Abstract
Evidence that pre-mRNA processing events are temporally and, in some cases, mechanistically coupled to transcription has led to the proposal that RNA polymerase II (Pol II) recruits pre-mRNA splicing factors to active genes. Here we address two key questions raised by this proposal: (i) whether the U1 snRNP, which binds to the 5′ splice site of each intron, is recruited cotranscriptionally in vivo and, (ii) if so, where along the length of active genes the U1 snRNP is concentrated. Using chromatin immunoprecipitation (ChIP) in yeast, we show that elevated levels of the U1 snRNP were specifically detected in gene regions containing introns and downstream of introns but not along the length of intronless genes. In contrast to capping enzymes, which bind directly to Pol II, the U1 snRNP was poorly detected in promoter regions, except in genes harboring promoter-proximal introns. Detection of the U1 snRNP was dependent on RNA synthesis and was abolished by intron removal. Microarray analysis revealed that intron-containing genes were preferentially selected by ChIP with the U1 snRNP. Thus, U1 snRNP accumulation at genes correlated with the presence and position of introns, indicating that introns are necessary for cotranscriptional U1 snRNP recruitment and/or retention.
Pre-mRNA splicing is a two-step transesterification reaction carried out by the spliceosome, a large and dynamic multicomponent RNA-protein complex (52). The first steps in the assembly of the spliceosome on pre-mRNA involve the recognition of the 5′ and 3′ ends of each intron (5′ and 3′ splice sites) by small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP splicing factors. Regulation of this process determines splice site usage in alternative pre-mRNA splicing (50). A report that 40 to 60% of human genes are alternatively spliced to produce multiple gene products (26) underscores the importance of understanding splice site recognition and subsequent spliceosome assembly. Although much progress has been made in recent years toward understanding the biochemical activities of many splicing regulators, it has been difficult to establish systems for examining the roles of such regulators on endogenous pre-mRNAs in vivo and the mechanisms by which they are recruited.
An important clue to understanding how splicing factors might initially assemble on pre-mRNA is provided by observations that splicing begins and is sometimes completed cotranscriptionally (for a review, see reference 39). For a number of genes, intron removal has been detected in nascent RNAs still tethered to the DNA axis by RNA polymerase II (Pol II) (3, 5, 42, 53, 54, 56). Evidence that transcription rates and promoter identity influence alternative splice site selection is consistent with a cotranscriptional splicing mechanism in humans (9, 21, 45) and yeast (K. J. Howe, C. M. Kane, and M. Ares, unpublished data). The findings that the C-terminal domain (CTD) of RNA Pol II is required for efficient capping, splicing, and polyadenylation of pre-mRNA (33) and specifically stimulates splicing in humans (14) have led to the proposal that Pol II itself recruits splicing factors to nascent RNA (4, 15, 31). Thus, splicing factors may resemble capping enzymes, which bind directly to Pol II via the CTD (7, 32) and do not appear to require RNA recognition for initial targeting to Pol II transcripts.
However, splicing need not always occur cotranscriptionally. A significant fraction of introns are excised after transcription termination (3, 54, 56, 57). Observations of recursive splicing, in which pre-mRNAs are spliced and then respliced, also indicate that not all splicing events are coupled directly to transcription (17, 29). Although cotranscriptional splicing in yeast is suggested by the kinetics of mRNA appearance (13), it has not been directly observed, and a report of recursive splicing has been used to argue against cotranscriptional splicing in yeast (29). Moreover, it is well known that purified pre-mRNAs synthesized by viral RNA polymerases can be spliced in vitro (25). Unlike the capping enzymes, many splicing regulators bind to sequence-specific elements in the pre-mRNA (50), suggesting that direct pre-mRNA binding may be sufficient for splicing in vivo. Thus, major questions in the field remain: to what extent are pre-mRNA splicing factors recruited cotranscriptionally and what are the requirements for pre-mRNA splicing factor recruitment in vivo?
Here we address these questions with respect to the U1 snRNP, the activity of which is required for pre-mRNA splicing in all species, from yeast to humans. The U1 snRNA base pairs with the 5′ splice site, thereby determining 5′ splice site usage, and the U1 snRNP is a component of the earliest biochemically defined splicing complexes (6, 36, 47-49, 58). Recently, it has been shown that the U1 snRNP-specific protein U1C also contacts the 5′ splice site in yeast (12). The U1 snRNP is not present in the active spliceosome, in which the U6 snRNA base pairs with the 5′ splice site to accomplish the catalytic steps (52). Although antigens shared among spliceosomal snRNPs have been detected by immunocytochemistry at active intron-containing transcription units in metazoans (22, 37), cotranscriptional U1 snRNP recruitment has never been specifically demonstrated or examined in detail.
Because nascent RNP complexes are likely to lie adjacent to the DNA axis (55), it seemed possible that splicing factors bound to nascent RNA could be detectable by chromatin immunoprecipitation (ChIP). Studies of chromatin and transcriptional regulation have been advanced by the ChIP technique, which has been used to localize specific factors and chromatin modifications to particular genomic regions (41). Because pre-mRNA splicing patterns are well understood in the yeast Saccharomyces cerevisiae and introns and exons annotated with respect to the complete genome sequence (30, 51), we chose to examine the association of the U1 snRNP with transcription units by ChIP in yeast cells. Here we show that the U1 snRNP accumulates on transcriptionally active intron-containing genes, a finding consistent with a cotranscriptional splicing mechanism in yeast cells. In contrast to capping enzymes, detectable at all promoters, the highest levels of the U1 snRNP coincided with regions of intron synthesis.
MATERIALS AND METHODS
Yeast strains, growth conditions, and tagging.
Yeast strains (Table 1) were grown in YP medium plus 2% glucose (YPD), YPD medium plus G418 (200 mg/ml), synthetic complete medium plus 2% glucose without histidine or uracil, or YP medium plus 2% galactose as necessary. Strains with protein A-tagged proteins Nam8p and Prp42p (BSY593 and BSY646) have been previously described (16, 43) and were a generous gift of Bertrand Seraphin. A PCR-based strategy (23) to epitope tag endogenous genes was used to generate strain YKK19, which has a hemagglutinin (HA) tag on the C terminus of Prp42p and a Myc-tag on the C terminus of Rpo21p, from strain LG1 and strains YKK20 and YIB37K, which both have an HA-tag on the C terminus of Prp42p, from DBY120 and YIB37, respectively. The hemagglutinin (HA)-tagged Prp42 was derived from pYM1 containing three copies of the HA epitope and KanMx6 selection marker. The Myc-tagged RPO21 was derived from pYM5 containing three copies of the Myc epitope and His selection marker. Tags were verified by Western blotting. Strain YGL130w contains a tandem affinity purification (TAP)-tagged Ceg1p (44) and was obtained from Cellzome. GAL1 gene expression was induced by growth in 2% galactose for 5 h. In the temperature shift assay, cells were grown at 24°C until an optical density at 600 nm of ∼0.600 was achieved; half of the the cells then continued growth at 24°C, while the remaining half were shifted to 37°C for 45 min.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference (isogenicity) |
|---|---|---|
| BSY593 | Previously described | Gottschalk et al. (16) |
| BSY646 | Previously described | Gottschalk et al. (16) |
| LG1 | MATaura3 his3-11 leu2-3,112 trp1-1 can1-100 | B. Byers |
| YKK19 | MATaura3 his3-11 leu2-3,112 trp1-1 can1-100 prp42::prp42HA/HIS3 rpo21::rpo21MYC/ kanMX6 | This study (isogenic with LG1) |
| DBY120 | MATaura3-52 rpb1-1 trp1::hisG | McNeil et al. (35) |
| YKK20 | MATaura3-52 rpb1-1 trp1::hisG prp42::prp42HA/kanMX6 | This study (isogenic with DBY120) |
| YIB37 | MATα ade2-1 ura3-1 leu2-3,112 trp1-1,15 his3-11,15 can1-100 DBP2::DBP2cDNA URA3 | Barta and Iggo (2) |
| YIB37K | MATα ade2-1 ura3-1 leu2-3,112 trp1-11,15 his3-11,15 can1-100 DBP2::DBP2cDNA URA3 prp42::prp42HA/kanMX6 | This study (isogenic with YIB37) |
| YGL130w | MATaade2 arg4 leu2-3,112 trp1-289 ura3-52 YGL130w::TAP-K.I.URA3 | Cellzome |
ChIP.
The ChIP was performed as described previously (18). Rabbit immunoglobulin G (IgG)-agarose beads (Sigma) were used with protein A-tagged proteins and TAP-tagged Ceg1p; anti-HA monoclonal antibody (MAb) 12CA5 (Boehringer Mannheim), followed by the addition of gamma-bind G Sepharose beads (Amersham) with HA-tagged proteins; anti-Myc MAb 9E10 (Santa Cruz) with Myc-tagged proteins, and MAb 8WG16 (Babco) against Pol II itself. In ChIPs of strains tagged with protein A tags or TAP tags, Cl-4b beads (Sigma) were used as negative control, while rabbit IgG-agarose beads (Sigma) were used to specifically retrieve these tags.
DNA was analyzed by PCR with multiplex primer sets (sequences available upon request) along the genes of interest. Cycling was for 3 min at 94°C, followed by 24 cycles with 1 min at 94°C, 1 min at 56°C, 2 min at 72°C, and then finally 7 min at 72°C. Three concentrations of each template were used over a 30-fold range, and PCR products were distinguished on high-resolution 2.3% MetaPhor agarose gels (BioWhittaker) and stained with Gelstar. Lanes were chosen for quantitation and figures, based on the intensity of the signal lying in the linear range. Negative control lanes represent PCRs in which the amount of template matched the amount of experimental template used. When results were quantified (ImageQuant software; Molecular Dynamics), the gel background was subtracted, and signals were normalized for the intensity of bands generated from the input material to adjust for differences in PCR efficiency. Signals in intron and exon 2 regions are expressed relative to promoter levels. Note that Prp42p ChIP signals at the promoter are very close to background; therefore, background signal from the control ChIPs was not subtracted.
Genome localization analysis.
Cy3- and Cy5-labeled probes were prepared by linker-mediated PCR of the HA-Prp42p and Myc-Pol II ChIP templates (see above) and sonicated genomic DNA fragments present in the starting extract according to the published protocol (44). Two-color competitive hybridization experiments with S. cerevisiae cDNA microarrays were performed at the Fred Hutchison Cancer Research Center (Seattle, Wash.). Microarray construction, target labeling, and hybridization protocols were adapted from those described previously (10). Yeast microarrays were constructed employing a set of 6229 open reading frame (ORF)-specific PCR primer pairs (Research Genetics, Huntsville, Ala.), which were used to amplify ≤1-kb 3′-end portions of each ORF. Individual PCR products were verified as unique via gel electrophoresis and purified by using ArrayIt 96-well PCR purification kits (TeleChem International, Sunnyvale, Calif.). Purified PCR products were mechanically “spotted” in 3× SSC (450 mM sodium cloride and 45 mM sodium citrate; pH 7.0) onto polylysine-coated microscope slides by using an OmniGrid high-precision robotic gridder (GeneMachines, San Carlo, Calif.). Probes were cohybridized to microarrays for 16 h at 63°C and sequentially washed at room temperature in 1× SSC-0.03% sodium dodecyl sulfate for 2 min, 1× SSC for 2 min, 0.2× SSC with agitation for 20 min, and 0.05× SSC with agitation for 10 min. Arrays were immediately centrifuged until dry and scanned by using a GenePix 4000 scanner (Axon Instruments, Union City, Calif.). Image analysis was performed by using GenePix Pro 3.0. In each independent experiment, data points were eliminated if there were defects over particular spots or if fluorescence intensities in either channel were unreliably low relative to the local background. To identify ORFs enriched by HA-Prp42p and Myc-Pol II ChIP procedures, the ratios of Cy3 and Cy5 fluorescence intensities were expressed as a ChIP score of log10(median intensity fluorochrome 1/median intensity fluorochrome 2)/√2̄. Frequency histograms of the ChIP scores indicated a normal distribution of values for genomic-genomic hybridizations and a major peak with an outlying second peak for HA-Prp42-genomic and Myc-Pol II-genomic data sets. The means and standard deviations (SD) of all ChIP scores in each experiment were determined, and ORFs with ChIP scores that were >2 SD away from the mean were chosen for further analysis. Outlying ORFs selected by these criteria from five HA-Prp42p ChIP experiments (three with Cy5-Prp42 probes and two with Cy3-Prp42 probes) and three Myc-Pol II ChIP experiments (two with Cy3-Pol II probes and one with Cy5-Pol II probes) were pooled and analyzed for the reproducibility of their identification and the presence of introns within the genomic segment. Note that, because the probes were double stranded, ORFs may be hit on either DNA strand. Therefore, in scoring for introns, we considered any ORF to be intron containing if the ORF itself has an intron or if the ORF is within 500 nucleotides (nt) of an intron-containing gene on either strand. Because of the density of ORFs within the yeast genome, this occurred fairly frequently (see Results).
RESULTS
To determine whether the U1 snRNP accumulates cotranscriptionally and is detectable by ChIP, we constructed strain YKK19 (Table 1), harboring tagged copies of endogenous Pol II (Myc-Rpo21p) and Prp42p (HA-Prp42p), a U1 snRNP-specific protein (34, 43). Figure 1A shows schematically the proposed binding of the tagged U1 snRNP to nascent RNA, which is in turn tethered to the DNA axis by Pol II. Tagging of both essential proteins had no effect on the growth rate of the strain (data not shown), indicating that the normal functions of the Prp42p and Pol II were not disrupted. Two endogenous, intron-containing genes, ASC1 and DBP2 (Fig. 1B and C), were initially selected for analysis by ChIP because they are highly transcribed and have relatively large first exons (>500 bp) (19, 51). The DNA shearing procedure abolished PCR detection of ≥1-kb stretches along the genes of interest while preserving detection of ≤400-bp stretches (data not shown). Therefore, if the U1 snRNP associates with these genes, the system should resolve signals before and after intron synthesis.
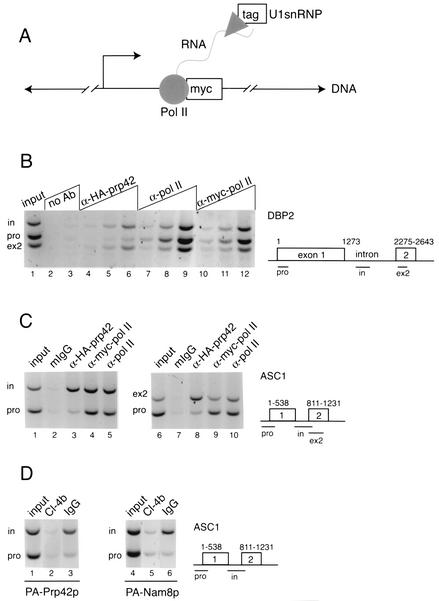
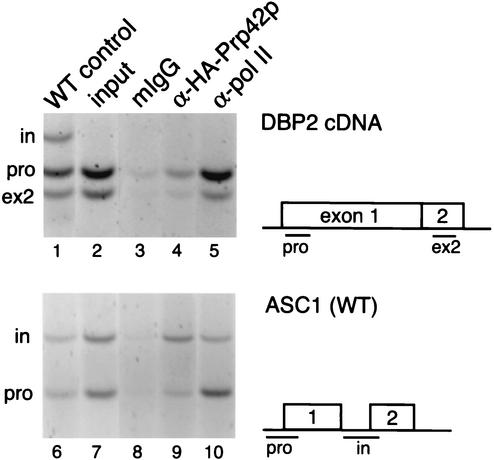
FIG. 1.
Detection of the U1 snRNP at intronic and downstream segments of DBP2 and ASC1 genes by ChIP. (A) Experimental design. In an endogenous transcription unit, the tagged U1 snRNP (Prp42p or Nam8p, triangle) is tethered to the chromatin by nascent RNA via Myc-tagged RNA Pol II (Rpo21p, circle). After formaldehyde cross-linking, random shearing of DNA (hatch marks), and specific immunoprecipitation for the tag, coimmunoprecipitated DNA will be detected by PCR. (B) PCR detection of specific regions within the DBP2 gene in the starting YKK19 extract (input, lane 1), control beads (no Ab, lanes 2 and 3), and ChIPs carried out with antibodies specific for the HA-tagged Prp42p (lanes 4 to 6), Pol II itself (8WG16, lanes 7 to 9), and Myc-tagged Pol II (lanes 10 to 12). A schematic diagram of DBP2 shows the layout of the ORF with respect to introns and exons, as well as the locations of predicted PCR products representing the promoter (pro), intron (in), and second exon (ex2) regions. A titration of ChIP templates used for PCR is shown, in which 0.3 μl (lanes 4, 7, and 10), 1.0 μl (lanes 2, 5, 8, and 11), and 3.0 μl (lanes 3, 6, 9, and 12) were added. Data shown here is representative of four independent ChIP experiments. (C) PCR detection of promoter (pro), intron (in), and exon2 (ex2) regions of the ASC1 gene in the starting extract (input, lanes 1 and 6), mouse nonimmune IgG control ChIP (mIgG, lanes 2 and 7), and ChIPs carried out with antibodies specific for the HA-tagged Prp42p (lanes 3 and 8), Myc-tagged Pol II (lanes 4 and 9), and Pol II itself (8WG16, lanes 5 and 10). A schematic diagram of ASC1 shows the layout of the ORF and the locations of predicted PCR products. The data shown here are representative of three independent ChIP experiments. In this experiment, Prp42p ChIP templates yielded intron DNA levels 4.6-fold and ex2 DNA levels 7.4-fold above promoter levels, relative to Pol II. (D) Detection of elevated U1 snRNP levels within the ASC1 intron in strains harboring protein A-tagged Prp42p or Nam8p, after ChIP with rabbit IgG-coated beads. The data shown here are representative of three (PA-Prp42p) and two (PA-Nam8p) independent ChIP experiments.
Three gene regions from both DBP2 and ASC1 were well represented in the ChIPs of Pol II, whether the anti-Myc tag antibody (Fig. 1B, lanes 10 to 12; Fig. 1C, lanes 4 and 9) or an antibody against Pol II itself (Fig. 1B, lanes 7 to 9; Fig. 1C, lanes 5 and 10) was used. In agreement with previous results with tagged versions of various subunits of Pol II (24, 46), the distribution of Pol II along both genes was found to be fairly uniform, usually decreasing slightly downstream from the promoter. PCR products specifically resulted from ChIP of Pol II, since the same products were detected only very weakly in the nonimmune control ChIP (Fig. 1B, lanes 2 and 3; Fig. 1C, lanes 2 and 7), and Pol II was not detected at either the transcriptionally repressed GAL1 gene or the untranscribed telomere VI R (see Fig. 3B and 4). In contrast, HA-Prp42p ChIP templates yielded only very low levels of PCR product corresponding to either of the promoter regions, whereas intron and exon 2 regions of both genes were well detected (Fig. 1B, lanes 4 to 6; Fig. 1C, lanes 3 and 8). Quantitation of PCR products within the linear range (see Materials and Methods) revealed that Prp42p ChIPs contained 6.4 ± 1.1 (mean ± SEM, n = 4 independent experiments)-fold-higher levels of DBP2 intron DNA than promoter-proximal DNA compared to Pol II ChIP templates prepared in parallel. Similarly, the DBP2 second exon was detected at 6.3 ± 2.4 (n = 4)-fold-higher levels than the promoter region. We conclude that Prp42p is concentrated in downstream regions of both DBP2 and ASC1 genes both during and after intron synthesis.
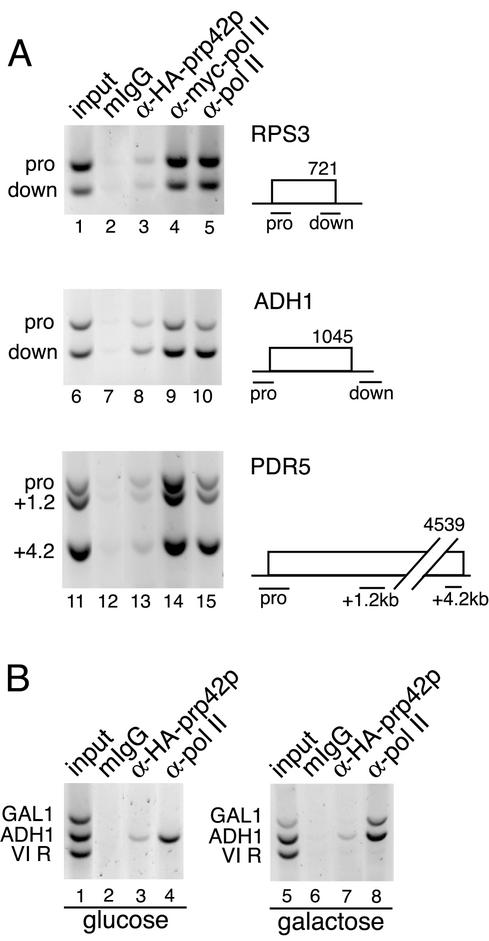
FIG. 3.
U1 snRNP levels remain constant along intronless genes. (A) ChIP analysis of three representative intronless genes—RPS3, ADH1, and PDR5—comparing promoter regions with downstream regions in the YKK19 strain. The data shown here are representative of two (RPS3 and PDR5) and three (ADH1) independent ChIP experiments. In these experiments, the ratio of Prp42p to Pol II signals at the downstream positions relative to promoters were 1.09 for RPS3, 0.88 for ADH1, and 0.93 and 0.62 for the two downstream positions in PDR5, which are ∼1.2 and ∼4.0 kb away from the promoter, respectively. (B) Activation of GAL1 by growth in galactose (lanes 5 to 8) causes the accumulation of Pol II but not Prp42p at the promoter. When grown in glucose (lanes 1 to 4), Pol II was strongly and Prp42p was weakly detected only on ADH1. Neither protein was detected at the telomere VI R. The data shown here are representative of four independent ChIP experiments.
FIG. 4.
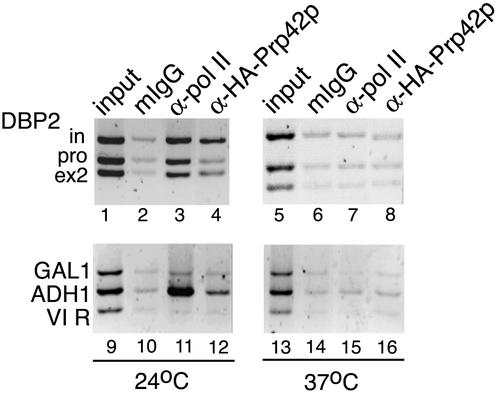
Accumulation of the U1 snRNP on DBP2 depends on active transcription. Strain YKK20 contains a temperature-sensitive Pol II (rpb1-1) and an HA-tagged prp42p. When ChIP was performed on YKK20 grown at permissive temperature (24°C), the distribution of Pol II and Prp42p was determined as shown on DBP2 (lanes 1 to 4) and on ADH1, GAL1, and the telomere VI R (lanes 9 to 12). When the temperature was shifted to a nonpermissive level (37°C), Pol II no longer accumulated along DBP2 (lane 7) or ADH1 (lane 15), and Prp42p was not detected above control levels on DBP2 (lane 8) or ADH1 (lane 16). Neither protein was detected on GAL1 or the telomere VI R. The data shown here are representative of four independent ChIP experiments.
To confirm that Prp42p detection reflects the cotranscriptional accumulation of the U1 snRNP, we performed ChIP experiments with strains harboring other tagged versions of U1 snRNP-specific proteins. First, we examined protein A-tagged Prp42p and Nam8p, a second U1-specific protein (16). These strains were used previously to show that both PA-Prp42p and PA-Nam8p associate with the U1 snRNA by immunoprecipitation (16) and to study the interaction of PA-Nam8p with pre-mRNA in commitment complex formation (43). Figure 1D shows that ChIP with rabbit IgG-coated beads preferentially selects the ASC1 intron region relative to the promoter in both strains. This experiment also indicates that the HA tag introduced into YKK19 does not produce different results from other tagged versions. Second, HA-tagged Prp40p, another U1 snRNP component which has been shown to bind hyperphosphorylated Pol II CTD by Far Western analysis (38), was detected on DBP2 and ASC1 in an identical pattern to Prp42p (data not shown).
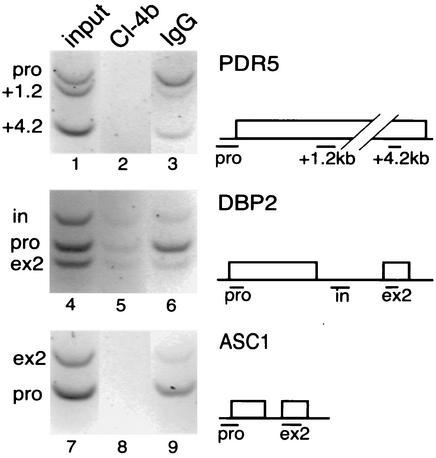
The present observation of U1 snRNP accumulation in downstream regions of ASC1 and DBP2 contrasts strongly with previous studies of the capping enzymes Ceg1p, Cet1p, and Abd1p, which have been detected at promoter regions of transcriptionally active genes (24, 46). Note that, unlike Abd1p, which remains associated with downstream regions, Ceg1p and Cet1p are preferentially concentrated in promoter regions (24, 46). To facilitate a direct comparison between U1 snRNP and capping enzyme dynamics on ASC1 and DBP2, ChIP was performed by using a strain containing TAP-tagged Ceg1p, the mRNA guanylyltransferase (Fig. 2). As expected, TAP-Ceg1p was highly concentrated at the promoter regions of the intronless gene PDR5 assayed in a previous study (24). Similarly, TAP-Ceg1p was concentrated on both ASC1 and DBP2 promoter regions, verifying the differential distribution of capping and splicing factors on two intron-containing genes.
FIG. 2.
Ceg1p is concentrated at the promoters of PDR5, ASC1, and DBP2. The distribution of the TAP-tagged mRNA guanylyltransferase, Ceg1p, was compared in promoter and downstream regions of three genes (see gene diagrams). Input lanes 1, 4, and 7 show the starting extract from the TAP-tagged Ceg1p yeast strain, and Cl-4b lanes 2, 5, and 8 show no antibody control. High levels of Ceg1p were detected on promoter (pro) regions when the immunoprecipitation was performed with rabbit IgG-coated beads (lanes 3, 6, and 9), and the downstream regions of all three genes (+1.2kb and +4.2kb for PDR5, in and ex2 for DBP2, and ex2 for ASC1) showed a decrease in Ceg1p. The data shown here are representative of two (PDR5 and ASC1) and three (DBP2) independent ChIP experiments.
To address the possibility that U1 snRNP may accumulate in downstream regions of all genes, we assayed U1 snRNP and Pol II distributions along three intronless genes. ChIP with anti-Myc-Pol II or anti-Pol II showed robust signals for promoter and downstream regions in RPS3, ADH1, and PDR5 genes (Fig. 3A, lanes 4, 5, 9, 10, 14, and 15). In contrast, HA-Prp42p was detected at very low levels relative to the control at every position along RPS3 and PDR5, while somewhat higher levels of HA-Prp-42p were observed on ADH1 (Fig. 3A lanes 3, 8, and 13). Relative to levels obtained with Pol II ChIP templates, signals from HA-Prp42p ChIP either decreased or remained the same along the length of each gene. Therefore, U1 snRNP accumulation on DBP2 or ASC1 downstream regions cannot be attributed to generic changes in affinity for elongating Pol II, nonspecific binding to nascent RNA, or recruitment by the 5′ cap.
Because a low level of U1 snRNP association was detected within intronless genes relative to the nonimmune controls (Fig. 3A, compare lanes 2 and 3, 7 and 8, and 12 and 13), we sought to determine whether gene induction is sufficient for U1 snRNP accumulation. GAL1 gene transcription was induced and changes in Prp42p association with the GAL1 promoter were determined. When YKK19 was grown in glucose, Pol II was detected at the ADH1 promoter but not on GAL1 or a transcriptionally inactive telomeric region on chromosome VI-R (Fig. 3B, lane 4). Low levels of HA-Prp42p were detected at ADH1 only (Fig. 3B, lane 3), as expected (see Fig. 3A). After 5 h of growth in galactose, Pol II was present on both ADH1 and GAL1, but Prp42p was not detected on the GAL1 promoter (Fig. 3B, lanes 7 and 8) or in the downstream region (data not shown) relative to the nonimmune control. This suggests that U1 snRNP detection on chromatin is not due to transcriptional activity per se.
If U1 snRNP accumulation reflects specific binding to cognate sites in nascent RNA as proposed (Fig. 1A), then it is expected to depend on RNA synthesis. To test this prediction, we introduced an HA tag into the endogenous copy of Prp42p in strain DBY120, harboring the temperature-sensitive rpb1-1 allele of the Pol II large subunit (35, 40). A previous study showed that in this strain the Pol II holoenzyme dissociates from previously active transcription units when the cells are shifted to the nonpermissive temperature (46). As expected, Pol II was detected at the ADH1 promoter and along DBP2 in DBY120 cells grown at 24°C but not at GAL1 or the telomere VI R (Fig. 4). After 45 min of growth at 37°C, Pol II was not detectable above background levels at any gene region tested (Fig. 4, lanes 7 and 15). Similarly, the U1 snRNP was detectable on the intron and exon 2 regions of DBP2 and ADH1 promoter in DBY120 cells grown at 24°C but was undetectable after the shift to 37°C (Fig. 4, lanes 4, 8, 12, and 16). A similar loss of Pol II and U1 snRNP detection at 37°C was observed for ASC1 (data not shown). These data indicate that active transcription is required for U1 snRNP accumulation and that the U1 snRNP is not associated with chromatin independent of transcriptional activity.
To test whether association of the U1 snRNP with downstream regions depends on the intron, we assayed U1 snRNP levels along the DBP2 gene in a strain lacking the DBP2 intron. In this strain, the endogenous DBP2 ORF was replaced by the DBP2 cDNA; transcription levels of this intronless gene were previously found to be twofold higher than wild-type levels (2). We had previously detected elevated U1 snRNP levels on both the intron and second exon of wild-type DBP2 (see Fig. 1). Figure 5 shows that removal of the intron abolishes accumulation of the U1 snRNP on downstream DNA corresponding to the second exon. Normalizing to Pol II ChIP signals, the ratio of Prp42p downstream to Prp42p at the promoter was only 0.68 (versus 6.3-fold in wild-type). Thus, despite Pol II-driven expression, elevated U1 snRNP levels were not observed at the DBP2 allele lacking its intron.
FIG. 5.
Deletion of the DBP2 intron abolishes cotranscriptional U1 snRNP accumulation. Prp42p was tagged at the C terminus with an HA tag in strain yIB37 that contains the DBP2 cDNA in place of the endogenous intron-containing ORF (2). As expected, no intron product was observed in the input (lane 2) compared to input from strain YKK19, which is wild type for DBP2 (lane 1). Equivalent amounts of template for mIgG control, anti-HA, or anti-Pol II were used for PCR (lanes 3 to 5). As a positive control, ChIP analysis is shown for ASC1 (lanes 6 to 10), which is wild type in this strain. The data shown here are representative of four independent ChIP experiments.
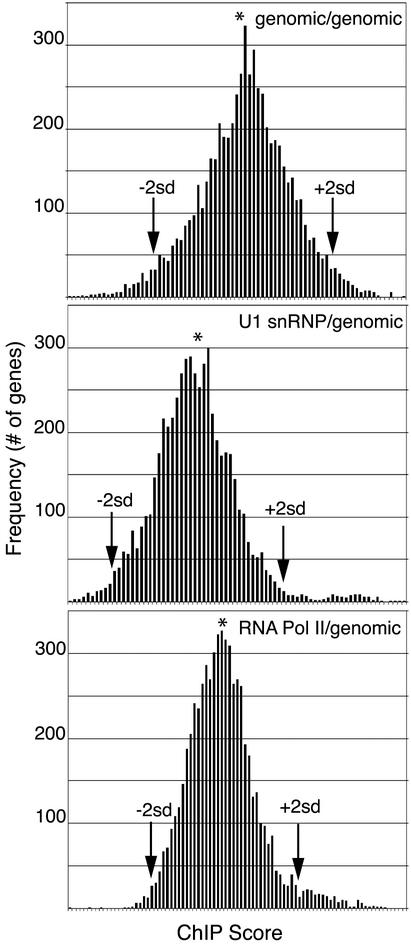
Higher levels of U1 snRNP were detected on the DBP2 gene when the allele contained an intron (Fig. 1 and 5). Therefore, we postulated that intron-containing genes, constituting only ∼5% of yeast ORFs, might accumulate more U1 snRNP than intronless genes. To test this directly, we performed genome localization analysis (20, 44). Cy3- and Cy5-labeled probes were generated from sheared YKK19 genomic DNA and HA-Prp42p and Myc-Pol II ChIP templates by linker-mediated PCR and hybridized with microarrays representing 6,229 yeast ORFs. The ratios of Cy3 and Cy5 median fluorescence intensities were used to analyze the data (see Materials and Methods). Microarrays hybridized with Cy3-genomic DNA versus Cy5-genomic DNA revealed a normal distribution of the data, as expected (Fig. 6, top panel). Interestingly, the data obtained from microarrays probed with HA-Prp42p ChIP templates versus genomic DNA yielded a major peak and a minor second peak of several hundred ORFs in each experiment, reflecting relatively higher scores for the labeled HA-Prp42p ChIP template probe (Fig. 6, center panel). Similarly, a second outlying peak was obtained from microarrays probed with Myc-Pol II ChIP templates and genomic DNA (Fig. 6, bottom panel). The striking difference between the shapes of the curves obtained with both ChIP templates compared to the genomic-genomic distribution indicates that specific sets of ORFs were selected by HA-Prp42p and Myc-Pol II ChIPs.
FIG. 6.
Genome localization analysis of HA-Prp42 and Myc-Pol II. Cy3- and Cy5-labeled probes were synthesized from sheared genomic DNA or ChIP templates by linker-mediated PCR and hybridized with microarrays representing 6,229 yeast ORFs. Frequency histograms of ChIP scores [log10(median intensity fluorochrome 1/median intensity fluorochrome 2)/√2̄] for representative experiments (Cy3-genomic/Cy5-genomic probes, top panel; Cy3-U1 snRNP/Cy5-genomic probes, middle panel; Cy3-Pol II/Cy5-genomic probes, bottom panel) show a normal distribution, with outlying second peaks in the U1 snRNP and Pol II experiments.
It was likely that the second outlying peaks obtained with HA-Prp42p and Myc-Pol II ChIP templates represented the population of ORFs associated with relatively high concentrations of Prp42p and Pol II. The mean values and the SD were determined for each experiment, and datum points >2 SD away from the mean were selected for further analysis. Results from five experiments comparing HA-Prp42p-genomic DNA and three experiments comparing Myc-Pol II-genomic DNA showed a high degree of reproducibility in the ORFs identified, by using the “2 SD criterion.” A list of HA-Prp42 ORF hits is provided in Table A1. Of the 388 ORFs hit in the HA-Prp42p arrays, 77 ORFs were hit every time (100%) and 161 ORFs were hit more than once (≥40%, Table 2). Of 373 ORFs hit in the Myc-Pol II arrays, 234 occurred more than once (≥67%, Table 2). A comparison of the Myc-Pol II hits with transcriptional frequency data (19) revealed that the outlying peak indeed contained highly transcribed ORFs (data not shown).
TABLE 2.
ORFs selected by genome localization analysis
| ChIP template(s) in microarraya | % Expts hitb | No. of ORFs | No. of ORFs with introns (%)c |
|---|---|---|---|
| U1 snRNP vs genomic DNA | 100 | 77 | 71 (92) |
| ≥80 | 103 | 93 (90) | |
| ≥60 | 125 | 104 (83) | |
| ≥40 | 161 | 118 (73) | |
| RNA Pol II vs genomic DNA | 100 | 141 | 72 (51) |
| ≥67 | 234 | 103 (44) |
Indicates Cy3- and Cy5-labeled template pairs applied to microarray.
Indicates the percentage of experiments (five for U1 snRNP, three for RNA Pol II) in which an ORF received a score >2 SD away from the mean of the population.
Indicates the number of selected ORFs which either contain introns or are ≤500 nt away from an intron-containing ORF on the same or opposite strand.
To determine whether the outlying peak observed with the HA-Prp42p ChIP templates was enriched in intron-containing ORFs, all ORFs were evaluated according to the yeast introndatabase (http://www.cse.ucsc.edu/research/compbio/yeast_introns.html). Because genome localization analysis has the potential to detect ORFs on either the Watson or the Crick strand and because the resolution of the ChIP assay is ∼400 bp, each ORF hit was also examined with respect to its position within the genome to determine whether introns occurred <500 nt away from the ORF, on either the same or opposite strand of DNA. If an ORF hit contained an intron or was found to be proximal to an intron-containing gene by the above criteria, the hit was scored as intron containing. Table 2 shows the results obtained for all of the ORFs hit more than once in the HA-Prp42p and Myc-Pol II microarrays. For the ORFs hit in every HA-Prp42p experiment, 92% were intron containing compared to 51% for Myc-Pol II. In contrast, only 4.2% of the ORFs distributed outside 2 SD for the genomic-genomic distribution were intron containing, reflecting the fact that only 5% of the yeast ORFs contain introns. Thus, we conclude that the HA-Prp42p ChIP template is highly enriched for intron-containing ORFs with respect to the genome overall.
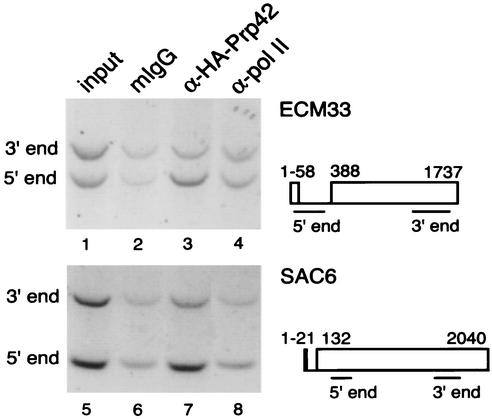
The yeast genome is predicted to contain 239 to 255 spliceosomal intron-containing ORFs (http://www.cse.ucsc.edu/research/compbio/yeast_introns.html and http://www-db.embl-heidelberg.de/jss/servlet/de.embl.bk.wwwTools.GroupLeftEMBL/ExternalInfo/seraphin/yidb.html). Al-though we detected 118 intron-containing genes by genome localization of Prp42p, we did not detect all of them. Because U1 snRNP accumulation is transcription dependent (Fig. 4), some intron-containing genes may not be expressed at levels high enough to be detected in the outlying peak. However, because the array was produced by using oligonucleotide pairs to amplify ≤1 kb of the 3′ end of each ORF (see Materials and Methods), we also considered the possibility that genes containing relatively long second exons may exhibit diminished U1 snRNP accumulation in the 3′ regions represented on the arrays, either because the U1 snRNP has already left the nascent mRNP due to spliceosome assembly or because the tags become inaccessible to antibodies in downstream gene regions. To address this concern, we examined U1 snRNP accumulation on upstream and downstream regions of two such genes, ECM33 and SAC6. Both of these genes contain introns very close to their promoters (Fig. 7), and indeed significant U1 snRNP accumulation was detected in promoter-proximal regions by ChIP (Fig. 7, lanes 3 and 7). Interestingly, HA-Prp42p detection was reduced by ∼70% in downstream regions of both genes. Neither ORF was well represented in the HA-Prp42p microarray results (Table 3), suggesting that other ORFs with relatively long second exons might not have been detected by the microarray analysis performed here.
FIG. 7.
Reduced detection of the U1 snRNP on downstream regions of intron-containing genes with long second exons. PCR detection of the 5′ and 3′ ends of ECM33 and SAC6 genes in the YKK19 strain comparing levels of Pol II and Prp42p. A schematic diagram of each gene shows the layout of each ORF and the locations of 5′ and 3′ primer products. Levels of Pol II (lanes 4 and 8) were evenly distributed along the length of both genes. Prp42p levels (lanes 3 and 7) showed accumulation of the U1 snRNP on the 5′ end of both genes and decreasing levels in the 3′ regions. The data shown here are representative of four (ECM33) and three (SAC6) independent ChIP experiments.
TABLE 3.
Genome localization data for genes examined in this study
| Gene | Intron | % Expts hit by:
|
mRNA/hb | |
|---|---|---|---|---|
| U1 snRNPa | RNA Pol IIa | |||
| DBP2 | + | 80 | 100 | 61.5 |
| ASC1 | + | 60 | 100 | 124.0 |
| ECM33 | + | 20 | 67 | 18.6 |
| SAC6 | + | 0 | 0 | 6.1 |
| PDR5 | − | 40 | 100 | 30.6 |
| ADH1 | − | 100 | 100 | 125.6 |
| RPS3 | − | 0 | 100 | 144.4 |
| GAL1 | − | 0 | 0 | NA |
Indicates the percentage of experiments (five for U1 snRNP, three for RNA Pol II) in which an ORF received a score >2 SD away from the mean of the population.
Data taken from Holstege et al. (19).
DISCUSSION
The earliest events in the life of an RNA are difficult to study in vivo. Nascent RNA represents only a tiny fraction of any given RNA, and it is difficult to establish an order of events from the detection of rare RNA species within the greater pool. A recent view of RNA Pol II transcription units holds that, in addition to being the site of RNA synthesis, they are also RNA processing units at which 5′-end capping, pre-mRNA splicing, and polyadenylation occur while the pre-mRNA is still being synthesized by Pol II (39). However, it has only been possible to demonstrate cotranscriptional RNA processing for a limited number of genes, species, and biochemical events. Here we report the use of ChIP for the detection of U1 snRNP levels at active yeast transcription units. The data provide evidence that (i) the U1 snRNP is recruited cotranscriptionally; (ii) U1 snRNP accumulation along the length of intron-containing transcription units is dynamic, with high concentrations detectable in regions of intron synthesis; and (iii) the U1 snRNP is preferentially concentrated on intron-containing genes with respect to the largely intronless yeast genome.
In the present study, yeast strains harboring specific epitope tags on three endogenous U1 snRNP components—Prp42p, Nam8p, and Prp40p—were used to monitor the sites of accumulation of the U1 snRNP within transcription units. Using ChIP followed by PCR for the detection of specific gene regions, we found that all three U1 snRNP proteins are highly enriched in the downstream regions of two intron-containing genes: ASC1 and DBP2. The resolution of the assay is sufficient to detect differences in U1 snRNP accumulation at the promoters versus intron-containing regions of ASC1 and DBP2, and indeed ∼6.5-fold increases in the levels of Prp42p were detected downstream of the 5′ splice site. The distribution of the U1 snRNP in gene regions distal to the promoter contrasts dramatically with that of the capping enzyme Ceg1p, which was detected preferentially at both promoters. Because Prp42p detection on downstream regions of ASC1 and DBP2 in a conditionally mutant Pol II (rpb1-1) strain was abolished upon shift to the nonpermissive temperature, we conclude that U1 snRNP accumulation is dependent on active transcription. Moreover, neither Prp42p nor Pol II was detected on transcriptionally inactive chromatin, such as the telomere VI R or the GAL1 gene under conditions of glucose repression.
Two hypotheses explaining the observed increase in U1 snRNP in downstream regions of ASC1 and DBP2 were tested. First, events occurring at all active Pol II transcription units were considered. These include changes in the Pol II itself as it progresses 5′ to 3′ (e.g., changes in the CTD phosphorylation state, association of elongation factors, and/or association of a network of other factors, such as polyadenylation factors [4]), the affinity of the U1 snRNP for the 5′ cap (8, 27), or possible nonspecific association with nascent RNA which becomes progressively longer toward the 3′ end of any gene. These possibilities were addressed by examining the highly transcribed intronless genes RPS3, ADH1, and PDR5. As expected, Pol II was detected at high levels relative to the background along the lengths of all three genes. Prp42p was detected at low levels above background, with no detectable changes in levels along the lengths of any of the three genes, even though PDR5 (4,539 nt) is 1,896 nt longer than DBP2 and 3,308 nt longer than ASC1 (see Fig. 3). Therefore, we tested the second possibility: that U1 snRNP accumulation was specified by the presence of the intron. Indeed, removal of the DBP2 intron abolished accumulation of the U1 snRNP on the downstream region (see Fig. 5). These data indicate that the enhanced detection of the U1 snRNP at downstream regions of ASC1 and DBP2 reflects the presence of an intron in the gene rather than events common to all Pol II transcription units.
As a definitive test of the proposal that the U1 snRNP accumulates cotranscriptionally on intron-containing genes, genome localization studies were carried out. In this approach, fluorescently labeled probes are synthesized from the ChIP templates and hybridized with microarrays containing 6,229 confirmed and predicted ORFs in the yeast genome. Genome localization analysis has been previously used to identify gene targets of a variety of transcription factors (20, 44). Several hundred ORFs were enriched in the HA-Prp42p ChIP template compared to sheared genomic DNA, producing an outlying second peak of the ratio of median intensities (see Fig. 6). Up to 92% of the ORFs reproducibly identified in five microarray experiments were intron containing (see Table 2 and Results). As a positive control for the assay, microarrays were also hybridized with probes synthesized from the Myc-Pol II ChIP templates; several hundred ORFs were identified in the outlying population, and up to 51% of these were intron containing. Although only ∼5% of ORFs in the yeast genome contain introns, these genes tend to be among the most highly transcribed (1). Thus, the prevalence of intron-containing genes detected in the Pol II genome localization analysis was expected. The fact that a specific set of transcriptionally active genes is reproducibly identified by HA-Prp42p genome localization analysis confirms that U1 snRNP accumulation is cotranscriptional.
Close examination of the ORFs identified by Prp42p genome localization analysis raises several noteworthy points. First, because cotranscriptional U1 snRNP accumulation depends on transcription of the gene in the first place, the use of genome localization analysis to identify RNA processing targets is likely to be biased by transcriptional activity of a given gene relative to the rest of the transcriptome. This prediction is borne out by the present data, in which the ORFs identified by HA-Prp42p ChIP were found to be highly transcribed (see Tables 3 and A1). Transcriptionally inactive intron-containing genes, such as those induced in meiosis, were not detected. It is likely that other intron-containing genes may not be expressed highly enough to be represented in the outlying second peak.
Second, a number of intronless ORFs were consistently identified by HA-Prp42p as well as Myc-Pol II (see Tables 2, 3, and A1). This suggests that either U1 snRNP specifically accumulates at some intronless genes or that the U1 snRNP nonspecifically accumulates at highly transcribed genes. Interestingly, ADH1 was among the ORFs identified by both HA-Prp42p and Myc-Pol II ChIP templates in the genome localization analysis. We had observed in previous experiments that Prp42p was reproducibly and evenly detected along the length of ADH1 relative to the nonimmune control ChIP (see Fig. 3). ADH1 does not contain any consensus or nonconsensus sequence that has been shown to support U1 snRNA base pairing in the context of splicing (51). Thus, we cannot exclude the possibility that the U1 snRNP has a role in ADH1 gene expression through an unknown mechanism.
Finally, the absence of some highly transcribed intron-containing ORFs in the set of ORFs identified by HA-Prp42p genome localization led us to examine U1 snRNP distribution in ORFs containing a very short first exon followed by a very long second exon. We found that two such genes, ECM33 and SAC6, exhibit a high level of Prp42p at their 5′ ends and a reduced level at their 3′ ends (see Fig. 7). Possible explanations include either the loss of the U1 snRNP due to spliceosome assembly or the inaccessibility of the tags further downstream in the ORF. Thus, in spite of U1 snRNP accumulation at their 5′ ends, these ORFs were not reproducibly identified in the genome localization analysis, probably because the array used here was biased toward the 3′ end of each ORF. We conclude that genome localization analysis for RNA processing factors is feasible and informative, but it is more complex than paradigms based on DNA binding, as in the case of transcription factors. Genes not identified as targets of RNA processing factors should not be eliminated as candidates until they are analyzed in more detail.
Taken together, the results presented here support the conclusion that the U1 snRNP is recruited cotranscriptionally to intron-containing genes in yeast. If pre-mRNA splicing catalysis occurs cotranscriptionally in yeast, as it does in other species, then additional splicing factors, such as those recognizing the 3′ splice site and components of the spliceosome, may also accumulate cotranscriptionally. Recent models have emphasized the importance of coupling between transcription and pre-mRNA splicing, focusing on the CTD of Pol II as a platform for direct molecular interactions (4, 15, 31). However, the CTD is not required for efficient splicing in yeast (11, 28). Indeed, our data indicate that RNA Pol II is not sufficient for U1 snRNP accumulation; Pol II is abundantly detectable along intron-containing, as well as intronless genes, and yet U1 snRNP levels do not correlate with the distribution of Pol II. Instead, elevated U1 snRNP levels within transcription units coincide with sites of intron RNA synthesis, suggesting a dominant role for RNA splicing signals in U1 snRNP accumulation. The observation that uniform and usually reduced levels of the U1 snRNP were detected on intronless genes suggests that the U1 snRNP may have low affinity for some additional element(s) present at transcription units, which may contribute to either initial recruitment and/or retention of the U1 snRNP at intron-containing genes. One candidate for such an interaction may indeed be the CTD of Pol II, since it has been shown that Prp40p, another component of the U1 snRNP, binds the phosphorylated CTD in vitro (38). It is interesting that Prp40p was not detected at promoters but mirrored Prp42p in its distribution (data not shown), suggesting that Prp40p may bind the CTD in vivo only after U1 snRNP recruitment to intron-containing gene regions. Thus, it is clear that the mechanism of U1 snRNP recruitment is fundamentally different from the more straightforward case of the capping enzymes, which bind to the hyperphosphorylated CTD to ensure capping of every Pol II transcript. The demonstration that intronless genes accumulate relatively less U1 snRNP overall than intron-containing genes in the transcriptome suggests a more complex and dynamic mechanism for recruitment which may support greater plasticity in splicing, particularly in higher organisms where alternative splicing is a common mode of gene regulation.
Acknowledgments
We thank L. Goetsch, B. Seraphin, and R. Iggo for strains; E. Young and W. Zacharaie for the tagging plasmids; and M. Ares for communicating unpublished results. We are grateful to the Gottschling lab for sharing their ChIP protocol, J. Delrow and J. Howard for help with the microarray analysis, and A. Hopper, D. Stanek, F. Stewart, J. Valcarcel, and A. Weiner for discussions and comments on the manuscript.
K.M.K. is the recipient of a predoctoral fellowship from Boehringer Ingelheim Fonds. Supported by the Max Planck Gesellschaft and a Research Project Grant (RPG-00-110-01-MGO) from the American Cancer Society.
APPENDIX
The ORFs selected more than once by HA-Prp42p genome localization analysis were determined and are presented in Table A1.
TABLE A1.
List of all ORFs selected more than once by HA-Prp42p genome localization analysis
| ORF | Gene | % Prp42 expts hita | % Pol II expts hitb | No. of introns in ORFc | Introns across or nearbyd |
|---|---|---|---|---|---|
| YAL003W | EFB1 | 100 | 100 | 1 | |
| YBL027W | RPL19B | 100 | 100 | 1 | |
| YBL072C | RPS8A | 100 | 100 | 1 | |
| YBR048W | RPS11B | 100 | 100 | 1 | |
| YBR084C-A | RPL19A | 100 | 100 | 1 | |
| YBR181C | RPS6B | 100 | 67 | 1 | |
| YBR189W | RPS9B | 100 | 100 | 1 | |
| YBR191W | RPL21A | 100 | 100 | 1 | |
| YCR031C | RPS14A | 100 | 100 | 1 | |
| YDL082W | RPL13A | 100 | 100 | 1 | |
| YDL130W | RPP1B | 100 | 100 | 1 | |
| YDL136W | RPL35B | 100 | 100 | 1 | |
| YDL191W | RPL35A | 100 | 100 | 1 | |
| YDR025W | RPS11A | 100 | 67 | 1 | |
| YDR064W | RPS13 | 100 | 100 | 1 | |
| YDR447C | RPS17B | 100 | 100 | 1 | |
| YDR450W | RPS18A | 100 | 67 | 1 | |
| YDR471W | RPL27B | 100 | 100 | 1 | |
| YDR500C | RPL37B | 100 | 33 | 1 | |
| YER102W | RPS8B | 100 | 100 | 1 | |
| YER117W | RPL23B | 100 | 100 | 1 | |
| YFL034C-A | RPL22B | 100 | 100 | 1 | |
| YFR031C-A | RPL2A | 100 | 100 | 1 | |
| YGL030W | RPL30 | 100 | 100 | 1 | |
| YGL076C | RPL7A | 100 | 100 | 2 | |
| YGL189C | RPS26A | 100 | 100 | 1 | |
| YGR118W | RPS23A | 100 | 100 | 1 | |
| YGR148C | RPL24B | 100 | 33 | 1 | |
| YGR214W | RPS0A | 100 | 100 | 1 | |
| YHL001W | RPL14B | 100 | 100 | 1 | |
| YHR010W | RPL27A | 100 | 100 | 1 | |
| YHR141C | RPL42B | 100 | 67 | 1 | |
| YHR203C | RPS4B | 100 | 100 | 1 | |
| YIL052C | RPL34B | 100 | 100 | 1 | |
| YIL069C | RPS24B | 100 | 67 | 1 | |
| YIL133C | RPL16A | 100 | 100 | 1 | |
| YIL148W | RPL40A | 100 | 100 | 1 | |
| YJL191W | RPS14B | 100 | 100 | 1 | |
| YJR094W-A | RPL43B | 100 | 100 | 1 | |
| YJR145C | RPS4A | 100 | 100 | 1 | |
| YKL006W | RPL14A | 100 | 100 | 1 | |
| YKR094C | RPL40B | 100 | 100 | 1 | |
| YLR185W | RPL37A | 100 | 100 | 1 | |
| YLR333C | RPS25B | 100 | 100 | 1 | |
| YLR367W | RPS22B | 100 | 100 | 2 | |
| YLR448W | RPL6B | 100 | 100 | 1 | |
| YML024W | RPS17A | 100 | 100 | 1 | |
| YMR142C | RPL13B | 100 | 100 | 1 | |
| YMR194W | RPL36A | 100 | 100 | 1 | |
| YNL096C | RPS7B | 100 | 67 | 1 | |
| YNL162W | RPL42A | 100 | 67 | 1 | |
| YNL301C | RPL18B | 100 | 100 | 1 | |
| YNL302C | RPS19B | 100 | 67 | 1 | |
| YOL120C | RPL18A | 100 | 100 | 1 | |
| YOL121C | RPS19A | 100 | 67 | 1 | |
| YOL127W | RPL25 | 100 | 100 | 1 | |
| YOR096W | RPS7A | 100 | 100 | 1 | |
| YOR234C | RPL33B | 100 | 0 | 1 | |
| YOR293W | RPS10A | 100 | 100 | 1 | |
| YOR312C | RPL20B | 100 | 67 | 1 | |
| YPL079W | RPL21B | 100 | 67 | 1 | |
| YPL081W | RPS9A | 100 | 100 | 1 | |
| YPL090C | RPS6A | 100 | 67 | 1 | |
| YPL143W | RPL33A | 100 | 67 | 1 | |
| YPL198W | RPL7B | 100 | 67 | 2 | |
| YPR132W | RPS23B | 100 | 100 | 1 | |
| YGL102C | 100 | 100 | 0 | RPL28 | |
| YHL033C | RPL8A | 100 | 100 | 0 | No |
| YJL188C | BUD19 | 100 | 100 | 0 | RPL39 |
| YLL044W | 100 | 100 | 0 | No | |
| YLL045C | RPL8B | 100 | 100 | 0 | No |
| YLR062C | BUD28 | 100 | 100 | 0 | RPL22A |
| YLR441C | RPS1A | 100 | 100 | 0 | No |
| YMR242C | RPL20A | 100 | 67 | 0 | No |
| YOL086C | ADH1 | 100 | 100 | 0 | No |
| YPL142C | 100 | 67 | 0 | RPL33A | |
| YPR044C | 100 | 67 | 0 | RPL43A | |
| YBL087C | RPL23A | 80 | 100 | 1 | |
| YBL092W | RPL32 | 80 | 100 | 1 | |
| YBR082C | UBC4 | 80 | 0 | 1 | |
| YER056C-A | RPL34A | 80 | 67 | 1 | |
| YER074W | RPS24A | 80 | 67 | 1 | |
| YER131W | RPS26B | 80 | 0 | 1 | |
| YGL031C | RPL24A | 80 | 33 | 1 | |
| YGL103W | RPL28 | 80 | 100 | 1 | |
| YGR027C | RPS25A | 80 | 67 | 1 | |
| YGR034W | RPL26B | 80 | 67 | 1 | |
| YHR021C | RPS27B | 80 | 33 | 1 | |
| YIL018W | RPL2B | 80 | 100 | 1 | |
| YJL136C | RPS21B | 80 | 0 | 1 | |
| YLR048W | RPS0B | 80 | 0 | 1 | |
| YLR061W | RPL22A | 80 | 67 | 1 | |
| YLR344W | RPL26A | 80 | 67 | 1 | |
| YML073C | RPL6A | 80 | 100 | 1 | |
| YMR230W | RPS10B | 80 | 67 | 1 | |
| YNL112W | DBP2 | 80 | 100 | 1 | |
| YOR122C | PFY1 | 80 | 0 | 1 | |
| YPL249C-A | RPL36B | 80 | 100 | 1 | |
| YLR167W | RPS31 | 80 | 33 | 0 | No |
| YLR355C | ILV5 | 80 | 100 | 0 | No |
| YML063W | RPS1B | 80 | 100 | 0 | No |
| YNL190W | 80 | 33 | 0 | No | |
| YPL197C | 80 | 100 | 0 | RPL7B | |
| YBL040C | ERD2 | 60 | 0 | 1 | |
| YDL061C | RPS29B | 60 | 0 | 1 | |
| YDL083C | RPS16B | 60 | 0 | 1 | |
| YJL189W | RPL39 | 60 | 100 | 1 | |
| YKL180W | RPL17A | 60 | 100 | 1 | |
| YLR406C | RPL31B | 60 | 100 | 1 | |
| YMR116C | ASC1 | 60 | 100 | 1 | |
| YNR053C | 60 | 67 | 1 | ||
| YPR043W | RPL43A | 60 | 33 | 1 | |
| YBR118W | TEF2 | 60 | 100 | 0 | MUD1 |
| YHR143W | 60 | 100 | 0 | No | |
| YJL190C | RPS22A | 60 | 100 | 0 | RPS14B and RPL39 |
| YKL060C | FBA1 | 60 | 100 | 0 | No |
| YKL153W | 60 | 100 | 0 | No | |
| YKR042W | UTH1 | 60 | 100 | 0 | No |
| YLR009W | RLP24 | 60 | 33 | 0 | No |
| YLR044C | PDC1 | 60 | 100 | 0 | No |
| YMR217W | GUA1 | 60 | 67 | 0 | No |
| YMR303C | ADH2 | 60 | 67 | 0 | No |
| YMR307W | GAS1 | 60 | 100 | 0 | No |
| YNL066W | SUN4 | 60 | 67 | 0 | No |
| YPR080W | TEF1 | 60 | 100 | 0 | No |
| YBL018C | POP8 | 40 | 0 | 1 | |
| YDR381W | YRA1 | 40 | 100 | 1 | |
| YFL039C | ACT1 | 40 | 0 | 1 | |
| YHR039C-B | VMA10 | 40 | 0 | 1 | |
| YJL001W | PRE3 | 40 | 0 | 1 | |
| YKL006C-A | SFT1 | 40 | 33 | 1 | |
| YKL081W | TEF4 | 40 | 100 | 1 | |
| YLR388W | RPS29A | 40 | 0 | 1 | |
| YML026C | RPS18B | 40 | 67 | 1 | |
| YMR079W | SEC14 | 40 | 0 | 1 | |
| YOR182C | RPS30B | 40 | 0 | 1 | |
| YBR009C | HHF1 | 40 | 100 | 0 | No |
| YBR016W | 40 | 0 | 0 | No | |
| YBR062C | 40 | 33 | 0 | No | |
| YBR196C | PGI1 | 40 | 0 | 0 | No |
| YDR033W | MRH1 | 40 | 67 | 0 | No |
| YDR224C | HTB1 | 40 | 100 | 0 | No |
| YEL034W | HYP2 | 40 | 100 | 0 | No |
| YGL106W | MLC1 | 40 | 0 | 0 | No |
| YGR082W | TOM20 | 40 | 0 | 0 | No |
| YGR192C | TDH3 | 40 | 100 | 0 | No |
| YGR234W | YHB1 | 40 | 100 | 0 | No |
| YHL015W | RPS20 | 40 | 100 | 0 | No |
| YIL053W | RHR2 | 40 | 100 | 0 | RPL34B |
| YJR009C | TDH2 | 40 | 100 | 0 | No |
| YLR340W | RPP0 | 40 | 100 | 0 | No |
| YML106W | URA5 | 40 | 33 | 0 | No |
| YMR193C-A | 40 | 0 | 0 | RPL36A | |
| YNL030W | HHF2 | 40 | 100 | 0 | No |
| YNL043C | 40 | 0 | 0 | YIP3 | |
| YNL255C | GIS2 | 40 | 33 | 0 | No |
| YNL282W | POP3 | 40 | 0 | 0 | No |
| YOL039W | RPP2A | 40 | 67 | 0 | No |
| YOR153W | PDR5 | 40 | 100 | 0 | No |
| YLR076C | 40 | 100 | 0 | No | |
| YLR110C | CCW12 | 40 | 100 | 0 | No |
Indicates the percentage of microarray experiments (out of five) in which the ORF was selected by HA-Prp42 ChIP by using the >2 SD criterion.
Indicates the percentage of microarray experiments (out of three) in which the ORF was selected in parallel Myc-Pol II ChIPs using the >2 SD criterion.
Indicates the number of introns contained in the ORF selected.
Indicates the name of the intron-containing ORF(s) occurring on the strand opposite or within ≤500 bp of the selected intronless ORF. Because the ChIP template probes are double stranded and up to 400 bp long, ORFs may be detected due to Prp42p accumulation on ORFs nearby or on the opposite DNA strand.
REFERENCES
- 1.Ares, M., Jr., L. Grate, and M. H. Pauling. 1999. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA 5:1138-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barta, I., and R. Iggo. 1995. Autoregulation of expression of the yeast Dbp2p “DEAD-box” protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 14:3800-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauren, G., and L. Wieslander. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76:183-192. [DOI] [PubMed] [Google Scholar]
- 4.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. [DOI] [PubMed] [Google Scholar]
- 5.Beyer, A. L., and Y. N. Osheim. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2:754-765. [DOI] [PubMed] [Google Scholar]
- 6.Black, D. L., B. Chabot, and J. A. Steitz. 1985. U2 as well as U1 small nuclear ribonucleoproteins are involved in pre-messenger RNA splicing. Cell 42:737-750. [DOI] [PubMed] [Google Scholar]
- 7.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colot, H. V., F. Stutz, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 10:1699-1708. [DOI] [PubMed]
- 9.Cramer, P., C. G. Pesce, F. E. Baralle, and A. R. Kornblihtt. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 11.Dower, K., and M. Rosbash. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419:86-90. [DOI] [PubMed] [Google Scholar]
- 13.Elliott, D. J., and M. Rosbash. 1996. Yeast pre-mRNA is composed of two populations with distinct kinetic properties. Exp. Cell Res. 229:181-188. [DOI] [PubMed] [Google Scholar]
- 14.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstrohm, A. C., A. L. Greenleaf, and M. A. Garcia-Blanco. 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31-47. [DOI] [PubMed] [Google Scholar]
- 16.Gottschalk, A., J. Tang, O. Puig, J. Salgado, G. Neubauer, H. V. Colot, M. Mann, B. Seraphin, M. Rosbash, R. Luhrmann, and P. Fabrizio. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4:374-393. [PMC free article] [PubMed] [Google Scholar]
- 17.Hatton, A. R., V. Subramaniam, and A. J. Lopez. 1998. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon-exon junctions. Mol. Cell 2:787-796. [DOI] [PubMed] [Google Scholar]
- 18.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304:399-414. [DOI] [PubMed] [Google Scholar]
- 19.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 20.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 21.Kadener, S., P. Cramer, G. Nogues, D. Cazalla, M. de La Mata, J. P. Fededa, S. E. Werbajh, A. Srebrow, and A. R. Kornblihtt. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20:5759-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiseleva, E., T. Wurtz, N. Visa, and B. Daneholt. 1994. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J. 13:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 24.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krainer, A. R., and T. Maniatis. 1985. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell 42:725-736. [DOI] [PubMed] [Google Scholar]
- 26.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10:1683-1698. [DOI] [PubMed] [Google Scholar]
- 28.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002.. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed]
- 29.Lopez, P. J., and B. Seraphin. 2000. Uncoupling yeast intron recognition from transcription with recursive splicing. EMBO Rep. 1:334-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez, P. J., and B. Seraphin. 2000. YIDB: the Yeast Intron DataBase. Nucleic Acids Res. 28:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 32.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 34.McLean, M. R., and B. C. Rymond. 1998. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 18:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeil, J. B., H. Agah, and D. Bentley. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12:2510-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaud, S., and R. Reed. 1991. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 5:2534-2546. [DOI] [PubMed] [Google Scholar]
- 37.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3:697-705. [DOI] [PubMed] [Google Scholar]
- 38.Morris, D. P., and A. L. Greenleaf. 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 275:39935-39943. [DOI] [PubMed] [Google Scholar]
- 39.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115:3865-3871. [DOI] [PubMed] [Google Scholar]
- 40.Nonet, M., C. Scafe, J. Sexton, and R. Young. 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 42.Osheim, Y. N., O. L. Miller, Jr., and A. L. Beyer. 1985. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell 43:143-151. [DOI] [PubMed] [Google Scholar]
- 43.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 44a.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, G. C., C. Gooding, H. Y. Mak, N. J. Proudfoot, and C. W. Smith. 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 26:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seraphin, B., L. Kretzner, and M. Rosbash. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 7:2533-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59:349-358. [DOI] [PubMed] [Google Scholar]
- 49.Siliciano, P. G., and C. Guthrie. 1988. 5′ Splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 2:1258-1267. [DOI] [PubMed] [Google Scholar]
- 50.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 51.Spingola, M., L. Grate, D. Haussler, and M. Ares, Jr. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 53.Tennyson, C. N., H. J. Klamut, and R. G. Worton. 1995. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 9:184-190. [DOI] [PubMed] [Google Scholar]
- 54.Wetterberg, I., G. Bauren, and L. Wieslander. 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2:641-651. [PMC free article] [PubMed] [Google Scholar]
- 55.Wetterberg, I., J. Zhao, S. Masich, L. Wieslander, and U. Skoglund. 2001. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 20:2564-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wuarin, J., and U. Schibler. 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 14:7219-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zachar, Z., J. Kramer, I. P. Mims, and P. M. Bingham. 1993. Evidence for channeled diffusion of pre-mRNAs during nuclear RNA transport in metazoans. J. Cell Biol. 121:729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhuang, Y., and A. M. Weiner. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46:827-835. [DOI] [PubMed] [Google Scholar]