Abstract
Myotubularin is a dual-specific phosphatase that dephosphorylates phosphatidylinositol 3-phosphate and phosphatidylinositol (3,5)-bisphosphate. Mutations in myotubularin result in the human disease X-linked myotubular myopathy, characterized by persistence of muscle fibers that retain an immature phenotype. We have previously reported the identification of the 3-phosphatase adapter protein (3-PAP), a catalytically inactive member of the myotubularin gene family, which coprecipitates lipid phosphatidylinositol 3-phosphate-3-phosphatase activity from lysates of human platelets. We have now identified myotubularin as the catalytically active 3-phosphatase subunit interacting with 3-PAP. A 65-kDa polypeptide, coprecipitating with endogenous 3-PAP, was purified from SDS/PAGE, subjected to trypsin digestion, and analyzed by collision-induced dissociation tandem MS. Three peptides derived from human myotubularin were identified. Association between 3-PAP and myotubularin was confirmed by reciprocal coimmunoprecipitation of both endogenous and recombinant proteins expressed in K562 cells. Recombinant myotubularin localized to the plasma membrane, causing extensive filopodia formation. However, coexpression of 3-PAP with myotubularin led to attenuation of the plasma membrane phenotype, associated with myotubularin relocalization to the cytosol. Collectively these studies indicate 3-PAP functions as an “adapter” for myotubularin, regulating myotubularin intracellular location and thereby altering the phenotype resulting from myotubularin overexpression.
Phosphoinositide 3-kinase-derived molecules regulate diverse signaling functions including vesicular trafficking, actin cytoskeletal rearrangement, apoptosis, and cell proliferation (1, 2). The signaling molecule phosphatidylinositol 3-phosphate [PtdIns(3)P] binds and localizes FYVE, Phox, and pleckstrin homology domain-containing proteins to specific subcellular compartments such as early and late endosomes and thereby regulates endosomal and other vesicular trafficking pathways (3–5).
Myotubularin is a dual-specificity protein tyrosine phosphatase-like enzyme, which contains a HCX5R catalytic motif and functions as a PtdIns(3)P and phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2] 3-phosphatase (6–11, 12). The myotubularin gene locus, MTM1, is mutated in X-linked myotubular myopathy, a severe congenital disorder characterized by neonatal generalized muscle weakness, hypotonia, and death early in life (13). Myotubularin defines the largest family of lipid phosphatases in the human genome, which includes eight catalytically active enzymes, and in addition six putative inactive homologues of unknown function, which contain mutations within the HCX5R catalytic motif (14–16). Mutations in the myotubularin-related gene MTMR2, which encodes a PtdIns(3)P and PtdIns(3,5)P2 3-phosphatase, are responsible for the neurodegenerative disorder, type 4B1 Charcot Marie Tooth syndrome, a hereditary demyelinating peripheral neuropathy (17, 18).
The catalytically inactive myotubularin family members include SET-binding factor 1 (MTMR5), LIP-STYX (MTMR9), MTMR10, the 3-phosphatase adapter protein (3-PAP) (MTMR12), and MTMR13 (SET-binding factor 2) (7, 14–16, 19, 20). The function of these catalytically inactive phosphatases is unknown. Very recently, mutations in MTMR13 have been described in type 4B2 Charcot Marie Tooth syndrome (19, 20). Also, male mice deficient in SET-binding factor 1 are azoospermatic and infertile (21). We originally purified 3-PAP from rat brain as a heterodimer with an unidentified 65-kDa PtdIns(3)P 3-phosphatase (22). The human 3-PAP cDNA had sequence and domain homology with myotubularin, but lacked the HCX5R catalytic motif (23). Recombinant 3-PAP did not dephosphorylate PtdIns(3)P; however, 3-PAP immunoprecipitates from human platelet cytosol exhibited PtdIns(3)P 3-phosphatase activity, suggesting 3-PAP functions as an adapter subunit to an unidentified PtdIns(3)P 3-phosphatase (23).
In this study we have identified myotubularin as a catalytic subunit that complexes with 3-PAP. Furthermore, 3-PAP determines myotubularin cytosolic localization, and thereby alters the phenotype of myotubularin-overexpressing cells.
Materials and Methods
Materials. Restriction and DNA-modifying enzymes were obtained from Promega. Antibodies against myotubularin have been described (24). Antibody to the FLAG epitope (mAb KM5–1C7) was obtained from the Walter and Eliza Hall Institute of Medical Research (Melbourne). mAb to the hemagglutinin (HA) epitope used to probe immunoblots was obtained from Silenus (Novi, MI). Polyclonal antibody to the HA epitope used in microscopy was obtained from Upstate Biotechnology (Lake Placid, NY). All fluorochrome-tagged secondary antibodies (Alexa Fluor) were supplied by Molecular Probes, and horseradish peroxidase-tagged secondary antibodies for immunoblot analysis were obtained from DAKO. The pCGN vector was a gift from Tony Tiganis (Monash University), pEFBOS-FLAG vector was a gift from Tracey Willson (Walter and Eliza Hall Institute of Medical Research), and the myotubularin and MTMR2 cDNA were from G. S. Taylor (University of Michigan Medical School, Ann Arbor). All other reagents were from Sigma-Aldrich unless otherwise stated.
Cloning of MTMR6 cDNA. The MTMR6 cDNA was amplified by PCR, using the human brain cDNA library (Marathon Ready cDNA kit, CLONTECH) as a template and ligated into a pCMV5-HA expression vector. The MTMR6 cDNA isolated matched the sequence of GenBank accession no. NM_004685.
Production of Antipeptide Antibodies. Rabbits were immunized with the synthetic peptide (Chiron). VRPEEIHT-NEKEVTEKEC, which includes 3-PAP residues Val-24–Glu-40 (Fig. 1A), conjugated to diphtheria toxoid through the C-terminal Cys residue. Affinity-purified antipeptide antibody (anti-3-PAP antibody) was obtained by chromatography of immune sera on the specific peptide coupled to thiopropyl-Sepharose 6B resin. After extensive washing, antibodies were eluted from the column with 0.1 M glycine·HCl, pH 2.5, and neutralized with Tris, pH 8.0.
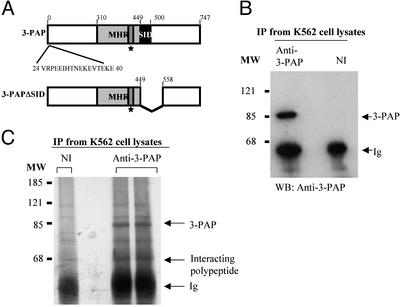
Fig. 1.
Immunoprecipitation of 3-PAP and a 65-kDa polypeptide. (A) Modular structure of 3-PAP and 3-PAPΔSID. The extent of the myotubularin homology region and the SID is shown. The box identified by a star represents the mutated catalytic motif. Amino acids 24–40 represent the immunogenic peptide used to raise the polyclonal rabbit anti-3-PAP antibody. The amino acids deleted in 3-PAPΔSID variant are shown. (B) Proteins were immunoprecipitated (IP) from 107 K562 cells, using 5 μg of either anti-3-PAP antibody (anti-3-PAP) or nonimmune sera (NI), and examined by immunoblot (WB) analysis probed with the anti-3-PAP antibody. The migration of 3-PAP and Ig heavy chain (Ig) is marked by arrows. MW indicates the migration of molecular mass markers (kDa). (C) Immunoprecipitates prepared as above were pooled and separated by SDS/PAGE and visualized by silver stain. The migration of the 65-kDa polypeptide that coimmunoprecipitates with 3-PAP is indicated.
Immunoprecipitation of Endogenous 3-PAP and Epitope-Tagged Recombinant Myotubularin, 3-PAP, 3-PAPΔ-SET-Interacting Domain (SID), MTMR2, and MTMR6. Endogenous 3-PAP was immunoprecipitated from a pellet of 107 K562 cells, treated with lysis buffer [50 mM Tris, pH 8.0/150 mM NaCl/1% (vol/vol) Triton X-100/1 mM benzamidine/2 mM PMSF/2 μg/ml leupeptin/2 μg/ml aprotinin) for 1 h at 4°C. Lysates were centrifuged at 15,400 × g for 10 min to obtain the Triton X-100–soluble supernatant, which was mixed with 5 μg of anti-3-PAP antibody, or nonimmune sera, incubated with rocking at 4°C for 2 h, followed by addition of 50 μl of a 50% slurry of protein A-Sepharose in Tris·NaCl, pH 8.0, and overnight incubation. The protein A-Sepharose was pelleted, washed six times with Tris·NaCl, pH 8.0, containing 1% (vol/vol) Triton X-100, resolved by SDS/PAGE, analyzed by immunoblot, and probed with anti-3-PAP antibody. Immunoprecipitates from K562 cell lysates by using anti-3-PAP or nonimmune antibody were resolved by SDS/PAGE and visualized by silver stain (Bio-Rad) using the manufacturer's protocol.
COS-7 cells were transiently transfected with HA-myotubularin, FLAG-3-PAP, or FLAG-3-PAPΔSID (Fig. 1 A) by electroporation and harvested 48 h later. Lysates were prepared, and immunoprecipitation was as described above. Epitope-tagged recombinant proteins were immunoprecipitated with 1–2 μg of the appropriate anti-HA or anti-FLAG antibody. All immunoblots were probed with goat anti-rabbit or goat anti-mouse secondary antibodies conjugated to horseradish peroxidase and detected by enhanced chemiluminescent substrate (Pierce).
Recombinant FLAG-tagged MTM1, MTMR2, and HA-tagged MTMR6 were coexpressed with Myc-tagged 3-PAP in COS-7 cells. Myc-tagged 3-PAP was immunoprecipitated with anti-Myc polyclonal antibody and probed by using anti-Flag antibody for MTM1 and MTMR2 and anti-HA antibody for MTMR6. Immunoprecipitates of Myc-3-PAP were also probed with anti-Myc antibody to ensure equal loading of recombinant 3-PAP.
Identification of Myotubularin in the Immunoprecipitates of Endogenous 3-PAP. Immunoprecipitates from 5 × 108 K562 cells (performed in batches and pooled), using 250 μg of anti-3-PAP antibody and 1,000 μl of slurry of protein A-Sepharose in Tris·NaCl, were separated by 1D SDS/PAGE (7.5% acrylamide) and stained with Coomassie blue R250. The polypeptide migrating at 65 kDa was excised and digested in situ with 0.5 μg of trypsin (25). Generated peptides were resolved by using capillary reversed-phase HPLC (26) and identified by collision-induced dissociation tandem MS using a quadrupole ion-trap mass spectrometer (model LCQ, Finnigan-MAT, San Jose, CA) equipped with electrospray ionization (27). The sequences of uninterpreted collision-induced dissociation spectra were identified by using the sequest algorithm and confirmed by manual de novo interpretation of their b- and y-type production series (28).
PtdIns(3)P 3-Phosphatase Assays. PtdIns(3)32P was generated as described (23). Immunoprecipitates prepared from parental K562 cell lysates by using anti-3-PAP, or nonimmune sera, and immunoprecipitates made with anti-FLAG antibody from lysates of K562 cells transfected with FLAG-3-PAP, FLAG-3-PAPΔSID, or vector alone were washed and incubated with PtdIns(3)32P. Lipids were extracted, resolved by TLC, visualized by autoradiography, excised, and measured by liquid scintillation counting (23). The residual PtdIns(3)32P extracted after phosphatase assay was inversely proportional to PtdIns(3)P 3-phosphatase activity exhibited by the immunoprecipitates.
Cell Microscopy. COS-7 cells expressing HA-myotubularin (0.5–5 μg) and FLAG-3-PAP or FLAG-3-PAPΔSID (5 μg of each) were washed with PBS, and then fixed and permeabilized for 10 min in PBS with 3.7% formaldehyde and 0.2% Triton X-100. Expression of FLAG and HA-tagged proteins was localized by using anti-FLAG antibody (mAb KM5–1C7) and anti-HA polyclonal antibody (07-221, Upstate Biotechnology) and detected by using Alexa Fluor 594 goat anti-mouse IgG (for FLAG tag) and Alexa Fluor 488 goat anti-rabbit IgG (for HA tag). Coverslips were mounted by using SlowFade (Molecular Probes) and visualized by confocal microscopy.
Results
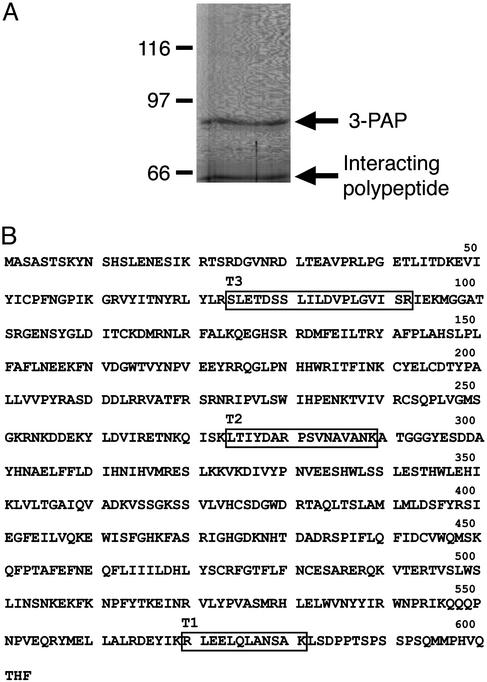
Identification of Myotubularin as the Catalytic Subunit Associated with 3-PAP. Polyclonal antibodies were developed to a peptide unique to 3-PAP (amino acids 24–40) (Fig. 1 A), which immunoprecipitated and immunoblotted endogenous 3-PAP from K562 cell lysates (Fig. 1B), as shown by the presence of a single polypeptide species of ≈86 kDa, in immune but not in nonimmune immunoprecipitates, consistent with 3-PAP molecular mass (23). Large-scale 3-PAP immunoprecipitates of K562 cell lysates were also visualized by silver staining of preparative SDS/PAGE gels and demonstrated in addition to ≈86 kDa 3-PAP, an ≈65-kDa polypeptide in immune, but not nonimmune, immunoprecipitations (Fig. 1C). The coprecipitating 65-kDa polypeptide, this band was excised from a preparative gel (Fig. 2A) and digested with trypsin in situ, and the resulting peptides were analyzed by tandem MS. The amino acid sequences of these peptides (Fig. 2B) were determined either by manual interpretation of the collision-induced spectra of the major peptide ion or computer-aided fragment-matching algorithms (sequest). Three peptides were identified as tryptic peptides from human myotubularin (Swissprot accession no. Q13496). These three peptides are derived from the amino-terminal, carboxyl-terminal, and central regions of myotubularin (Fig. 2B) and represent 7.8% of the protein sequence.
Fig. 2.
Identification of myotubularin as the 65-kDa polypeptide interacting with 3-PAP. (A) Proteins were immunoprecipitated from Triton X-100-soluble lysates prepared from 5 × 108 K562 cells using 250 μg of anti-3PAP polyclonal antibody, separated by SDS/PAGE, and visualized with Coomassie blue staining. The 65-kDa polypeptide (arrow) was extracted from the gel for tandem MS analysis. The migration of the 3-PAP polypeptide is indicated; MW indicates molecular mass markers in kDa. (B) Amino acid sequence of human myotubularin (Swissprot accession no. Q13496). Human myotubularin peptide sequences identified by mass spectroscopic sequencing are boxed.
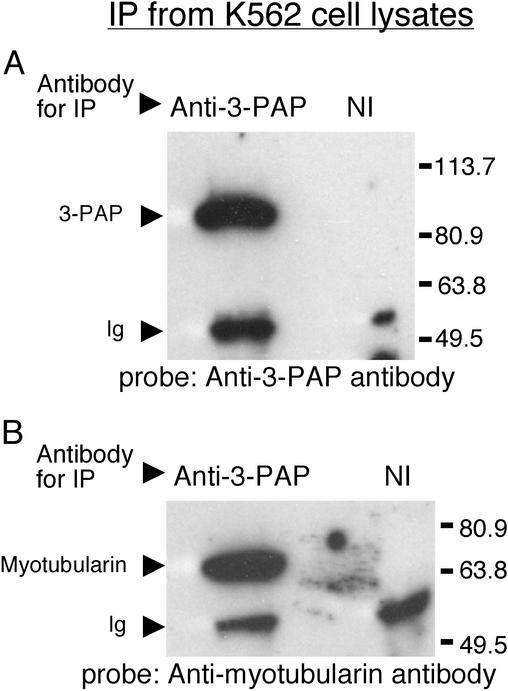
Association of endogenous 3-PAP and myotubularin was also shown by coimmunoprecipitation and immunoblot analysis. Control studies demonstrated that 3-PAP was present in the K562 cell-derived immunoprecipitates made with the 3-PAP antibody, but not in the immunoprecipitates made with nonimmune sera (Fig. 3A). Immunoprecipitates of 3-PAP from parental K562 cells were also immunoblotted with antimyotubularin antibodies. A single ≈65-kDa polypeptide was detected in the anti-3-PAP antibody immunoprecipitates, but not in the nonimmune immunoprecipitates (Fig. 3B). Immunoblot analysis of myotubularin immunoprecipitates with anti-3-PAP antibody revealed a single 86-kDa polypeptide consistent with 3-PAP (data not shown).
Fig. 3.
Interaction between endogenous 3-PAP and myotubularin. Proteins were immunoprecipitated from 107 K562 cells using 5 μg of either anti-3-PAP antibody or equivalent nonimmune sera (NI) and examined by immunoblot probed with anti-3-PAP antibody (A) or antimyotubularin antibody (B). Arrows indicate migration of 3-PAP, the 65-kDa polypeptide identified as myotubularin and Ig heavy chain (Ig). MW represents migration of molecular mass markers in kDa.
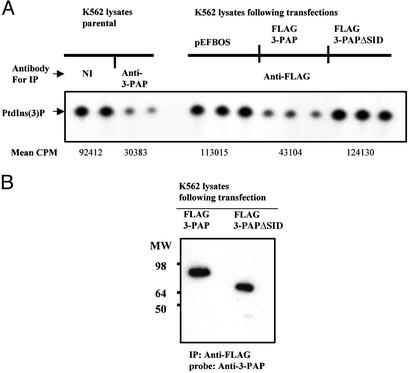
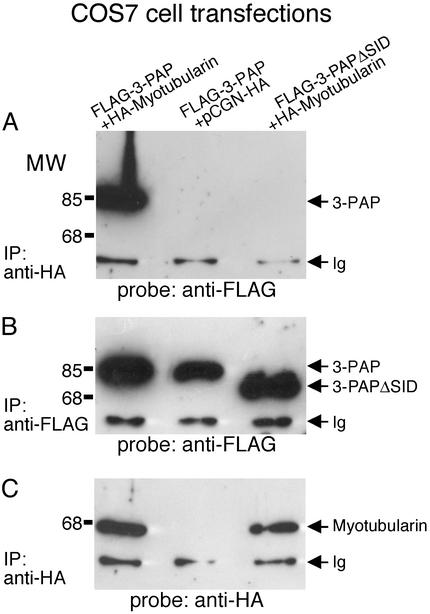
3-PAPΔSID and Interaction with Myotubularin. The SID is a region of significant amino acid conservation in the myotubularin gene family (14, 15). We identified a transcriptional variant of 3-PAP, 3-PAPΔSID, which lacks amino acids 449–558 including the SID; however, no characterization of the tissue or cellular distribution and/or function of this putative spliced variant has been determined (Fig. 1 A) (23). To investigate whether the transcriptional variant 3-PAPΔSID complexes with myotubularin, FLAG-tagged 3-PAP versus FLAG-3-PAPΔSID was immunoprecipitated from transfected K562 cells, and immunoprecipitated PtdIns(3)P 3-phosphatase activity was measured by the dephosphorylation of D3-labeled 32P from PtdIns(3)32P (Fig. 4A). The D3-phosphatase activity of FLAG-3-PAP immunoprecipitates was 2.5-fold greater (0.6 SD, n = 6) than that of FLAG-3-PAPΔSID. The activity displayed by FLAG-3-PAPΔSID immunoprecipitates was similar to that of control immunoprecipitates from K562 mock transfections with empty vector. Control studies demonstrated equivalent immunoprecipitation of FLAG-3-PAP and FLAG-3-PAPΔSID (Fig. 4B). Furthermore, endogenous 3-PAP immunoprecipitated from parental K562 cell lysates, exhibited PtdIns(3)P 3-phosphatase activity, similar to FLAG-3-PAP immunoprecipitates (Fig. 4A). To further investigate the myotubularin interaction with 3-PAP, FLAG 3-PAP versus FLAG 3-PAPΔSID were cotransfected with HA-myotubularin in COS-7 cells, and reciprocal immunoprecipitation and immunoblot analysis was performed. 3-PAP, but not 3-PAPΔSID, coprecipitated with HA-myotubularin (Fig. 5A). Prolonged exposure of immunoblots in repeated experiments did reveal coprecipitation of 3-PAPΔSID in some experiments, but at insignificant levels compared with 3-PAP. In control studies equal coprecipitation of FLAG-3-PAP and FLAG-3-PAPΔSID was demonstrated in FLAG immunoprecipitates (Fig. 5B) and equal HA-myotubularin was coprecipitated with the anti-HA antibody (Fig. 5C). These studies demonstrate intact 3-PAP, but not 3-PAPΔSID, complexes with myotubularin and suggests that this interaction may be facilitated by 3-PAP-SID. However, equally we cannot exclude the possibility that deletion of the SID results in a change in the 3-PAP conformation, which prevents association with myotubularin.
Fig. 4.
3-PAP but not 3-PAPΔSID associates with a PtdIns(3)P-3-phosphatase catalytic subunit. (A) Lysates from parental K562 cells and K562 cells transfected with FLAG epitoped-tagged 3-PAP, FLAG epitope-tagged 3-PAPΔSID, or empty vector (pEFBOS) were immunoprecipitated (IP) with nonimmune (NI) sera, anti-3-PAP antibody, or anti-FLAG antibody as indicated. Immunoprecipitates were analyzed for PtdIns(3)P 3-phosphatase activity. Immunoprecipitates of FLAG-3-PAP or FLAG-3-PAPΔSID from the above experiment were also immunoblotted with anti-FLAG antibody to demonstrate equivalent levels of FLAG-3-PAP and FLAG-3-PAPΔSID recombinant proteins in the immunoprecipitates used for 3-phosphatase assays.
Fig. 5.
3-PAP but not 3-PAPΔSID complexes with myotubularin. Lysates of COS-7 cells cotransfected with either FLAG epitope-tagged 3-PAP or FLAG epitope-tagged 3-PAPΔSID together with HA epitope-tagged myotubularin or empty vector (pCGN-HA) were subjected to immunoprecipitation (IP) followed by examination by immunoblot using antibodies to each tag as indicated. (A) Immunoprecipitation with anti-HA and probed with anti-FLAG. (B) Immunoprecipitation and probe with anti-FLAG. (C) Immunoprecipitation and probe with anti-HA. Ig indicates the Ig heavy chain, and MW indicates the migration of molecular mass markers in kDa.
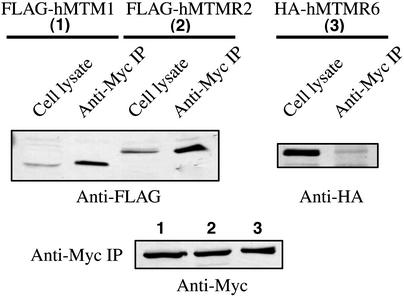
Recent reports have revealed MTMR2, but not MTM1, interacts with the catalytically inactive MTMR5 (29). We also investigated whether 3-PAP complexed with MTMR2. Cells were cotransfected with Myc-tagged 3-PAP and empty vector, FLAG-MTM1, FLAG-MTMR2, or HA-MTMR6 and immunoprecipitated with anti-Myc antibody to immunoprecipitate Myc-3-PAP and immunoblotted with anti-FLAG, anti-HA, or anti-Myc antibodies. In these studies 3-PAP coimmunoprecipitated with MTM1 and MTMR2 but not MTMR6 (Fig. 6 Upper). Control studies revealed equal immunoprecipitation of 3-PAP in all cotransfections (Fig. 6 Lower).
Fig. 6.
Association of 3-PAP with myotubularin (MTM1) and MTMR2 but not MTMR6. Recombinant FLAG-tagged MTM1 (no. 1), MTMR2 (no. 2), and HA-tagged MTMR6 (no. 3) were coexpressed with Myc-tagged 3-PAP in COS-7 cells. (Upper) Myc-tagged 3-PAP was immunoprecipitated with anti-Myc polyclonal antibody and probed by using anti-Flag antibody for MTM1 and MTMR2 and anti-HA antibody for MTMR6. (Lower) Western blots of the Myc immunoprecipitates probed with anti-Myc to demonstrate equal precipitation of 3-PAP.
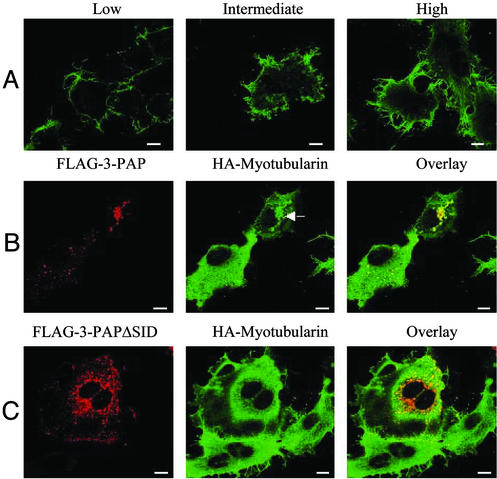
Interaction with 3-PAP Modulates Myotubularin Intracellular Localization. Recent studies have shown myotubularin, but not MTMR2, can modulate an endosomal pool of PtdIns(3)P in intact cells, as shown by the loss of localization of GST-FYVE fusion protein to endosomes (11). Myotubularin localizes in intact cells to the plasma membrane and overexpression of myotubularin causes extensive plasma membrane remodeling, membrane ruffling, and extensive filopodia projections (11, 24). Myotubularin plasma membrane localization and induction of plasma membrane remodeling and filopodial projections is independent of both catalytic PtdIns(3)P 3-phosphatase activity and a direct interaction with the actin cytoskeleton (24). We performed dose-dependent myotubularin overexpression in COS-7 cells. Expression of moderate to high levels of WT myotubularin initiated a phenotype as previously reported with membrane ruffling and extensive filopodia, which correlated with the level of myotubularin expression (Fig. 7A). Myotubularin localized predominantly at the plasma membrane in both low- and high-expressing COS-7 cells. However, FLAG-3-PAP and FLAG-3-PAPΔSID localized in the cytosol in punctuate vesicles and no plasma membrane localization was demonstrated (Fig. 7 B and C, respectively, Left). Coexpression of FLAG-3-PAP with HA-myotubularin completely inhibited the myotubularin-induced membrane remodeling phenotype, and under these conditions, cells exhibited a uniform plasma membrane with absence of membrane projections and filopodia (Fig. 7B Center). Myotubularin was no longer at the plasma membrane, but rather on 3-PAP coexpression, it was localized predominantly to the cytoplasm. In some cells, coexpression of 3-PAP with myotubularin resulted in myotubularin localization to punctate vesicles, in addition to cytosolic staining (Fig. 7B Center, see arrow). The localization of 3-PAP to vesicles was not affected by coexpression of myotubularin. It was noteworthy that 3-PAPΔSID coexpression did not alter either the plasma membrane phenotype of myotubularin-expressing cells, including filopodial projections, or myotubularin plasma membrane localization (Fig. 7C Center).
Fig. 7.
Myotubularin-induced plasma membrane remodeling is inhibited by 3-PAP. (A) Overexpression of low, intermediate, and higher levels of WT HA-tagged myotubularin in COS7 cells. Myotubularin localization was determined by anti-HA indirect immunofluorescence. (B) Localization of FLAG-3-PAP (Left, detected with anti-FLAG) coexpressed with HA-myotubularin (Center, detected with anti-HA) is shown. Arrow indicates the localization of HA-myotubularin to punctate vesicles in 3-PAP coexpressing cells. Colocalization is shown in the overlay images (Right). (C) Localization of FLAG-3-PAPΔSID (Left, detected with anti-FLAG) coexpressed with HA-myotubularin (Center, detected with anti-HA) is shown. Colocalization is shown in the overlay images (Right). (Scale bars: 10 μm.)
Discussion
This study has shown that myotubularin complexes in vivo with 3-PAP, a highly related but catalytically inactive myotubularin homologue. This association serves to relocalize myotubularin from the plasma membrane to the cytosol and reverses the cellular phenotype resulting from myotubularin overexpression. These studies indicate a novel role for 3-PAP in regulating myotubularin.
Direct detection of endogenous myotubularin has not been reported; however, the localization of recombinant tagged myotubularin has been published by several laboratories and is consistent with a dual location in the cytosol and at the plasma membrane (11, 24). Furthermore, overexpression of myotubularin at high levels produces a phenotype of extension of numerous filopodia and membrane ruffling (11, 24). Although the molecular mechanisms mediating this phenotype have not been determined, it is noteworthy that the phenotype does not require myotubularin catalytic activity (24). Given myotubularin's plasma membrane localization, it has been proposed that myotubularin may regulate a plasma membrane pool of PtdIns(3)P, whereas MTMR2 would regulate a perinuclear subpool (24, 30). We have shown that 3-PAP reverses the filopodial and plasma membrane phenotype of myotubularin-expressing cells, correlating with loss of myotubularin expression at the plasma membrane. Therefore it is possible that given normal endogenous myotubularin levels, there would be sufficient 3-PAP to localize myotubularin to the cytosol.
Myotubularin has recently been shown to hydrolyze PtdIns(3,5)P2 in addition to PtdIns(3)P (12). PtdIns(3)P is found on early and late endosomes (31). Although the precise localization of PtdIns(3,5)P2 has not been determined, based on the localization of the PIKfyve kinase this lipid is predicted to localize to late endosomes (32). Recombinant 3-PAP localizes to a vesicular compartment, and the identification of the nature of these vesicles is the subject of ongoing studies.
We previously reported the purification to homogeneity of a type I 65-kDa PtdIns(3)P 3-phosphatase from rat brain, which formed a homodimer, and a type II enzyme, which comprised a 65- and 78-kDa heterodimer (22). The rat type II 3-phosphatase 65/78-kDa heterodimer hydrolyzed PtdIns(3)P at three times the rate measured for the 65-kDa enzyme (22). Biochemical analysis and V8 and trypsin peptide maps indicated the two 65-kDa subunits were identical, whereas amino acid sequencing and cloning of the 78-kDa polypeptide identified 3-PAP (22, 23). The studies reported here showing that myotubularin complexes with 3-PAP in vivo are consistent with the contention that myotubularin was the 65-kDa polypeptide, which we originally copurified with 3-PAP in the type II enzyme complex (22). Both myotubularin and the type I 3-phosphatase have the same molecular mass of 65 kDa and show similar substrate specificity, hydrolyzing PtdIns(3)P, forming PtdIns. Although we have identified only myotubularin in complex with endogenous 3-PAP in vivo in K562 cells, it does not exclude a role for 3-PAP in complexing with other members of the myotubularin family. In this regard, we have shown that recombinant 3-PAP complexes with recombinant MTMR2 in coexpression studies.
We have shown a previously uncharacterized transcriptional variant of 3-PAP lacking the SID, 3-PAPΔSID, demonstrates decreased association with myotubularin, compared with intact 3-PAP. The SID is a 51-aa residue that represents a putative paired amphiphatic helix and was designated the SID, based on the ability of this region to interact with SET domains (7). The only member of the myotubularin family shown to interact with SET domains is the catalytically inactive SET-binding factor 1 (MTMR5), which was identified on the basis of an interaction with the Hrx SET domain, the mammalian homologue of the Drosphilia trithorax (7). The role of the SID in the MTM family members, including 3-PAP, requires further investigation.
The role of the catalytically inactive 3-phosphatases such as MTMR5 (SET-binding factor 1), MTMR9 (LIP-STYX), and other myotubularin homologues have yet to be defined. The current work showing a complex between 3-PAP and myotubularin, plus the recent study showing a complex between MTMR2 and MTMR5 (29), suggests that the inactive myotubularin-related proteins may function as adapters for the active forms. Dixon and colleagues (29) have shown that MTMR2, but not myotubularin, interacts with MTMR5, a catalytically inactive phosphatase via each proteins coil–coil domain. This interaction was shown via overexpression of recombinant MTMR2 and immunoprecipitation, followed by sequencing of interacting polypeptides. However, coimmunoprecipitation of endogenous proteins was not demonstrated (29). We have shown that 3-PAP also interacts with MTMR2, but not MTMR6, in cotransfected cells, suggesting that the catalytically dead myotubularin homologues have the potential to interact with more than one lipid 3-phosphatase.
Acknowledgments
We thank Drs. L. Ooms and J. Dyson for helpful discussions. This work was supported by the National Health and Medical Research Council of Australia (Grant 143582 to H.H.N. and C.A.M.), the National Institutes of Health (Grants HL55772 and HL16634 to P.W.M.), Association Française contre les Myopathies, Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and the Hôpital Universitaire de Strasbourg.
Abbreviations: PtdIns(3)P, phosphatidylinositol 3-phosphate; PtdIns(3,5)P2, phosphatidylinositol 3,5-bisphosphate; 3-PAP, 3-phosphatase adapter protein; HA, hemagglutinin; SID, SET-interacting domain.
References
- 1.Rameh, L. E. & Cantley, L. C. (1999) J. Biol. Chem. 274, 8347–8350. [DOI] [PubMed] [Google Scholar]
- 2.Czech, M. P. (2000) Cell 100, 603–606. [DOI] [PubMed] [Google Scholar]
- 3.Patki, V., Lawe, D. C., Corvera, S., Virbasius, J. V. & Chawla, A. (1998) Nature 394, 433–434. [DOI] [PubMed] [Google Scholar]
- 4.Kanai, F., Liu, H., Field, S. J., Akbary, H., Matsuo, T., Brown, G. E., Cantley, L. C. & Yaffe, M. B. (2001) Nat. Cell Biol. 3, 675–678. [DOI] [PubMed] [Google Scholar]
- 5.Dowler, S., Currie, R. A., Campbell, D. G., Deak, M., Kular, G., Downes, C. P. & Alessi, D. R. (2000) Biochem. J. 351, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denu, J. M. & Dixon, J. E. (1998) Curr. Opin. Chem. Biol. 2, 633–641. [DOI] [PubMed] [Google Scholar]
- 7.Cui, X., De Vivo, I., Slany, R., Miyamoto, A., Firestein, R. & Cleary, M. L. (1998) Nat. Genet. 18, 331–337. [DOI] [PubMed] [Google Scholar]
- 8.Laporte, J., Blondeau, F., Buj-Bello, A., Tentler, D., Kretz, C., Dahl, N. & Mandel, J.-L. (1998) Hum. Mol. Genet. 7, 1703–1712. [DOI] [PubMed] [Google Scholar]
- 9.Taylor, G. S., Maehama, T. & Dixon, J. E. (2000) Proc. Natl. Acad. Sci. USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blondeau, F., Laporte, J., Bodin, S., Superti-Furga, G., Payrastre, B. & Mandel, J. L. (2000) Hum. Mol. Genet. 9, 2223–2229. [DOI] [PubMed] [Google Scholar]
- 11.Kim, S.-A., Taylor, G. S., Torgersen, K. M. & Dixon, J. E. (2002) J. Biol. Chem. 277, 4526–4531. [DOI] [PubMed] [Google Scholar]
- 12.Schaletzky, J., Dove, S. K., Short, B., Lorenzo, O., Clague, M. J. & Barr, F. A. (2003) Curr. Biol. 13, 504–509. [DOI] [PubMed] [Google Scholar]
- 13.Laporte, J., Hu, L. J., Kretz, C., Mandel, J. L., Kioschis, P., Coy, J. F., Klauck, S. M., Poustka A. & Dahl, N. (1996) Nat. Genet. 13, 175–182. [DOI] [PubMed] [Google Scholar]
- 14.Laporte, J., Blondeau, F., Buj-Bello, A. & Mandel, J.-L. (2001) Trends Genet. 17, 221–228. [DOI] [PubMed] [Google Scholar]
- 15.Nandurkar, H. H. & Huysmans, R. (2002) IUBMB Life 53, 37–43. [DOI] [PubMed] [Google Scholar]
- 16.Wishart, M. J. & Dixon, J. E. (2002) Trends Cell Biol. 12, 579–585. [DOI] [PubMed] [Google Scholar]
- 17.Bolino, A., Muglia, M., Conforti, F. L., LeGuern, E., Salih, M. A., Georgiou, D. M., Christodoulou, K., Hausmanowa-Petrusewicz, I., Mandich, P., Schenone, A., et al. (2000) Nat. Genet. 25, 17–19. [DOI] [PubMed] [Google Scholar]
- 18.Berger, P., Bonneick, S., Willi, S., Wymann, M. & Suter, U. (2002) Hum. Mol. Genet. 11, 1569–1579. [DOI] [PubMed] [Google Scholar]
- 19.Senderek, J., Bergmann, C., Weber, S., Ketelsen, U. P., Schorle, H., Rudnik-Schoneborn, S., Buttner, R., Buchheim, E. & Zerres, K. (2003) Hum. Mol. Genet. 12, 349–356. [DOI] [PubMed] [Google Scholar]
- 20.Azzedine, H., Bolino, A., Taieb, T., Birouk, N., Di Duca, M., Bouhouche, A., Benamou, S., Mrabet, A., Hammadouche, T., Chkili, T., et al. (2003) Am. J. Hum. Genet. 72, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firestein, R., Nagy, P. L., Daly, M., Huie, P., Conti, M. & Cleary, M. L. (2002) J. Clin. Invest. 109, 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caldwell, K. K., Lips, D. L., Bansal, V. S. & Majerus, P. W. (1991) J. Biol. Chem. 266, 18378–18386. [PubMed] [Google Scholar]
- 23.Nandurkar, H. H., Caldwell, K. K., Whisstock, J. C., Layton, M. J., Gaudet, E. A., Norris, F. A., Majerus, P. W. & Mitchell, C. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9499–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laporte, J., Blondeau, F., Gansmuller, A., Lutz, Y., Vonesch, J. L. & Mandel, J.-L. (2002) J. Cell. Sci. 115, 3105–3117. [DOI] [PubMed] [Google Scholar]
- 25.Moritz, R. L., Eddes, J. S., Reid, G. E. & Simpson, R. J. (1996) Electrophoresis 17, 907–917. [DOI] [PubMed] [Google Scholar]
- 26.Moritz, R. L. & Simpson, R. J. (1992) J. Chromatogr. 599, 119–130. [DOI] [PubMed] [Google Scholar]
- 27.Reid, G. E., Rasmussen, R. K., Dorow, D. S. & Simpson, R. J. (1998) Electrophoresis 19, 946–955. [DOI] [PubMed] [Google Scholar]
- 28.Roepstorff, P. & Fohlman, J. (1984) Biomed. Mass Spectrom. 11, 601–602. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S.-A., Vacratsis, P. O., Firestein, R., Cleary, M. L. & Dixon, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laporte, J., Liaubet, L., Blondeau, F., Tronchere, H., Mandel, J.-L. & Payrastre, B. (2002) Biochem. Biophys. Res. Commun. 291, 305–312. [DOI] [PubMed] [Google Scholar]
- 31.Gillooly, D. J., Simonsen, A. & Stenmark, H. (2001) Biochem. J. 355, 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shisheva, A. (2001) Cell Biol. Int. 25, 1201–1206. [DOI] [PubMed] [Google Scholar]