Abstract
Plants can sense the direction of gravity and change the growth orientation of their organs. The molecular mechanisms of gravity sensing and signal transduction during gravitropism are not well known. We have isolated several shoot gravitropism (sgr) mutants of Arabidopsis. The sgr3-1 mutant exhibits a reduced gravitropic response in the inflorescence stems. In the inflorescence stems of Arabidopsis, gravity is sensed in endodermal cells that contain sedimentable amyloplasts. In sgr3-1, some amyloplasts in the endodermis failed to sediment in the direction of gravity. SGR3 encodes a syntaxin, AtVAM3, which had previously been cloned as a homologue of yeast Vam3p. AtVAM3 is localized to the prevacuolar compartment and vacuole and is suggested to function in vesicle transport to the vacuole. We have also cloned another soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE), ZIG/AtVTI11, a mutation that causes abnormal gravitropism. This mutant displayed an abnormal distribution of amyloplasts in the endodermal cells similar to that in sgr3-1. Endodermis-specific expression of SGR3 and ZIG by using the SCR promoter could complement the abnormal shoot gravitropism of each mutant. Protein–protein interaction between AtVAM3 and AtVTI11 in the endodermal cells was detected immunologically. The sgr3-1 mutation appeared to reduce the affinity of AtVAM3 for AtVTI11 or SYP5. These results suggest that vesicle transport to the prevacuolar compartment/vacuole in the endodermal cells, mediated by a specific SNARE complex containing AtVAM3 and AtVTI11, plays an important role in shoot gravitropism.
Because higher plants are sessile throughout their lifetime, they have evolved many mechanisms to sense and adapt to various environmental changes. Gravitropism, the alignment of growth relative to gravity, is one of these important environmental responses. Shoots and roots of land plants generally show negative and positive gravitropism, respectively. Consequently, these organs are able to reorient themselves to a position appropriate for their functions. The gravitropic response is composed of the following sequential steps: sensing of the direction of gravity, conversion of physical stimuli into biochemical signals, transmission of these signals to the responsive tissue, and differential growth of organs, resulting in curvature (1, 2).
Molecular genetic approaches using Arabidopsis thaliana have provided strong evidence that supports classical hypotheses. Isolation and analysis of mutants defective in auxin-related genes have given strong support to the Cholodny–Went hypothesis, which proposed that asymmetric auxin distribution causes the differential growth of tropism (3, 4). The starch–statolith hypothesis has suggested that sedimentable amyloplasts containing starch granules act in sensing the direction of gravity in specialized cells (5). This hypothesis is also strongly supported by studies of starchless and amyloplastless mutants (6–9).
We have isolated a number of shoot gravitropism (sgr) mutants of Arabidopsis, which are deficient in the gravitropic response of inflorescence stems (10, 11). The inflorescence stem consists of an epidermis, cortex, endodermis, and stele (containing the vascular tissues), which are arranged concentrically from the outside to the inside of the stem. The endodermal cells of hypocotyls and stems contain amyloplasts that sediment in the direction of gravity. The sgr1/scr (scarecrow) and sgr7/shr (short-root) mutants lack a normal endodermis in all organs and show an agravitropic response in shoots but not roots. These results indicate that endodermal cells containing amyloplasts are essential for gravity sensing in shoots (10, 12).
Recent studies of sgr mutants have given rise to the concept that the vacuole and/or vesicle transport to the vacuole in the endodermal cells are involved in gravity sensing (13–15). Both sgr2 and zig(zigzag)/sgr4 mutants show little gravitropic response in inflorescence stems. The SGR2 (At1g31480) gene encodes a phospholipase A1-like protein localized to the vacuolar membrane, although its biochemical function is not yet known. Inflorescence stems of the zig/sgr4 mutant show not only little gravitropic response but also a characteristic aberrant morphology; they bend at nodes, and thus elongate in a zigzag fashion (11). The ZIG (At5g39510) gene encodes a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE; AtVTI11), which is localized to the trans-Golgi network and prevacuolar compartment (PVC) and is suggested to be involved in vesicle transport (16). Abnormal vesicular/vacuolar structures were observed in several tissues, including the endodermis, in both mutants. In addition, many endodermal amyloplasts did not sediment and almost all of them appeared to stick to the cell periphery in the mutants (14). In each mutant, endodermis-specific expression, the wild-type gene could complement the gravitropism defect. These results suggest that the formation and function of the vacuole in the endodermal cells are important in the early processes of shoot gravitropism (14).
Intracellular protein trafficking and membrane fusion are mediated by the evolutionarily conserved SNARE superfamily (17, 18). SNAREs carried on transport vesicles (v-SNAREs) interact with those on the target compartment (t-SNAREs) to form stable trans-SNARE complexes (19). Three or four SNAREs assemble into a complex involving the formation of a four-helical bundle that brings the bilayers into close proximity and then leads to membrane fusion (20, 21). It has been demonstrated that AtVTI11 interacts with other SNAREs, such as AtVAM3, SYP (syntaxin of plant) 21, and SYP5 (22). This observation raises questions about the involvement of these cognate SNAREs of AtVTI11 in gravitropism.
In this work, through genetic and biochemical analysis of sgr3-1, we found that one of the cognate SNAREs of AtVTI11, AtVAM3, is involved in shoot gravitropism, and we suggest that vesicle transport to the vacuole in the endodermis is important for shoot gravitropism.
Methods
Plant Materials and Growth Conditions. The Columbia (Col) ecotype of A. thaliana was used as the wild type. The sgr3-1 mutant was isolated from the M2 population of Col that had been mutagenized with EMS. The screening strategy used to isolate these mutants has been described (10). Gravitropism of inflorescence stems was assayed by using plants grown on soil under constant white light at 23°C.
zig-1 can be regarded as a null allele because a transmembrane domain and a part of the SNARE motif of the AtVTI11 protein were deleted (13).
Gravitropism Assay. To examine the gravitropic responses of inflorescence stems, intact plants with primary stems 4–8 cm in length were used. To measure the gravitropic response of the stems, plants were placed horizontally in darkness at 23°C, as described (13). The curvature of the stem was measured every 30 min as the angle formed between the growing direction of the apex and the horizontal base line. At least 20 individuals of each genotype were examined.
Histological Analysis. Stem segments cut from primary inflorescence stems were fixed in 10% (vol/vol) formaldehyde, 5% (vol/vol) acetic acid, and 50% (vol/vol) ethanol in 0.2-ml tubes under vacuum. The growth orientation of the stem segments was maintained during fixation. After dehydration in a series of ethanol, samples were embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. Sections (3 μm) were stained with toluidine blue.
Electron microscopic observation was performed as described (14). In brief, stem segments (1–2 cm below the apex) from young primary stems 4–8 cm in length were fixed with 2% glutaraldehyde and postfixed with 1% osmium tetroxide. The growth orientation of the stem segments was maintained during fixation. The samples were dehydrated, then embedded in Spurr's resin. Ultrathin sections (70–90 nm) were cut longitudinally, stained with uranyl acetate and lead citrate, and then observed under transmission electron microscopy (H-7100, Hitachi, Tokyo).
Mapping of SGR3. The sgr3-1 homozygous mutant was crossed to Landsberg erecta wild-type plants to generate a mapping population. SGR3 was mapped to the middle of chromosome 5. For fine-scale mapping, DNA was prepared from ≈1,100 F2 progeny.
Polymorphisms between Col and Landsberg erecta were identified, which allowed PCR markers to be designed based on the chromosome 5 sequence data from the Arabidopsis Genome Initiative supplied by the Kazusa group. The resulting cleaved–amplified polymorphic sequence markers were used to map the recombination breakpoints by PCR and restriction digestion.
Cloning of SGR3 and Plasmid Construction. A 4.0-kb genomic DNA fragment from BAC clone MSD23 was subcloned into the binary vector pBIN19 (pBIN_gSGR3).
Total RNA was isolated from etiolated hypocotyls with the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). cDNAs were synthesized by using SuperScript II reverse transcriptase (Invitrogen). PCR amplification of SGR3 cDNA was performed with the following primer set: cSGR3-F (5′-GCGGATCCAGGGCCACGATCCACGCCTTGACC-3′) and cSGR3-R (5′-CGGGATCCTCAAACCGAACTGAACCAACCC-3′). The PCR products were cloned into pBlueScript (SK+). ZIG cDNA had already been cloned by our laboratory as described (13). To tag AtVTI11 protein with the T7-epitope at the N-terminal end, the cDNA was cloned into the BamHI site of pET21-a (Novagen). The SCR promoter (kindly donated by Dr. Philip N. Benfey) was already inserted into the pBI121 plasmid, which lacks the uidA gene (pBI121del_pSCR). The cSGR3 and T7-ZIG genes were inserted into the pBI121del_pSCR plasmid, downstream of the SCR promoter. All of these constructs were transformed into Agrobacterium tumefaciens strain MP90 and then introduced into the sgr3-1 or zig-1 plants (23). T1 plants were selected for resistance to kanamycin. The presence of the transgene in these plants was tested by PCR. Segregation of the transgene in the T2 generation was confirmed.
Antibody Production. The cytoplasmic region of SYP51 (residues, Ala-2-Ser-204; At1g16240) was amplified by PCR with the primers AtVAM7N (5′-ATAGGATCCTGGCGTCTTCATCGGATTCATGG-3′) and AtVAM7C (5′-AGAGGATCCTTAACTTCTCATATTCTTGTTCAT-3′) and then subcloned into the BamHI site of pGEX-3T. The GST-SYP51 fusion protein was produced in Escherichia coli NM-522. The fusion proteins were purified on glutathione-Sepharose 4B columns and used for immunizing rabbits. To test specificity of the antibody, plant extracts from Col and a T-DNA insertion mutant of SYP51 (Δsyp5; Torrey Mesa Research Institute, San Diego) were used for protein blotting and were probed with the anti-SYP51 antiserum. A strong band was detected at ≈30 kDa in wild-type Columbia. Although the intensity was greatly reduced in the Δsyp5 mutant, a faint band was still detected at the same position as SYP51. The antiserum possibly cross-reacts with SYP52, a paralog of SYP51, as described by Sanderfoot et al. (22). Because SYP51 and SYP52 showed an identical expression pattern and biochemical behavior, in addition to a high degree of sequence similarity (22), our antiserum is sufficiently specific to detect SYP5 family proteins in our experiments. The anti-AtVAM3 antibody was described previously (24, 25).
Protein Extraction and Immunoprecipitation. Immunoprecipitation of detergent extracts from the shoots was carried out as described in Zheng et al. (16), with minor modifications. Three grams of shoots from 21-day-old plants was homogenized on ice in 6 ml of extraction buffer (50 mM Hepes-KOH, pH 6.5/10 mM potassium acetate/100 mM sodium chloride/5 mM EDTA/0.4 M sucrose) with a protease inhibitor mixture (Sigma). The homogenate was passed through Miracloth (Calbiochem) to remove debris. This extract was used for Western blot analysis after adding an equal volume of Laemmli buffer.
To perform the immunoprecipitation analysis, the extract was centrifuged at 10,000 × g to generate a membrane pellet. To solubilize the membrane proteins, the pellet was resuspended in 6 ml of extraction buffer containing 1% (vol/vol) Triton X-100 and protease inhibitor mixture and incubated at 4°C for 2 h. Insoluble material was repelleted at 10,000 × g. The supernatant (total protein extract) was incubated at 4°C for 6 h with 100 μl of protein A-Sepharose beads, to which was bound anti-AtVAM3 antiserum, anti-SYP5 antiserum, or anti-T7 monoclonal antibody (Novagen). The beads were then collected by centrifugation at 4°C, 500 × g, for 1 min and the supernatant was collected. The beads were washed five times in extraction buffer with 1% Triton X-100. Protein (immunoprecipitate) was then eluted from the beads in 100 μl of Laemmli buffer. Equal volumes of total protein extract, supernatant, or immunoprecipitate were separated by SDS/PAGE followed by immunoblotting with appropriate antibodies.
Results
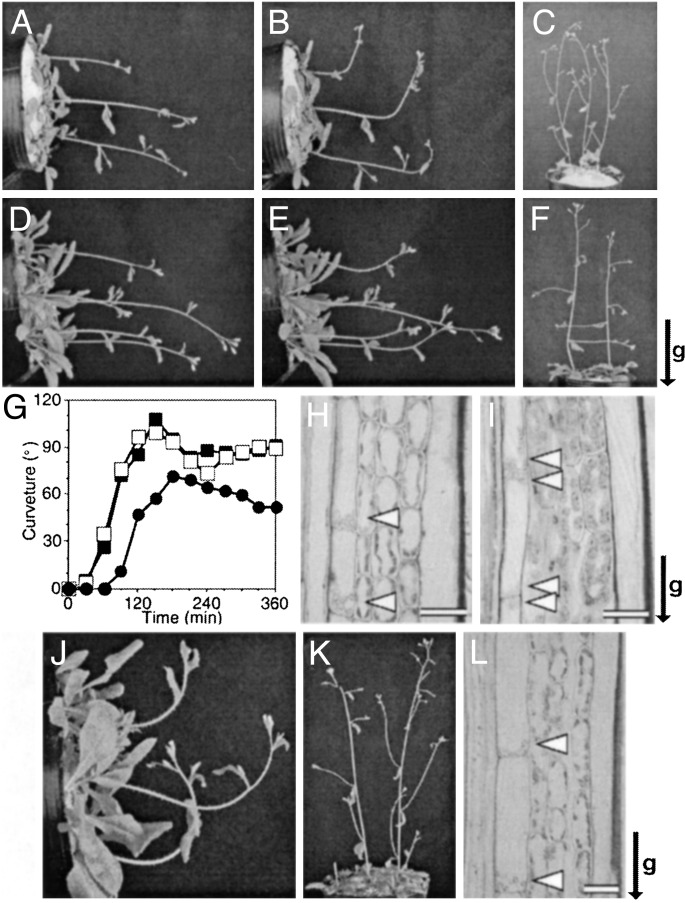
Phenotype of sgr3-1 Mutant. Arabidopsis wild-type (Col) inflorescence stems bent upward when they were gravistimulated by horizontal placement, and the curvature reached 90° in ≈90 min (Fig. 1 A, B, and G). In contrast, the curvature of the sgr3-1 inflorescence stems reached only 20–30° after 90 min and required ≈180 min to reach 90°, as reported in ref. 10 (Fig. 1 D,E, and G). On the other hand, etiolated hypocotyls of sgr3-1 grew uniformly upward, just as well as those of the wild type (data not shown). The roots of sgr3-1 also showed normal positive gravitropism (data not shown). Therefore, SGR3 is specifically involved in inflorescence stem gravitropism. The lateral shoots of the wild type grew upward, whereas those of sgr3-1 grew horizontally (Fig. 1 C and F) (10). No other morphological defects were detected in sgr3-1 plants.
Fig. 1.
Phenotype of sgr3-1. Gravitropic response of inflorescence stems of 4-week-old plants after 0 min (A and D) or 90 min (B, E, and J) of horizontal gravistimulation. (C, F, and K) Lateral shoots of 5-week-old plants. (G) Time course of gravitropic response of shoots: wild type (open squares), sgr3-1 (filled circles), and sgr3-1/pSCR::cSGR3 (filled squares). Four-week-old plants were gravistimulated by horizontal placement at 23°C in the dark. (H, I, and L) Amyloplast localization in the endodermal cells. Longitudinal sections through inflorescence stems (3–4 cm below the apex). The orientation of the gravity stimulus was maintained during fixation. Arrowheads indicate amyloplasts. (A–C and H) Wild type. (D–F and I) sgr3-1. (J–L) sgr3-1/pSCR::cSGR3. The arrow indicates the direction of gravity (g). (Scale bars = 20 μm.)
Abnormal Sedimentation of Amyloplasts in the Shoot Endodermis of sgr3-1. The Arabidopsis inflorescence stem usually has, from the outside in, one layer of epidermis, three or four layers of cortex, and one layer of endodermis. The endodermal cells contain sedimented amyloplasts (Fig. 1H). Although the tissue patterning of sgr3-1 was indistinguishable from that of the wild type, and some amyloplasts were located at the bottom edge of the endodermal cells, many amyloplasts remained at the top edge of the cells, as reported for zig-1 and sgr2–1 in ref. 14 (Fig. 1I). In contrast to the inflorescence stems, the endodermal amyloplasts in hypocotyls of sgr3-1 were sedimented just like those of the wild type (data not shown).
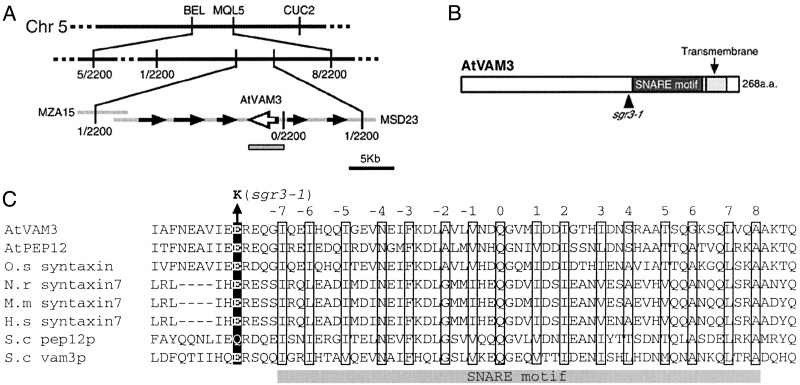
Map-Based Cloning of SGR3. The SGR3 locus mapped between the cleaved amplified polymorphic sequence markers BEL and MQL5 on chromosome 5. By analyzing ≈2,200 chromosomes, the locus was narrowed to an ≈35-kb region, containing six predicted ORFs (Fig. 2A). By sequencing sgr3-1 genomic DNA, a one-base change was found in one predicted ORF, AtVAM3 (Fig. 2 B and C). As reported by Sato et al. (24), AtVAM3 (At5g46860) encodes a t-SNARE homologous to yeast Vam3p, which is involved in protein trafficking to the vacuole (26). AtVAM3 was localized to the vacuole or PVC and may be involved in vesicle transport to these compartments (24, 27). The 4-kb wild-type genomic fragment containing the AtVAM3 ORF and predicted promoter region was cloned and introduced into sgr3-1 plants for a complementation test. Inflorescence stems of the transgenic plants bent to 90° in ≈90 min, like those of the wild type, and their lateral shoots grew upward (data not shown). In addition, all of the endodermal amyloplasts in the inflorescence stems were sedimented, as observed in wild-type stems (data not shown). These results indicate that AtVAM3 is the SGR3 gene.
Fig. 2.
Molecular cloning of the SGR3 gene. (A) The SGR3 locus was mapped to an ≈35-kb region between two cleaved amplified polymorphic sequence markers. The gray box indicates the genomic region cloned for complementation analysis. (B) The molecular lesion in sgr3-1. Black and gray boxes indicate the SNARE motif and the transmembrane domain of AtVAM3, respectively. (C) Alignment of amino acid sequences of the SNARE motifs from syntaxin homologues in plants (A. thaliana, NP568671, NP197185; Oryza sativa, BAA84625), mammals (Mus musculus, BAA90699; Norvegicus rattus, NP068641; Homo sapiens, NP003560), and Saccharomyces cerevisiae (AAB38370 and AAC49737). The central ionic layer of the SNARE motif is indicated as zero, and the numbers indicate adjacent hydrophobic layers. These layer assignments are based on the crystal structure of the core complex (20). The black box indicates the mutation site of sgr3-1.
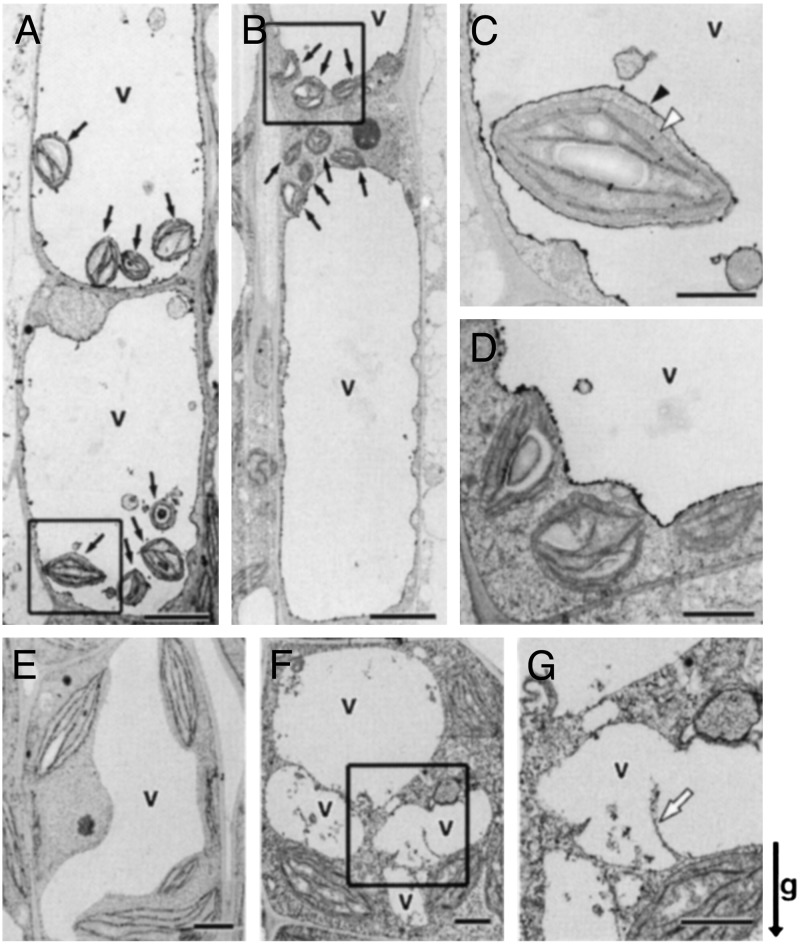
Ultrastructural Analysis of the Endodermal Cells of sgr3-1. To examine the effect of the sgr3-1 mutation at the subcellular level, inflorescence stems were observed by electron microscopy. As we reported (14), the sedimenting amyloplasts were surrounded by a thin cytoplasmic layer and by the vacuolar membrane in wild-type endodermal cells. They seemed to be trapped within the transvacuolar strands and suspended in the inside of the vacuole (Fig. 3 A and C). In sgr3-1, in contrast, all of the amyloplasts remained in the peripheral cytoplasmic areas at the top, bottom, and sides of the endodermal cells (Fig. 3 B and D). No amyloplasts were engulfed in the lumen of the vacuole.
Fig. 3.
Ultrastructure of the endodermal and cortical cells of the inflorescence stem. (A–D) The endodermal cell. Black arrows indicate amyloplasts. Black and white arrowheads indicate the vacuolar membrane and outer envelope of the amyloplast, respectively. (E–G) The cortical cell. The white arrow shows abnormal invaginated vacuolar membrane. (A, C, and E) Wild type. (B, D, F, and G) sgr3-1. C, D, and G are higher magnifications of boxed areas in A, B, and F, respectively. (Scale bars: A, B, E, and F = 5 μm; C, D, and G = 1 μm.) The arrow indicates the direction of gravity (g).
The cortex cells of sgr3-1 also showed a weak phenotype. In contrast to the smooth central vacuole of the wild type (Fig. 3E), the vacuolar membrane in the sgr3-1 mutants sometimes formed very irregular curves (Fig. 3 F and G). Deep cytoplasmic projections into the vacuolar lumen and abnormal membranous structures in the vacuole were observed more frequently in the mutant than in the wild type.
SGR3 Expressed in Endodermal Cells Is Required for Normal Shoot Gravitropism. A remarkable feature of the sgr3-1 phenotype is that only inflorescence stems show abnormal gravitropism, which is probably caused by abnormal sedimentation of amyloplasts in the endodermis. However, it has been shown that SGR3/AtVAM3 is expressed in all organs examined (24), and abnormal vacuolar structure was also observed in the cortex of mutant inflorescence stems (Fig. 3 F and G). To elucidate the function of AtVAM3 in the endodermis, SGR3 cDNA was expressed in the endodermal cells of sgr3-1 by using the SCR (SCARECROW) promoter. The SCR gene is specifically expressed in the endodermal cell layer of the root, hypocotyl, and inflorescence stem (28, 29).
Inflorescence stems of sgr3-1 plants transformed with pSCR::SGR3 showed gravitropic responses with kinetics similar to those of the wild type (Fig. 1 G and J), and their lateral shoots grew upward (Fig. 1K). All amyloplasts in the endodermis consistently sedimented in the direction of gravity in the transgenic plants (Fig. 1L). These results suggest that the expression of SGR3 in the endodermal cells is sufficient for the recovery of shoot gravitropism.
Interaction Between SGR3 and ZIG. SGR3 encodes a t-SNARE homologous to yeast Vam3p, as described above. It has been suggested that AtVAM3 functions in vesicle transport to the vacuole by forming a SNARE complex with other SNAREs (22, 24, 27). The gene responsible for another of our sgr mutants, ZIG, encodes the v-SNARE AtVTI11 (13). AtVTI11 localizes to the trans-Golgi network and the PVC (16) and interacts with AtVAM3 (27) and SYP5 (22).
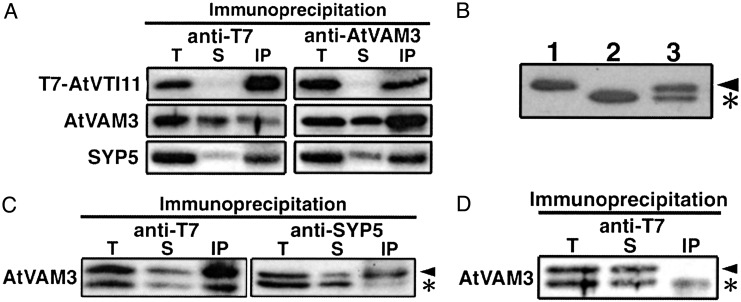
To examine whether these SNAREs form a complex in the endodermal cells, we created transgenic zig-1 plants that expressed AtVTI11 tagged with T7 epitope (T7-AtVTI11) only in the endodermis (zig-1/pSCR::T7-ZIG). Because the transgenic plants responded to gravity normally, T7-AtVTI11 protein is presumably functional (data not shown). Extracts of the transgenic plants were immunoprecipitated with anti-T7 monoclonal antibody or anti-AtVAM3 antisera, and then these immunoprecipitates were probed with the antibody or antisera indicated in Fig. 4A. AtVAM3 and T7-AtVTI11 were coimmunoprecipitated with anti-T7 antibody (Fig. 4A Left). A similar pattern was observed after immunoprecipitation with anti-AtVAM3 antiserum (Fig. 4A Right). In addition, SYP5 also coimmunoprecipitated with T7-AtVTI11 and AtVAM3 (Fig. 4A). These results suggest that AtVTI11, AtVAM3, and SYP5 formed a complex in the endodermal cells.
Fig. 4.
Interaction between AtVAM3 and AtVTI11. (A) Extract from zig-1/pSCR::T7-ZIG plants was immunoprecipitated with anti-T7 monoclonal antibody (Left) or anti-AtVAM3 antiserum (Right), and then probed with the antibody or antisera indicated to the left. (B) Total extracts from wild-type (lane 1), sgr3-1/- homozygous (lane 2), and sgr3-1/+ heterozygous (lane 3) plants were probed with anti-AtVAM3 antiserum. (C) Interaction between mutant AtVAM3 protein and partner SNAREs. Extract from sgr3-1/+ zig-1/pSCR::T7-ZIG was immunoprecipitated with anti-T7 antibody or anti-SYP5 antiserum and then probed with anti-AtVAM3 antiserum. (D) A mixture of sgr3-1 zig-1/pSCR::T7-ZIG and zig-1 homozygous plant extracts was immunoprecipitated with anti-T7 antibody and then probed with anti-AtVAM3 antiserum. T, total protein extract; S, supernatant; IP, immunoprecipitate. The arrowhead and asterisk indicate wild-type and mutant AtVAM3 protein, respectively.
sgr3-1 Mutation Reduces Affinity with the Cognate SNAREs. AtVAM3 shares some conserved domains with SNARE homologues of other species (Fig. 2 B and C). One of these domains is the SNARE motif, which is required to form tight SNARE complexes (20, 30). The sgr3-1 mutation causes an E-to-K amino acid substitution adjacent to the SNARE motif. It was suspected that this mutation could affect the ability of AtVAM3 to interact with other SNAREs. At first, we tried to detect the mutant AtVAM3 protein in sgr3-1 by protein blot. Surprisingly, the mutant protein migrated faster in SDS-polyacrylamide gels than the wild-type protein, so we could distinguish the mutant and wild-type proteins even in sgr3-1/+ heterozygotes (Fig. 4B). The band intensities of the two proteins were almost the same in heterozygous plants, suggesting that the interactions of mutant and wild-type AtVAM3 with other SNAREs could be compared simultaneously in these plants.
sgr3-1 was crossed to zig-1/pSCR::T7-ZIG and sgr3-1/+ zig-1/pSCR::T7-ZIG plants were selected from among the F3 progeny. Extracts from these plants were immunoprecipitated with anti-T7 antibody or anti-SYP5 antibody and then probed with anti-AtVAM3 antibody. Less mutant AtVAM3 was coimmunoprecipitated with T7-AtVTI11 or SYP5 than wild-type AtVAM3, even though nearly equal amounts of the two proteins were present in the total extracts (Fig. 4C). These results indicate that the affinity between mutant AtVAM3 and AtVTI11 or SYP5 is reduced by the sgr3-1 mutation.
It has been shown that SNARE proteins can assemble spontaneously in detergent solutions (31, 32). It is crucial to know whether the complex we detected was present in the endodermal cells or had formed after extraction. Extract from sgr3-1 zig-1/pSCR::T7-ZIG plants, containing T7-AtVTI11 and mutant AtVAM3 protein, and extract from zig-1 plants, containing wild-type AtVAM3 and no AtVTI11 protein, were mixed and then immunoprecipitated with anti-T7 antibody (Fig. 4D). If SNARE complexes formed in detergent solution during our immunoprecipitation experiments, wild-type AtVAM3 protein would be expected to coprecipitate with T7-AtVTI11, because the mutant AtVAM3 protein has less affinity for T7-AtVTI11 than does wild-type AtVAM3. As shown in Fig. 4D, little wild-type AtVAM3 protein was coimmunoprecipitated with T7-AtVTI11. This result indicates that the SNARE complex detected in our experiments is formed in vivo in endodermal cells.
Discussion
Recently, we cloned ZIG, encoding the v-SNARE AtVTI11, as the gene responsible for the gravitropism defect of zig/sgr4 (13). SNARE partners of AtVTI11 would also be expected to be involved in the gravitropic response, because vesicle transport mediated by AtVTI11 is required for gravitropism and this process is presumably dependent on other SNARE molecules to form tight complexes and lead to membrane fusion. As expected, we found a syntaxin mutant, sgr3-1, among our sgr mutants. SGR3 encodes AtVAM3, previously cloned as a homologue of the yeast syntaxin Vam3p (24). Vam3p is involved in vacuole fusion, protein trafficking to the vacuole, and autophagy (26, 33, 34). AtVAM3 can interact with other SNAREs, such as AtVTI11 and SYP5, in root-cultured plants and has been suggested to be involved, together with AtVTI11, in vesicle transport to the PVC and/or vacuole (22).
The altered mobility of the mutant AtVAM3 protein on SDS-polyacrylamide gels implies that the sgr3-1 mutation deleteriously affects the chemical properties of the AtVAM3 protein. However, the mutant AtVAM3 protein must be at least partially functional because it has been reported that a knockout mutation of SGR3 is lethal (35). In addition, immunoprecipitation experiments indicate that the mutant AtVAM3 protein in sgr3-1 retains the ability to interact with AtVTI11 and SYP5, although with less affinity than the wild-type protein. Considering that the sgr3-1 mutation occurred adjacent to the SNARE motif of AtVAM3, it may interfere with interactions among SNAREs that involve the SNARE motif. Amino acid 182 E, which is outside of the typical SNARE motif, may be critical for the function of this type of SNARE, consistent with the high degree of conservation of this amino acid among various species (Fig. 2 B and C).
The sgr3-1 mutation in AtVAM3 is probably deleterious to vesicle transport to the PVC/vacuole or vacuole. Indeed, abnormal vacuole morphology was observed not only in the endodermis but also in the cortex (Fig. 3 D, F, and G). Regarding the gravitropic response, however, only AtVAM3 expressed in the endodermal cells is important (Fig. 1 J–L). Interaction between AtVAM3 and AtVTI11 was detected by immunoprecipitation in transgenic plants carrying pSCR::T7-ZIG (Fig. 4A). This interaction had occurred in vivo in the endodermal cells because it is unlikely that these proteins formed new complexes during immunoprecipitation (Fig. 4D). Taken together, these data indicate that vesicle transport to the vacuole of endodermal cells, probably by means of the PVC and mediated by the cognate SNARE partners AtVAM3 and AtVTI11, is important for gravitropism.
How, then, do AtVAM3 and AtVTI11 mediate the gravitropic response in the endodermal cells? Amyloplasts were distributed abnormally in the epidermal cells of sgr3-1, just as in zig-1 (14). Furthermore, a flexible vacuolar membrane structure was observed surrounding amyloplasts in the endodermal cells of wild type, whereas all of the amyloplasts in the endodermal cells of sgr3-1 remained in the peripheral cytoplasmic region (Fig. 3 A and B). These results strongly suggest that a defect in vacuole function or formation might interfere with amyloplast movement, resulting in an inability to sense changes in the gravity vector. However, sgr3-1, which has a weak graviresponse, and zig-1, which has almost no graviresponse, exhibited a similar distribution of amyloplasts in the endodermal cells of stem segments. To resolve this discrepancy, amyloplast movement should be observed in living cells in these mutants. Such experiments are in progress.
Hypocotyls of sgr3-1 exhibited a normal graviresponse, whereas those of zig-1 showed abnormal gravitropism (10, 11). Furthermore, sgr3-1 inflorescence stems grew straight (Fig. 1F), in contrast to the zigzag-shaped inflorescence stems of zig-1 (10, 11). Why do these mutants show different phenotypes? One possible explanation is that AtVAM3 and AtVTI11 have distinct functions and thus interact with distinct partner(s) in tissues other than the endodermis of inflorescence stems (such as that of hypocotyls). Indeed, it has been shown that AtVTI11 also can interact with AtPEP12, an AtVAM3 homologue (22, 27). Moreover, AtVTI11 exhibited tissue-specific differences in function, i.e., involvement in the gravitropic response in endodermal cells but in the determination of stem shape in other tissues (14). AtVTI11 might form SNARE complex(s) with SNARE(s) other than AtVAM3 for stem morphogenesis. Consistent with this possibility, a knockout mutant of SGR3 was lethal, in contrast to the viability of deletion mutants of ZIG (13, 35). Thus, AtVAM3 is predicted to have specific functions independent of AtVTI11. Recently, the SNAP-25-type SNAP-33 and v-SNARE NPSN11 (novel plant SNAREs) have been identified as partner SNAREs of KNOLLE, which is a cytokinesis-specific syntaxin required for cell plate membrane fusion (36–38). It should be noted that both SNAP-33 and NPSN11 were suggested to have other functions beyond their involvement in cytokinesis (37, 38). Divergent SNARE molecules could make various combinations for distinct functions.
Acknowledgments
We thank Dr. Philip N. Benfey (Duke University, Durham, NC) for providing the plasmid containing the SCR promoter, Dr. Takehide Kato for valuable discussion, and Ms. Seiko Ishihara for technical assistance. This work was supported by the Japan Society for the Promotion of Science (12740438 to M.T.M.) and the Ministry of Agriculture, Forestry and Fisheries of Japan in the framework of the Pioneering Research Project in Biotechnology (to M.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: sgr, shoot gravitropism; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptors; v-SNARE, vesicle-associated SNARE; t-SNARE, target membrane-associated SNARE; SYP, syntaxin of plant; PVC, prevacuolar compartment; ZIG, ZIG/AtVTI11; SGR3, SGR3/AtVAM3/SYP22; AtVAM3, a gene product of SGR3; AtVTI11, a gene product of ZIG.
References
- 1.Tasaka, M., Kato, T. & Fukaki, H. (1999) Trends Plant Sci. 4 103-107. [DOI] [PubMed] [Google Scholar]
- 2.Tasaka, M., Kato, T. & Fukaki, H. (2001) Int. Rev. Cytol. 206 135-154. [DOI] [PubMed] [Google Scholar]
- 3.Masson, P. H., Tasaka, M., Morita, M. T., Guan, C., Chen, R. & Boonsirichai, K. (2002) The Arabidopsis Book (Am. Soc. Plant Biol., Rockville, MD), www.aspb.org/publications/arabidopsis/toc.cfm. [DOI] [PMC free article] [PubMed]
- 4.Friml, J., Wisniewska, J., Benkova, E., Mendgen, K. & Palme, K. (2002) Nature 415 806-809. [DOI] [PubMed] [Google Scholar]
- 5.Sack, F. D. (1991) Int. Rev. Cytol. 127 193-252. [DOI] [PubMed] [Google Scholar]
- 6.Kiss, J. Z., Wright, J. B. & Caspar, T. (1996) Plant Physiol. 97 237-244. [DOI] [PubMed] [Google Scholar]
- 7.Kiss, J. Z., Guisinger, M. M., Miller, A. J. & Stackhouse, K. S. (1997) Plant Cell Physiol. 38 518-525. [DOI] [PubMed] [Google Scholar]
- 8.Weise, S. E. & Kiss, J. Z. (1999) Int. J. Plant Sci. 160 521-527. [DOI] [PubMed] [Google Scholar]
- 9.Fujihira, K., Kurata, T., Watahiki, K. M., Karahara, I. & Yamamoto, K. T. (2000) Plant Cell Physiol. 41 1193-1199. [DOI] [PubMed] [Google Scholar]
- 10.Fukaki, H., Fujisawa, H. & Tasaka, M. (1996) Plant Physiol. 110 944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamauchi, Y., Fukaki, H., Fujisawa, H. & Tasaka, M. (1997) Plant Cell Physiol. 38 530-535. [DOI] [PubMed] [Google Scholar]
- 12.Fukaki, H., Wysocka-Diller, J., Kato, T., Fujisawa, H., Benfey, P. N. & Tasaka, M. (1998) Plant J. 14 425-430. [DOI] [PubMed] [Google Scholar]
- 13.Kato, T., Morita, M. T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M. & Tasaka, M. (2002) Plant Cell 14 33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita, M. T., Kato, T., Nagafusa, K., Saito, C., Ueda, T., Nakano, A. & Tasaka, M. (2002) Plant Cell 14 47-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, T., Morita, M. T. & Tasaka, M. (2002) J. Plant Growth Regul. 21 113-119. [DOI] [PubMed] [Google Scholar]
- 16.Zehng, H., von Mollard, G. F., Kovaleva, V., Stevens, T. & Raikhel, N. V. (1999) Mol. Biol. Cell 10 2251-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weimbs, T., Mostov, K., Low, S. H. & Hoffmann, K. (1998) Trends Cell Biol. 8 260-262. [DOI] [PubMed] [Google Scholar]
- 18.Jahn, R. & Südhof, T. C. (1999) Annu. Rev. Biochem. 68 863-911. [DOI] [PubMed] [Google Scholar]
- 19.Jahn, R., Lang, T. & Südhof, T. C. (2003) Cell 112 519-533. [DOI] [PubMed] [Google Scholar]
- 20.Sutton, R. B., Fasshauer, D., Jahn, R. & Brunger, A. (1998) Nature 395 347-353. [DOI] [PubMed] [Google Scholar]
- 21.Antonin, W., Fasshauer, D., Becker, S., Jahn, R. & Schneider, T. R. (2002) Nat. Struct. Biol. 9 107-111. [DOI] [PubMed] [Google Scholar]
- 22.Sanderfoot, A. A., Kovaleva, V., Bassham, D. C. & Raikhel, N. V. (2001) Mol. Biol. Cell 12 3733-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clough, S. J. & Bent, A. F. (1998) Plant J. 16 735-743. [DOI] [PubMed] [Google Scholar]
- 24.Sato, M. H., Nakamura, N., Ohsumi, Y., Kouchi, H., Kondo, M., Nishimura, I. K., Nishimura, M. & Wada, Y. (1997) J. Biol. Chem. 272 24530-24535. [DOI] [PubMed] [Google Scholar]
- 25.Uemura, T., Yoshimura, S. H., Takeyasu, K. & Sato, M. H. (2002) Genes Cells 7 743-753. [DOI] [PubMed] [Google Scholar]
- 26.Darsow, T., Rieder, S. E. & Emr, S. D. (1997) J. Cell Biol. 138 517-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanderfoot, A. A., Kovaleva, V., Zheng, H. & Raikhel, N. V. (1999) Plant Physiol. 121 929-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M. G., Feldmann, K. A. & Benfey, P. N. (1996) Cell 86 423-433. [DOI] [PubMed] [Google Scholar]
- 29.Wysocka-Diller, J., Helariutta, Y., Fukaki, H., Malamy, J. E. & Benfey, P. N. (2000) Development (Cambridge, U.K.) 127 595-603. [DOI] [PubMed] [Google Scholar]
- 30.Antonin, W., Hoplroyd, C., Fasshauer, D., Pabst, S., von Mollard, G. F. & Jahn, R. (2000) EMBO J. 19 6453-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi, T., McMahon, H., Yamasaki, S., Binz, T., Hata, Y., Sudhof, T. C. & Niemann, H. (1994) EMBO J. 13 5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fasshauer, D., Otto, H., Eliason, W. K., Jahn, R. & Brunger, A. T. (1997) J. Biol. Chem. 272 28036-28041. [DOI] [PubMed] [Google Scholar]
- 33.Wada, Y., Nakamura, N., Ohsumi, Y. & Hirata, A. (1997) J. Cell Sci. 110 1299-1306. [DOI] [PubMed] [Google Scholar]
- 34.Ungermann, C., von Mollard, G. F., Jensen, O. N., Margolis, N., Stevens, T. H. & Wickner, W. (1999) J. Cell Biol. 145 1435-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanderfoot, A. A., Pilgrim, M., Adam, L. & Raikhel, N. V. (2001) Plant Cell 13 659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukowitz, W., Mayer, U. & Jürgens, G. (1996) Cell 84 61-71. [DOI] [PubMed] [Google Scholar]
- 37.Heese, M., Gansel, X., Sticher, L., Wick, P., Grebe, M., Granier, F. & Jürgens, G. (2001) J. Cell Biol. 155 239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng, H., Bednarek, S. Y., Sanderfoot, A. A., Alonso, J., Ecker, J. R. & Raikhel, N. V. (2002) Plant Physiol. 129 530-539. [DOI] [PMC free article] [PubMed] [Google Scholar]