Abstract
Despite the tremendous importance of secondary metabolites for humans as for the plant itself, plant secondary metabolism remains poorly characterized. Here, we present an experimental approach, based on functional genomics, to facilitate gene discovery in plant secondary metabolism. Targeted metabolite analysis was combined with cDNA-amplified fragment length polymorphism-based transcript profiling of jasmonate-elicited tobacco Bright yellow 2 cells. Transcriptome analysis suggested an extensive jasmonate-mediated genetic reprogramming of metabolism, which correlated well with the observed shifts in the biosynthesis of the metabolites investigated. This method, which in addition to transcriptome data also generates gene tags, in the future might lead to the creation of novel tools for metabolic engineering of medicinal plant systems in general.
Plants are capable of synthesizing an overwhelming variety of low-molecular-weight organic compounds called secondary metabolites, usually with unique and complex structures. Presently, ≈100,000 such compounds have been isolated from higher plants (1). Numerous plant secondary metabolites possess interesting biological activities and find applications, such as pharmaceuticals, insecticides, dyes, flavors, and fragrances. Although secondary metabolism offers attractive targets for plant breeding, the enormous biosynthetic potential of plant cells is still not being exploited. In sharp contrast, metabolism of microorganisms has been successfully engineered for increased production of pharmaceuticals or novel compounds (2, 3). Despite a few decades of research, plant secondary metabolism remains poorly characterized (4). Genetic maps of biosynthetic pathways are still far from complete, whereas knowledge on the regulation of these pathways is practically nonexistent. However, such knowledge is of crucial importance to bypass the low product yield of various secondary metabolites in plants or plant cell cultures.
Functional genomics approaches are powerful tools to accelerate comprehensive investigations of cellular metabolism in specialized tissues or whole organisms (5). Yet, related to plant secondary metabolism, only a few reports have been published on such studies, which include the use of comparative quantitative trait loci mapping (6), 2D gel electrophoresis-based proteomics (7), or transcript analysis tools, such as differential display (8, 9), EST databases (10–12), and microarrays (13, 14). Nevertheless, still little is known about the genetics that control quantitatively and qualitatively natural variation in secondary metabolism.
Because of the lack of extensive genomic data for the vast majority of medicinal plants, it is difficult to use the commonly used microarray-based approach for transcriptome analysis in these plant systems. Such an approach requires prior development of large EST or cDNA clone collections (13, 14). As such, the cDNA-amplified fragment length polymorphism (AFLP) technology (15–17) offers an attractive alternative to identify genes involved in plant secondary metabolism. This method provides quantitative expression profiles, does not require prior sequence information, and, most importantly, allows the identification of novel genes. Here, we applied this technology, in combination with targeted metabolite analysis, on jasmonate-elicited tobacco Bright yellow 2 (BY-2) cell cultures. A transcriptome of nearly 600 jasmonate-modulated genes could be composed that was compared with the observed jasmonate-induced shifts in biosynthesis of tobacco metabolites and similar transcriptome analyses previously reported for tobacco and Arabidopsis plants.
Materials and Methods
Maintenance and Methyl Jasmonate Elicitation of BY-2 Cells. Cultured suspensions of Nicotiana tabacum L. cv. BY-2 cells were maintained as described (18). For methyl jasmonate elicitation, a 6-day-old culture was diluted 10-fold in fresh medium without growth regulators and grown for 12 h. Subsequently, methyl jasmonate (Duchefa, Haarlem, The Netherlands) at a final concentration of 50 μM was added to the culture, or an equivalent amount of DMSO (the solvent of methyl jasmonate) as a control. For the transcript profiling analysis, samples were taken at 0, 1, 2, 4, 6, 8, 10, 12, 16, 24, and 48 h after jasmonate addition or at 0, 4, 8, 12, 24, and 48 h after DMSO addition. For metabolic analysis, samples were taken at 0, 12, 16, 20, 24, 36, 48, and 98 h, either after jasmonate or DMSO addition. Before further analysis, cells were washed, vacuum-filtered, and lyophilized.
cDNA-AFLP Transcript Profiling. cDNA-AFLP-based transcript profiling was performed as described (16, 17). On RNA extraction and cDNA synthesis, each sample was digested first with MseI, followed by BstYI (Mse/Bst), or vice versa (Bst/Mse). After selective preamplification, messengers were screened with two or three additional selective nucleotides for the Mse/Bst and Bst/Mse templates, respectively. All possible primer combinations (160 total) were used. As such, a high-resolution genome-wide transcript profile covering at least 60% of all mRNAs is obtained (17). Quantitative data analysis was performed with aflp-quantarpro software (Keygene, Wageningen, The Netherlands). For each transcript tag, an indicative variation value was calculated by dividing the maximum value over the time course by the minimum value. Expression profiles with an indicative value of <4.0 were considered to be constitutive throughout the time course. This cut-off value was chosen in conformity with visual scoring of cDNA-AFLP gels to avoid identifying false positives created by PCR artifacts. Subsequently, each expression profile of individual genes was variance-normalized by standard statistical approaches used for microarray-derived data (19). For each transcript, the mean expression value over the time course of the DMSO-treated samples was subtracted from each individual data point after which the obtained value was divided by the standard deviation. cluster and treeview software (20) were used for average linkage hierarchical clustering. To characterize the jasmonate-modulated cDNA-AFLP fragments, the sequences, obtained from either the reamplified PCR product or individual clones after cloning these PCR products in pGEM-T easy (Promega), were compared against nucleotide and protein sequences in the publicly available databases by BLAST sequence alignments (21). When available, tag sequences were replaced with longer EST or isolated cDNA sequences to increase the chance of finding significant homology.
Metabolite Analysis. Alkaloids were extracted for GC-MS analysis as follows. Cells were weighed, and an internal standard (5α-cholestan) was added. The aqueous samples were made alkaline with ammonia [10% (vol/vol)] and were extracted by vortexing with dichloromethane. After centrifugation (2,000 × g, 10 min), the lower organic layer was separated and concentrated to 50 μl. Aliquots (3 μl) were injected to a 5890 gas chromatograph (Hewlett–Packard) coupled to a 5970 quadrupole mass selective detector (Hewlett–Packard). The analyses were performed on an NB-54 fused silica capillary column (15 m, 0.20-mm inner diameter; HNU-Nordion, Helsinki) by using split sampling mode (50:1) and a programmed oven temperature from 100 to 250°C at 7°C/min. The injector and detector temperatures were 250°C, and the analysis time was 30 min. Polyamines were extracted and quantified as described (22). For detection with TLC, phenylpropanoids were extracted as described (23). TLC was performed with ethylacetate/methanol/H2O (75/15/10) running buffer and spots were visualized under UV254 either before or after spraying with 1% AlCl2 solution (24, 25). For further analysis, phenylpropanoids were extracted by two different methods (23, 26), after which samples were subjected to acidic hydrolysis by adding 20% (vol/vol) HCl and incubated for 1 h at 80°C. Flavonoid aglycones and simple coumarins containing hydroxyl groups were determined as trimethylsilyl derivatives by GC-MS on unpolar columns. Both cell and media samples were assayed. Sesquiterpenes were extracted and detected by TLC as described (27) and by GC-MS concurrent with nicotine alkaloid analysis. Samples for carotenoid and sterol analysis were prepared by saponifying 1 g of freeze-dried cells. The isolated unsaponified matter was subsequently extracted into diethylether. Carotenoids were detected on a Waters Spherisorb ODS-1 column (4 × 250 mm, 5 μm; Alltech Associates) at 453 nm using methanol-tetrahydrofurane (95:5) as an eluent (isocratic, 0.8 ml/min), whereas the presence of sterols was verified by GC-MS after trimethylsilylation.
Results
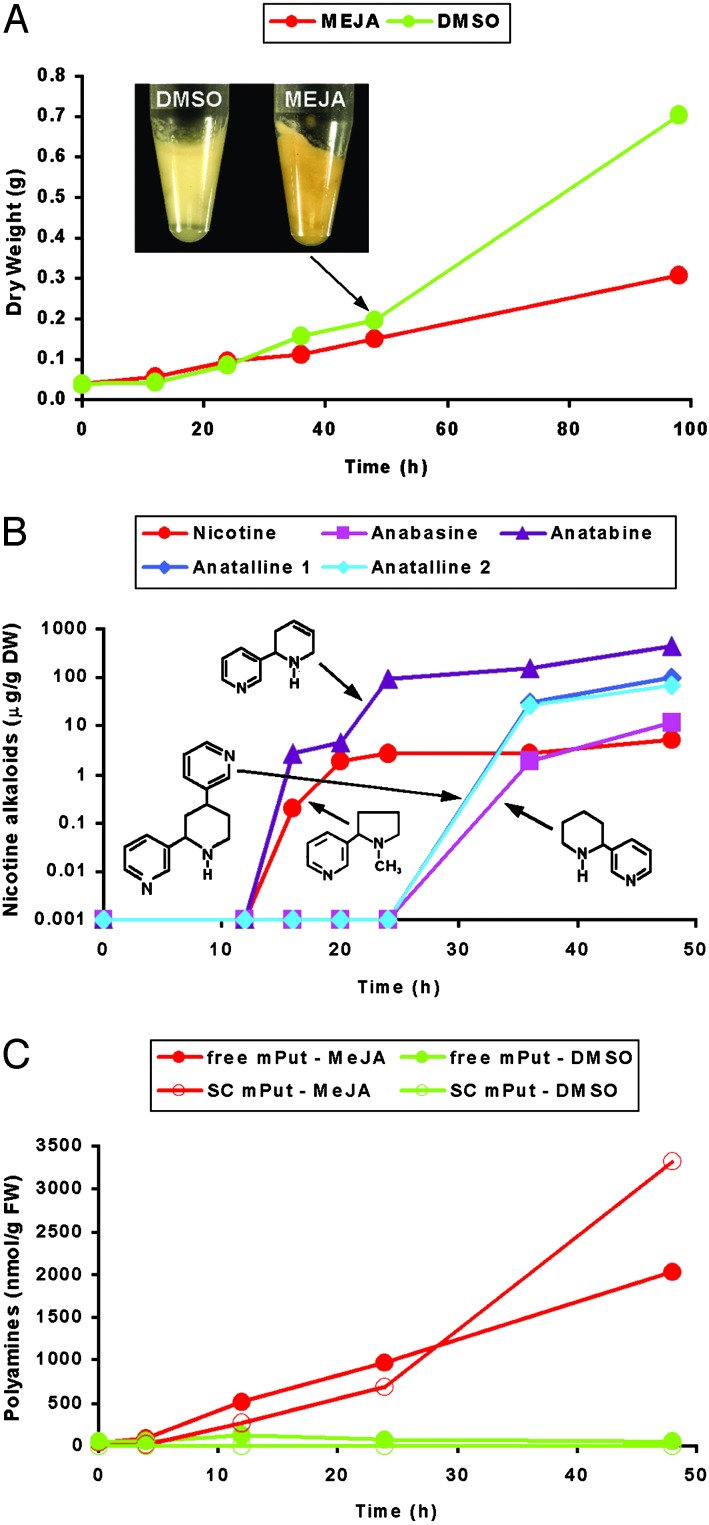
Jasmonate-Mediated Elicitation of Secondary Metabolism in Tobacco BY-2 Cell Cultures. Tobacco has an ample catalog of secondary metabolites (28), of which some are known to be elicitor-inducible in cell cultures. Here, we studied the change in the transcriptome of tobacco BY-2 cells as a consequence of elicitation. The most pronounced secondary metabolites of tobacco are the nicotine alkaloids, which were chosen as reference metabolites. The biosynthesis of nicotine is partially characterized and known to be elicited in BY-2 cells after removal of the growth hormone 2,4-D and the subsequent addition of methyl jasmonate, an oxylipin-type signaling molecule (29). The effects of methyl jasmonate treatment on BY-2 growth and the production of nicotine alkaloids are shown in Fig. 1 A and B. During the first 36 h, control cells and cells treated with jasmonate displayed similar growth rates, but after this initial period, cell growth was drastically reduced in jasmonate-treated cells when compared with nontreated cells. At this stage, jasmonate-treated BY-2 cells also exchanged their “bright yellow” appearance for a more “bright orange”-like color (Fig. 1 A). This color change was not further investigated considering the complexity of the observed jasmonate-mediated genetic reprogramming of tobacco metabolism (see below). The elicitation-dependent accumulation of nicotine was detectable after ≈12 h and reached rapidly its maximum level toward the end of the experimental period (48 h; Fig. 1B). For the genome-wide transcript profiling, samples were taken covering the 48-h experimental period, focusing on the initial 12 h.
Fig. 1.
Metabolite analysis in jasmonate-elicited BY-2 cells. (A) Growth curve of BY-2 cells treated with DMSO (green) and methyl jasmonate (MEJA; red). In the Inset the color shift from bright yellow to bright orange 48 h after MEJA elicitation is shown. Values are the mean of three replicates and have a 2–4% coefficient of variance (CV). (B) Accumulation of nicotine alkaloids in MEJA-treated BY-2 cells in micrograms per gram of dry weight. The chemical structures of nicotine, anatabine, anatalline, and anabasine are presented. Values are the mean of three replicates and have a CV of 2–10%. (C) Accumulation of polyamines in BY-2 cells treated with DMSO (green) and MEJA (red) in nanomoles per gram of fresh weight. Accumulation levels of mPut as free or soluble conjugated (SC) form are shown. Values are the mean of three replicates and have a CV of 2–20%. Elicitation was repeated three times, yielding similar experimental results.
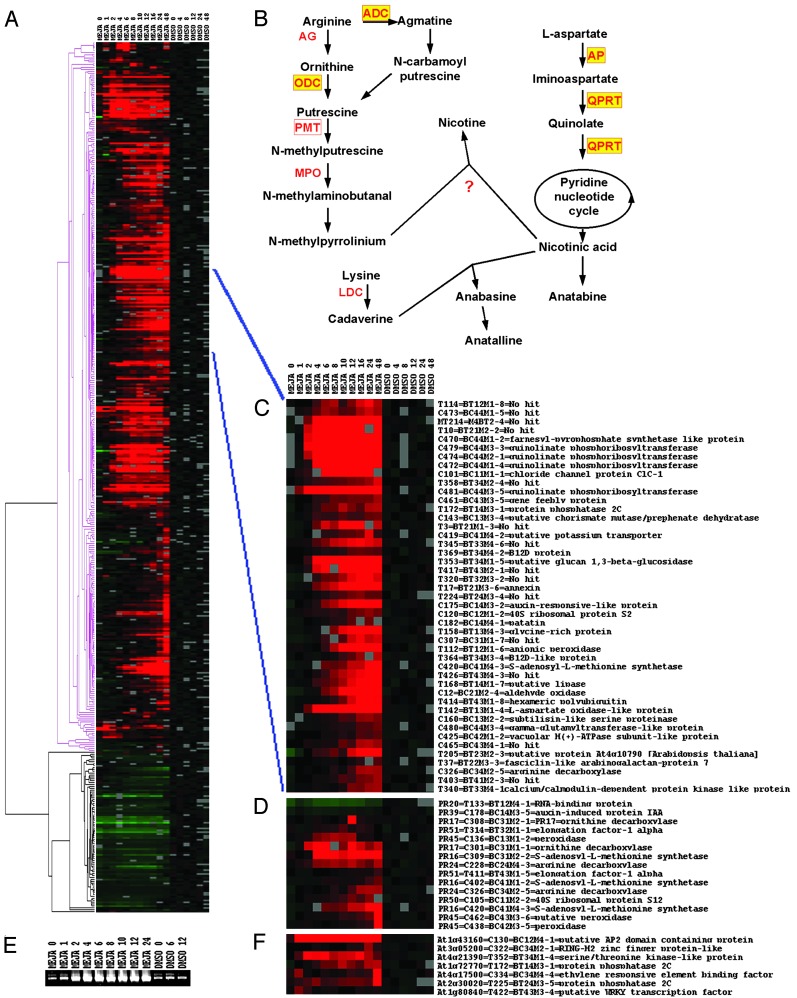
Quantitative and Qualitative Analysis of Jasmonate-Modulated Gene Expression. Quantitative temporal accumulation patterns of ≈20,000 transcript tags were determined and analyzed (see Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org). In total, 591 methyl jasmonate-modulated (MJM) transcript tags were isolated. Direct sequencing of the PCR products gave good-quality sequences for ≈78% of the fragments (see Table 2, which is published as supporting information on the PNAS web site). To the remaining 22%, no unique sequence could be attributed unambiguously. Cloning of these tags and sequencing of individual colonies revealed that the isolated bands consisted of two to four comigrating fragments of different genes (data not shown). Data of these multiple MJM tags were not incorporated for further discussion, because the corresponding expression profiles are the result of multiple superimposed expression profiles, and interpretation of such data consequently is prone to be erroneous. Homology searches with the sequences from the unique gene tags (459 in total, hereafter referred to as the MJM tags) revealed that 58% of these tags displayed similarity with genes of known functions and 16% with a gene without allocated function. In contrast, no homology to a known sequence was found for 26% of the tags (Table 1; see also Table 2). The similarity threshold maintained for BLAST searches was established at a value of 10e-3. However, due to the small size of some tags, in a few cases (see Table 2) a lower e value was accepted when unambiguous matches were observed. The MJM tags could be classified into nine functional groups (Table 1) according to the Munich Information Center for Protein Sequences (http://mips.gsf.de). The vast majority (82%) of jasmonate-modulated genes was up-regulated by jasmonate elicitation, of which ≈70% within 4 h after jasmonate elicitation. The group of jasmonate-repressed genes comprised 18% of the isolated gene tags (Fig. 2A; see also Table 2). Besides numerous novel genes, either with or without existing homologs of unknown function that were identified as jasmonate responsive, the generated gene platform in general correlated well with earlier reports on jasmonate- or wounding-modulated transcriptomes (30–33) as well as other jasmonate-modulated cellular and metabolic events (34, 35). For instance, genes involved in stress responses, such as cell wall modifications, oxidative burst, and production of defense molecules are clearly up-regulated (see Table 2). Another example of up-regulated genes was provided by a group of genes (putatively) involved in the biosynthesis of jasmonates (see Fig. 4, which is published as supporting information on the PNAS web site), an event, which is known to be subject to positive feedback regulation (36, 37). The predominance of jasmonate-up-regulated genes most probably reflects the basic metabolism and physiology, necessary and sufficient to sustain growth of a cell suspension culture, such as tobacco BY-cells (18).
Table 1. Classification of the MJM transcript tags in functional categories.
| Function | Tags, % | I, % | R, % |
|---|---|---|---|
| Metabolism/energy | 128 (27.9) | 116 (25.3) | 12 (2.6) |
| Transcription | 19 (4.1) | 11 (2.4) | 8 (1.7) |
| Protein synthesis/fate | 31 (6.8) | 21 (4.6) | 10 (2.2) |
| Signal transduction | 23 (5.0) | 22 (4.8) | 1 (0.2) |
| Transport facilitation | 18 (3.9) | 13 (2.8) | 5 (1.1) |
| Defense/cell organization | 47 (10.2) | 41 (8.9) | 6 (1.3) |
| Unclassified proteins | 72 (15.7) | 56 (12.2) | 16 (3.5) |
| No hits | 121 (26.4) | 95 (20.7) | 26 (5.7) |
| Total | 459 (100.0) | 375 (81.7) | 84 (18.3) |
The number and frequency (in parentheses) of all MJM tags with defined functional categories annotated is indicated. I, induced; R, repressed (according to average linkage hierarchical clustering)
Fig. 2.
Transcriptome of jasmonate-elicited BY-2 cells. (A) Average linkage hierarchical clustering of MJM gene tags. The time points for the cluster analysis are indicated (in hours) at the top. Red and green boxes reflect transcriptional activation and repression, respectively, relative to the average expression level in control (DMSO) cells. Gray boxes correspond to missing time points. Pink and black parts of the dendrogram indicate the clusters of activated and repressed genes, respectively. (B) Biosynthetic pathway of nicotine alkaloids. Enzymes involved in the metabolic conversions are in red, those for which the corresponding tobacco gene or orthologs from other plants have been cloned are boxed, and those for which a corresponding MJM tag has been identified in the transcript profiling analysis are in yellow. (C) Subcluster of the MJM transcriptome. Shown is a close-up of a subcluster comprising gene tags coregulated with genes known to be involved in nicotine biosynthesis. (D) Average linkage hierarchical clustering of tags common between the MJM transcriptome and the genes differentially expressed in tobacco roots (50). The corresponding protein (PR) code, as used previously (50), has been included. (E) Expression analysis of pmt. Expression of pmt was verified by RT-PCR analysis on the RNA prepared for cDNA-AFLP transcript profiling and using oligonucleotides spanning nucleotides 8–897 of the N. tabacum cv. Burley pmt ORF (38). Time points are indicated (in hours) at the top of the ethidium bromide-stained gel. (F) Average linkage hierarchical clustering of signal transducing genes common between the MJM transcriptome and the wound-induced Arabidopsis transcriptome (33). The corresponding Munich Information Center for Protein Sequences code has been included. ADC, arginine decarboxylase; AG, arginase; AP, aspartate oxidase; LDC, lysine decarboxylase; MEJA, methyl jasmonate; MPO, methylputrescine oxidase; ODC, ornithine decarboxylase; PMT, putrescine methyltransferase; QPRT, quinolinate phosphoribosyltransferase.
Nicotine Alkaloids and Polyamines. The MJM data set revealed the presence of five of the genes that are known to be involved in nicotine biosynthesis in Nicotiana species, most of which were early (within 1–2 h) induced by methyl jasmonate in tobacco BY-2 cells, and thus appear to be coregulated (Fig. 2 B–D; see also Table 2). Arginine decarboxylase and ornithine decarboxylase (structural genes for the biosynthesis of the N-methylpyrrolinium moiety), aspartate oxidase and quinolinate phosphoribosyltransferase (structural genes for the biosynthesis of the nicotinic acid moiety), and S-adenosyl-l-methionine synthetase (structural gene for the catalysis of the methyl-group donor for nicotine biosynthesis) responded. Another structural gene, responsible for the so-called “first committed step” in nicotine biosynthesis, i.e., putrescine methyl transferase (pmt), could not be identified with the cDNA-AFLP method used here because its nucleotide sequence does not harbor a BstYI restriction site (38, 39). Nonetheless, RT-PCR analysis clearly showed that expression of the pmt gene is also up-regulated as early as 1 h after jasmonate treatment (Fig. 2E) and thus demonstrates the coregulation of the pmt gene(s) with the other nicotine metabolic genes. These findings correlate well with earlier reports on the jasmonate inducibility of the genes encoding ornithine decarboxylase and PMT (29).
Elicitation of BY-2 cells by methyl jasmonate not only leads to accumulation of nicotine but also causes a marked increase in the content of various nicotinic acid-derived alkaloids, such as two anatalline isomers and anatabine (Fig. 1B). The GC-MS spectra of the alkaloids (see Table 3, which is published as supporting information on the PNAS web site) were similar to those obtained by either synthesis (40) or isolation from leaves and roots of Duboisia hopwoodii (41) or N. tabacum roots (42). Anabasine is a minor compound, which most probably is rapidly metabolized to anatalline (Fig. 2B). Similarly, nicotinic acid seems to be used effectively for the production of high amounts of anatabine, for which both heterocyclic rings are derived from nicotinic acid (Fig. 2B) (43). In contrast to the other nicotine alkaloids, anatalline had not been detected in BY-2 cells before. Although nicotine and anatabine started to accumulate after 12 h, the amounts of anabasine and anatalline increased after only 24 h (Fig. 1B). At later time points (98 h), all alkaloids reached their maximum levels but, in case of anatabine only, also accumulated in the culture medium (data not shown). In non-elicited BY-2 cells, trace amounts of nicotine were detected only after 98 h, whereas none of the other nicotine alkaloids could be perceived. In tobacco plants, either nicotine or nornicotine is the predominant alkaloid. In contrast, in elicited BY-2 cells, anatabine and anatalline are the main alkaloid components, whereas no nornicotine could be detected and nicotine was present as a minor compound only. To our knowledge, no genes have been identified that are involved in the biosynthesis of the nicotinic acid-derived compounds, other than nicotine. Functional analysis of novel MJM genes could resolve this issue.
Another striking observation was that methyl jasmonate induced a strong and rapid accumulation of N-methylputrescine (mPut), both as free and soluble-conjugated forms (Fig. 1C). Elicited cells also accumulated detectable amounts of insoluble conjugated mPut, but other remarkable differences in polyamine accumulation profiles (i.e., of putrescine, spermine, and spermidine) were not detected during the observed time period, neither in the free nor in the soluble and insoluble conjugated forms (data not shown). The accumulation of free mPut combined with its diversion toward conjugated forms and the concurrent activation of other alkaloid biosynthetic routes leading to anatalline and anatabine could suggest that a rate-limiting step in nicotine biosynthesis is the conversion of mPut to N-methylaminobutanal. This reaction is catalyzed by the enzyme methyl putrescine oxidase, of which the corresponding gene has not yet been identified. Again, functional analysis of novel genes, for instance putative oxidases, clustering together with known nicotine biosynthetic genes (Fig. 2C) might resolve this issue.
Phenylpropanoid, Isoprenoid, and Primary Metabolism. In addition to nicotine biosynthetic genes, the transcript profiling analysis also included a set of MJM gene tags, induced within 2–6 h, that show similarity to genes potentially encoding enzymes involved in phenylpropanoid metabolism (see Table 2 and Fig. 4). Indeed, TLC experiments suggested that jasmonates also induced the production of several phenylpropanoid-like compounds in tobacco BY-2 cells (see Fig. 5, which is published as supporting information on the PNAS web site). Various members of the phenylpropanoid family or structural genes of the phenylpropanoid pathway have previously been reported to be inducible by jasmonate or stress conditions (23, 33, 44, 45). After further analysis of elicited phenylpropanoid-like compounds, no clear-cut patterns of flavonoids, anthocyanins, lignans, or coumarins could be identified. However, the occurrence of these classes of compounds cannot fully be discarded because various trimethylsilyl derivatives of trace compounds were found at their typical mass range (between 350 and 550) (data not shown). Remarkably, although reported previously to be jasmonate responsive, also in tobacco cells (27), no increased accumulation of sesquiterpenes (for instance, capsidiol) could be detected in jasmonate-elicited BY-2 cells (data not shown). Moreover, metabolite analysis at later time points (96 h) showed a considerable decrease in intracellular levels of other isoprenoids, such as β-carotene (see Fig. 5) and campesterol (data not shown) in jasmonate-treated BY-2 cells. Again, this finding was surprising, because isoprenoid biosynthetic pathways have previously been reported to be jasmonate inducible (12, 46, 47). Accordingly, in the MJM data set, no genes proven to be involved in isoprenoid biosynthetic pathways were represented. Additionally, the transcript profiling indicated an extensive genetic reprogramming of primary metabolism, which might sustain secondary metabolic pathways (see Fig. 4). Analogous observations have been found in pathogen-stressed or UV-irradiated parsley cells (48, 49).
Comparison of the Jasmonate-Modulated BY-2 Transcriptome with Previously Reported Transcriptomes. The most relevant previously reported “transcriptome” that can be compared with the MJM data set was the set of genes differentially expressed in tobacco roots during early stages of alkaloid biosynthesis (50). This set consists of 60 cDNAs, whose sequence has been released for 45 that were isolated by subtractive hybridization screening of a tobacco root library. For eight of them, one or more homologs (lowest BLASTN value 1e-24) were found in the MJM data set, including all of the genes related to alkaloid biosynthesis (50), i.e., S-adenosyl-l-methionine synthetase, arginine decarboxylase, and ornithine decarboxylase (Fig. 2 B and D).
Another transcriptome of interest was the microarray survey (8,200 genes) of the transcriptional response to mechanical wounding of Arabidopsis plants (33), a response in which several plant hormones, among which jasmonates, are known to play an important role (34). Because of the qualitative differences in secondary metabolism between Arabidopsis and tobacco, the comparison is limited here to the Arabidopsis wound-responsive genes that encode signaling and regulatory proteins (129 in total; see Table 1 and ref. 33). Interestingly, seven MJM genes, all early induced by methyl jasmonate in tobacco BY-2 cells, had putative Arabidopsis counterparts (lowest BLASTX value 3e-7) that were also early induced by wounding (Fig. 2F). These genes code for putative AP2-domain and WRKY-type transcription factors, zinc finger proteins, and protein phosphatases and kinases; consequently, they are prime targets for further functional analysis to establish their potential role in jasmonate signaling and the regulation of plant secondary metabolism.
Comparison with the transcriptome analysis performed in tobacco BY-2 cells, but set up to identify plant cell-cycle-modulated genes (16), nicely illustrates the specificity of our experimental results. An overlap of 71 genes between the MJM gene tags (459 in total) and the cell-cycle-modulated gene tags (1,340 in total) was observed (lowest BLASTN value 3e-13). However, except for one of the tags corresponding to arginine decarboxylase, essentially none of the above-mentioned gene tags or those that encode genes clearly related to nicotine alkaloid biosynthesis (Fig. 2B) were present in this overlap (see Fig. 6, which is published as supporting information on the PNAS web site).
Discussion
Combining targeted metabolite analysis and cDNA-AFLP-based transcript profiling of jasmonate-elicited tobacco BY-2 cells yielded an ample inventory of known and novel genes (MJM genes), of which a substantial part is potentially involved, either structurally or regulatory, with tobacco secondary metabolism. In-depth comparisons with previously reported transcriptome analyses underscore the specificity and the potentially additive value of the cDNA-AFLP transcript profiling technology to unravel secondary metabolism in nonmodel plant systems. The gene inventory revealed the presence of all, except one, of the genes known to be involved in the biosynthesis of nicotine alkaloids in Nicotiana species. Most of these were induced early by methyl jasmonate in tobacco BY-2 cells (within 2 h) and clustered together with novel genes or genes encoding proteins with still unknown functions (Fig. 2C), which are probable candidates to encode the missing links in nicotine biosynthesis. Moreover, kinetic analysis of MJM genes in relation to the kinetics of appearance of the different nicotine-related alkaloids will allow selecting the most likely candidate genes involved in nicotine-related alkaloid biosynthesis.
The transcriptome also revealed numerous jasmonate-induced genes involved in signal transduction, such as transcription factors, GTP-binding proteins, receptors, kinases, and phosphatases (Fig. 2F; see also Table 2). Little is known about the regulation of secondary metabolite biosynthesis in plants and on the action mechanisms of jasmonates. Such a detailed analysis of jasmonate-regulated genes could shed new light on this interesting problem. For instance, the most represented family of transcription factors within the MJM data set is the AP2-domain family. Members of this family have been shown to be jasmonate responsive and to be involved in terpenoid indole alkaloid metabolism (51, 52). Remarkably, for all MJMs belonging to the AP2-domain family, up-regulation can be observed from the first time point in the experimental period (Fig. 2F; see also Table 2), thus preceding the up-regulation of genes involved in nicotine biosynthesis. This finding mirrors the potential role of the AP2 factors as activators of tobacco secondary metabolism.
In conclusion, the functional genomics approach described here could stimulate the further dissection and engineering of plant secondary metabolism. The extensive amounts of detailed kinetics data that are obtained can provide vital information to enable both biosynthetic pathway and signal cascade mapping. Importantly, the cDNA-AFLP-based transcript profiling technique can also discriminate between enzyme isoforms (17), which often play distinct roles in primary and secondary metabolism. However, the major advantage of the cDNA-AFLP-based transcript profiling is that no preexisting gene sequence databanks are required. Consequently, in principle this method can be applied on any plant or plant cell culture to unravel the biosynthesis of any metabolite. In the field of plant secondary metabolism this is an important asset because many highly interesting compounds are often produced by poorly characterized, rare, or exotic plant species.
Supplementary Material
Acknowledgments
We thank Debbie Rombaut, Geert Goeminne, and Kari Kammiovirta for excellent technical assistance; Wilson Ardiles-Diaz, Caroline Buysschaert, Jan Gielen, and Raimundo Villarroel for sequencing the MJM tags; and Prof. Mitsuo Okazaki (Shinshu University, Ueda, Japan) and Prof. Joe Chappell (University of Kentucky, Lexington) for kindly providing scopolin and capsidiol samples, respectively. This work was supported by research funds of the Ghent University Geconcerteerde Onderzoeksacties (No. 12051403 to A.G.), the Finnish National Technology Agency NeoBio Program (to K.-M.O.-C.), and the Italian Ministero dell'Istruzione dell'Università e della Ricerca (to S.B.). S.T.H. and V.D.S. are recipients of a predoctoral fellowship of the Finnish Graduate School of Applied Bioscience and a predoctoral fellowship of the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen, respectively.
Abbreviations: AFLP, amplified fragment length polymorphism; BY-2, Bright yellow-2; MJM, methyl jasmonate modulated; mPut, N-methylputrescine.
References
- 1.Verpoorte, R. (2000) in Metabolic Engineering of Plant Secondary Metabolism, eds. Verpoorte, R. & Alfermann, A. W. (Kluwer, Dordrecht, The Netherlands), pp. 1-29.
- 2.Hutchinson, C. R. (1994) Biotechnology 12 375-380. [DOI] [PubMed] [Google Scholar]
- 3.Chartrain, M., Salmon, P. M., Robinson, D. K. & Buckland, B. C. (2000) Curr. Opin. Biotechnol. 11 209-214. [DOI] [PubMed] [Google Scholar]
- 4.Verpoorte, R. & Memelink, J. (2002) Curr. Opin. Biotechnol. 13 181-187. [DOI] [PubMed] [Google Scholar]
- 5.Dixon, R. A. (2001) Nature 411 843-847. [DOI] [PubMed] [Google Scholar]
- 6.Kliebenstein, D., Pedersen, D., Barker, B. & Mitchell-Olds, T. (2001) Genetics 161 325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decker, G., Wanner, G., Zenk, M. H. & Lottspeich, F. (2000) Electrophoresis 21 3500-3516. [DOI] [PubMed] [Google Scholar]
- 8.Schoendorf, A., Rithner, C. D., Williams, R. M. & Croteau, R. B. (2001) Proc. Natl. Acad. Sci. USA 98 1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamazaki, M. & Saito, K. (2002) Cell. Mol. Life Sci. 59 1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lange, B. M., Wildung, M. R., Stauber, E. J., Sanchez, C., Pouchnik, D. & Croteau, R. (2000) Proc. Natl. Acad. Sci. USA 97 2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelton, D., Leach, D., Baverstock, P. & Henry, R. (2002) Plant Sci. 162 9-15. [Google Scholar]
- 12.Suzuki, H., Achnine, L., Xu, R., Matsuda, S. P. T. & Dixon, R. A. (2002) Plant J. 32 1033-1048. [DOI] [PubMed] [Google Scholar]
- 13.Aharoni, A., Keizer, L. C., Bouwmeester, H. J., Sun, Z., Alvarez-Huerta, M., Verhoeven, H. A., Blaas, J., van Houwelingen, A. M., De Vos, R. C., van der Voet, H., et al. (2000) Plant Cell 12 647-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guterman, I., Shalit, M., Menda, N., Piestun, D., Dafny-Yelin, M., Shalev, G., Bar, E., Davydov, O., Ovadis, M., Emanuel, M., et al. (2002) Plant Cell 14 2325-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breyne, P. & Zabeau, M. (2001) Curr. Opin. Plant Biol. 4 136-142. [DOI] [PubMed] [Google Scholar]
- 16.Breyne, P., Dreesen, R., Vandepoele, K., De Veylder, L., Van Breusegem, F., Callewaert, L., Rombauts, S., Raes, J., Cannoot, B., Engler, G., et al. (2002) Proc. Natl. Acad. Sci. USA 99 14825-14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breyne, P., Dreesen, R., Cannoot, B., Rombaut, D., Vandepoele, K., Rombauts, S., Vanderhaeghen, R., Inzé, D. & Zabeau, M. (2003) Mol. Genet. Genom. 269 173-179. [DOI] [PubMed] [Google Scholar]
- 18.Nagata, T., Nemoto, Y. & Hasezawa S. (1992) Int. Rev. Cytol. 132 1-30. [Google Scholar]
- 19.Tavazoie, S., Hughes, J. D., Campbell, M. J., Cho, R. J. & Church, G. M. (1999) Nat. Genet. 22 281-285. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman D. J. (1997) Nucleic Acids Res. 25 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biondi, S., Scaramagli, S., Capitani, F., Altamura, M. M. & Torrigiani, P. (2001) J. Exp. Bot. 52 231-242. [PubMed] [Google Scholar]
- 23.Sharan, M., Taguchi, G., Gonda, K., Jouke, T., Shimosaka, M., Hayashida, N. & Okazaki, M. (1998) Plant Sci. 132 13-19. [Google Scholar]
- 24.Gage, T. G., Douglas, C. D. & Wender, S. H. (1951) Anal. Chem. 23 1582. [Google Scholar]
- 25.Medic-Saric, M., Stanic, G. & Bosnjak, I. (2001) Pharmazie 56 156-159. [PubMed] [Google Scholar]
- 26.Keinänen, M., Oldham, N. J. & Baldwin, I. T. (2001) J. Agric. Food Chem. 49 3553-3558. [DOI] [PubMed] [Google Scholar]
- 27.Mandujano-Chávez, A., Schoenbeck, M. A., Ralston, L. F., Lozoya-Gloria, E. & Chappell, J. (2000) Arch. Biochem. Biophys. 381 285-294. [DOI] [PubMed] [Google Scholar]
- 28.Nugroho, L. H. & Verpoorte, R. (2002) Plant Cell Tissue Organ Cult. 68 105-125. [Google Scholar]
- 29.Imanishi, S., Hashizume, K., Nakakita, M., Kojima, H., Matsubayashi, Y., Hashimoto, T., Sakagami, Y., Yamada, Y. & Nakamura, K. (1998) Plant Mol. Biol. 38 1101-1111. [DOI] [PubMed] [Google Scholar]
- 30.Schenk, P. M., Kazan, K., Wilson, I., Anderson, J. P., Richmond, T., Somerville, S. C. & Manners, J. M. (2000) Proc. Natl. Acad. Sci. USA 97 11655-11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki, Y., Asamizu, E., Shibata, D., Nakamura, Y., Kaneko, T., Awai, K., Amagai, M., Kuwata, C., Tsugane, T., Masuda, T., et al. (2001) DNA Res. 8 153-161. [DOI] [PubMed] [Google Scholar]
- 32.Stintzi, A., Weber, H., Reymond, P., Browse, J. & Farmer, E. E. (2001) Proc. Natl. Acad. Sci. USA 98 12837-12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheong, Y. H., Chang, H. S., Gupta, R., Wang, X., Zhu, T. & Luan, S. (2002) Plant Physiol. 129 661-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creelman, R. A. & Mullet, J. E. (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 355-381. [DOI] [PubMed] [Google Scholar]
- 35.Wasternack, C. & Parthier, B. (1998) Trends Plant Sci. 2 302-307. [Google Scholar]
- 36.Avdiushko, S., Croft, K. P., Brown, G. C., Jackson, D. M., Hamilton-Kemp, T. R. & Hildebrand, D. (1995) Plant Physiol. 109 1227-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bate, N. J. & Rothstein, S. J. (1998) Plant J. 16 561-569. [DOI] [PubMed] [Google Scholar]
- 38.Hibi, N., Higashiguchi, S., Hashimoto, T. & Yamada, Y. (1994) Plant Cell 6 723-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riechers, D. E. & Timko, M. P. (1999) Plant Mol. Biol. 41 387-401. [DOI] [PubMed] [Google Scholar]
- 40.Glenn, D. F. & Edwards, W. B., III (1978) J. Org. Chem. 43 2860-2870. [Google Scholar]
- 41.Luanratana, O. & Griffin, W. J. (1982) Phytochemistry 21 449-451. [Google Scholar]
- 42.Kisaki, T., Mizusaki, S. & Tamaki, E. (1968) Phytochemistry 7 323-327. [Google Scholar]
- 43.Strunz, G. M. & Findlay, J. A. (1985) in The Alkaloids, Chemistry and Pharmacology, ed. Brossi, A. (Academic, Orlando, FL), Vol. 26, pp. 124-151. [Google Scholar]
- 44.Dixon, R. A. & Paiva, N. L. (1995) Plant Cell 7 1085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Memelink, J., Verpoorte, R. & Kijne, J. W. (2001) Trends Plant Sci. 6 212-219. [DOI] [PubMed] [Google Scholar]
- 46.Menke, F. L., Parchmann, S., Mueller, M. J., Kijne, J. W. & Memelink, J. (1999) Plant Physiol. 119 1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi, D., Bostock, R. M., Avdiushko, S. & Hildebrand, D. F. (1994) Proc. Natl. Acad. Sci. USA 91 2329-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somssich, I. E. & Hahlbrock, K. (1998) Trends Plant Sci. 3 86-90. [Google Scholar]
- 49.Logemann, E., Tavernaro, A., Schulz, W., Somssich, I. E. & Hahlbrock, K. (2000) Proc. Natl. Acad. Sci. USA 97 1903-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, J., Sheehan, M., Brookman, H. & Timko, M. P. (2000) Plant Sci. 158 19-32. [DOI] [PubMed] [Google Scholar]
- 51.Menke, F. L., Champion, A., Kijne, J. W. & Memelink, J. (1999) EMBO J. 18 4455-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Fits, L. & Memelink, J. (2000) Science 289 295-297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.