. 22 4202-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorentino, D. F. & Crabtree, G. R. (1997) Mol. Biol. Cell 8 2519-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braguglia, D., Heun, P., Pasero, P., Duncker, B. P. & Gasser, S. M. (1998) J. Mol. Biol. 281 631-649. [DOI] [PubMed] [Google Scholar]
- 19.Budd, M. E. & Campbell, J. L. (1997) Mol. Cell. Biol. 17 2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waga, S. & Stillman, B. (1994) Nature 369 207-212. [DOI] [PubMed] [Google Scholar]
- 21.Ishimi, Y., Claude, A., Bullock, P. & Hurwitz, J. (1988) J. Biol. Chem. 263 19723-19733. [PubMed] [Google Scholar]
- 22.Ireland, M. J., Reinke, S. S. & Livingston, D. M. (2000) Genetics 155 1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, J.-Y., Choi, E., Bae, S.-H., Lee, K.-H., Gim, B.-S., Kim, H.-D., Park, C., MacNeill, S. A. & Seo, Y.-S. (2000) Genetics 155 1055-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae, S. H., Bae, K.-H., Kim, J. A. & Seo, Y. S. (2001) Nature 412 456-461. [DOI] [PubMed] [Google Scholar]
- 25.Ayyagari, R., Gomes, X. V., Gordenin, D. A. & Burgers, P. M. J. (2003) J. Biol. Chem. 278 1618-1625. [DOI] [PubMed] [Google Scholar]
- 26.Jin, Y. H., Ayyagari, R., Resnick, M. A., Gordenin, D. A. & Burgers, P. M. (2003) J. Biol. Chem. 278 1626-1633. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Q., Choe, W. & Campbell, J. L. (2000) J. Biol. Chem. 275 1615-1624. [DOI] [PubMed] [Google Scholar]
- 28.Eki, T., Okamura, K., Shiratori, A., Abe, M., Nogami, M., Taguchi, H., Shibata, T., Murakami, Y. & Hanaoka, F. (1996) Genomics 37 408-410. [DOI] [PubMed] [Google Scholar]
- 29.Lee, K. H., Lee, M. H., Lee, T. H., Han, J. W., Park, Y. J., Ahnn, J., Seo, Y. S. & Koo, H. S. (2003) Mol. Cells 15 81-86. [PubMed] [Google Scholar]
- 30.Guthrie, C. & Fink, G. (1991) Guide to Yeast Genetics and Molecular Biology (Academic, New York).
- 31.Ellis, N. A., Groden, J., Ye, T. Z., Straughen, J., Lennon, D., Ciocci, S., Proytcheva, M. & German, J. (1995) Cell 83 655-666. [DOI] [PubMed] [Google Scholar]
- 32.Imamura, O., Ichikawa, K., Yamabe, Y., Goto, M., Sugawara, M. & Furuichi, Y. (1997) Genomics 41 298-300. [DOI] [PubMed] [Google Scholar]
- 33.Kitao, S., Ohsugi, I., Ichikawa, K., Goto, M., Furuichi, Y. & Shimamoto, A. (1998) Genomics 54 443-452. [DOI] [PubMed] [Google Scholar]
- 34.Budd, M. E., Choe, W.-C. & Campbell, J. L. (1995) J. Biol. Chem. 270 26766-26769. [DOI] [PubMed] [Google Scholar]
- 35.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104 397-408. [DOI] [PubMed] [Google Scholar]
- 36.Brosh, R. M., Jr., Driscoll, H. C., Dianov, G. & Sommers, J. A. (2002) Biochemistry 41 12204-12216. [DOI] [PubMed] [Google Scholar]
- 37.Brosh, J., Robert M., von Kobbe, C., Sommers, J. A., Karmakar, P., Opresko, P. L., Piotrowski, J., Dianova, I., Dianov, G. L. & Bohr, V. A. (2001) EMBO J. 20 5791-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lian, S., Graham, J. & Yan, H. (2000) Genes Dev. 14 2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bae, S.-H., Kim, D. W., Kim, J., Kim, J.-H., Kim, D.-H., Kim, H.-D., Kang, H.-Y. & Seo, Y.-S. (2002) J. Biol. Chem. 277 26632-26641. [DOI] [PubMed] [Google Scholar]
- 40.Hickson, I. D., Davies, S. L., Li, J.-L., Vevitt, N. C., Mohaghegh, P., North, P. S. & Wu, L. (2001) Biochem. Soc. Trans. 29 201-204. [DOI] [PubMed] [Google Scholar]
- 41.Franchitto, A. & Pichierri, P. (2002) J. Cell Biol. 157 15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sengupta, S., Linke, S. P., Pedeux, R., Yang, Q., Farnsworth, J., Garfield, S. H., Valerie, K., Shay, J. W., Ellis, N. A., Wasylyk, B. & Harris, C. C. (2003) EMBO J. 22 1210-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong, A. H. Y., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W. V., Bussey, H., et al. (2001) Science 294 2364-2368. [DOI] [PubMed] [Google Scholar]
- 44.Hand, R. & German, J. (1975) Proc. Natl. Acad. Sci. USA 72 758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hand, R. & German, J. (1977) Hum. Genet. 38 297-306. [DOI] [PubMed] [Google Scholar]
- 46.Sun, H., Karow, J. K., Hickson, I. D. & Maizels, N. (1998) J. Biol. Chem. 273 27587-27592. [DOI] [PubMed] [Google Scholar]
- 47.Huber, M. D., Lee, D. C. & Maizels, N. (2002) Nucleic Acids. Res. 30 3954-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weitao, T., Budd, M., Mays Hoopes, L. L. & Campbell, J. L. (2003) J. Biol. Chem. 278 22513-22522. [DOI] [PubMed] [Google Scholar]
- 49.Opresko, P. L., von Kobbe, C., Laine, J.-P., Harrigan, J., Hickson, I. D. & Bohr, V. A. (2002) J. Biol. Chem. 277 41110-41119. [DOI] [PubMed] [Google Scholar]
- 50.Stavropoulos, D. J., Bradshaw, P. S., Li, X., Pasic, I., Truong, K., Ikura, M., Ungrin, M. & Meyn, M. S. (2002) Hum. Mol. Genet. 11 3135-3134. [DOI] [PubMed] [Google Scholar]