Abstract
Interplay among GATA transcription factors is an important determinant of cell fate during hematopoiesis. Although GATA-2 regulates hematopoietic stem cell function, mechanisms controlling GATA-2 expression are undefined. Of particular interest is the repression of GATA-2, because sustained GATA-2 expression in hematopoietic stem and progenitor cells alters hematopoiesis. GATA-2 transcription is derepressed in erythroid precursors lacking GATA-1, but the underlying mechanisms are unknown. Using chromatin immunoprecipitation analysis, we show that GATA-1 binds a highly restricted upstream region of the ≈70-kb GATA-2 domain, despite >80 GATA sites throughout the domain. GATA-2 also binds this region in the absence of GATA-1. Genetic complementation studies in GATA-1-null cells showed that GATA-1 rapidly displaces GATA-2, which is coupled to transcriptional repression. GATA-1 also displaces CREB-binding protein (CBP), despite the fact that GATA-1 binds CBP in other contexts. Repression correlates with reduced histone acetylation domain-wide, but not altered methylation of histone H3 at lysine 4. The GATA factor-binding region exhibited cell-type-specific enhancer activity in transient transfection assays. We propose that GATA-1 instigates GATA-2 repression by means of disruption of positive autoregulation, followed by establishment of a domain-wide repressive chromatin structure. Such a mechanism is predicted to be critical for the control of hematopoiesis.
Homologous transcription factors with similar or identical DNA-binding specificities can activate distinct target genes and exert unique biological functions. The GATA family of zinc finger factors (GATA-1–GATA-6) exemplifies this scenario (1–3). GATA-1 is expressed in erythroid, megakaryocytic, and mast cells, as well as in the testis (4). GATA-2 is expressed in hematopoietic stem and progenitor cells, endothelial cells, and diverse tissues including the central nervous system, placenta, fetal liver, and fetal heart (5–8). Although GATA-2 controls early stages of hematopoiesis (5, 7) and pituitary (9), central nervous system, and urogenital development (10, 11), GATA-1 regulates terminal differentiation and function of erythroid and megakaryocytic cells (12–16) and early stages of eosinophil differentiation (17, 18). Ectopic GATA-2 expression in murine primitive hematopoietic cells suppresses hematopoiesis (19–21), and expression in ES cells increases primitive hematopoietic colony formation (22). A common theme is that enforced GATA-2 expression in progenitors affects differentiation, and therefore GATA-2 expression must be tightly regulated.
Despite the unique expression patterns and developmental functions of GATA-1 and GATA-2, considerable interplay exists between these factors. An important aspect of the interplay involves the transcriptional regulation of GATA-1 and GATA-2. Approximately 500 bp upstream of the erythroid-specific GATA-1 transcription start sites are two GATA sites flanking a CCAAC box, which are implicated in positive autoregulation of GATA-1 transcription (23, 24). The double GATA motif is critical for the generation of eosinophils but not for erythrocytes and megakaryocytes (17). An enhancer, hypersensitive site (HS)1, resides ≈3.9 kb upstream of the erythroid-specific IE GATA-1 promoter (25–27). HS1 contains a GATA-E-box motif, which mediates assembly of a complex containing GATA-1, TAL1, Lmo2, and Lbd1 (27, 28). Targeted deletion of HS1 revealed a critical role for GATA-1 expression during megakaryopoiesis (29), but it is unclear whether GATA-1 or -2 functions through HS1. Because GATA-2 is expressed in hematopoietic stem cells and progenitors (30–32), GATA-2 might activate GATA-1 transcription before autoregulation.
At early stages of hematopoiesis, bone morphogenetic protein 4 (BMP-4) signaling induces GATA-2 transcription (33–35). BMP-4-dependent GATA-2 transcription occurs in Xenopus without new protein synthesis (35), indicating a direct transcriptional mechanism. BMP-4 signals to Smad transcription factors (36), but Smad sites have not been delineated in the GATA-2 locus. Murine GATA-2 has alternate first exons with two promoters: 1S, which is hematopoietic-specific, and 1G, which has broader specificity (8) (Fig. 1). Chicken GATA-2 also has alternate first exons (37). Murine GATA-2 promoters contain GATA sites (8), but it is unknown whether they are functional and whether GATA-2 expression is autoregulated.
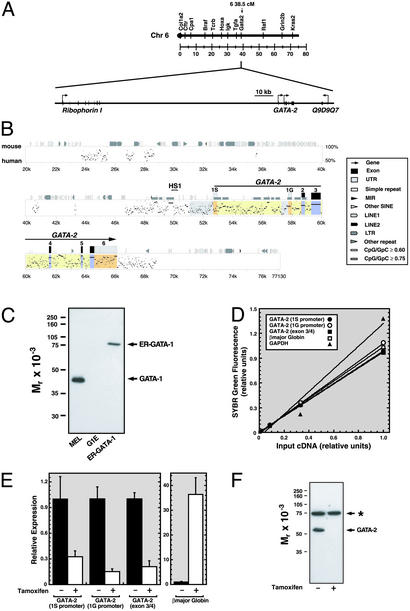
Fig. 1.
GATA-1-dependent repression of transcription from 1S and 1G GATA-2 promoters. (A Upper) Linkage map showing location of murine GATA-2 on chromosome 6. (A Lower) Map showing 150 kb surrounding GATA-2 including the 5′ flanking gene Ribophorin I and the putative 3′ flanking gene Q9D9Q7. (B) pipmaker (76) alignment of a 77-kb region of the mouse and human GATA-2 domains. Shaded regions indicate the following: promoters, gray; untranslated regions, orange; exons, blue; introns, yellow. HS1 denotes a DNaseI hypersensitive site mapped previously (71). (C) Western blot analysis of GATA-1 and ER-GATA-1 expression with anti-GATA-1 antibody in whole-cell lysates from DMSO-induced MEL cells, G1E, and G1E-ER-GATA cells after a 20-h treatment with 1 μM tamoxifen. (D) SYBR green fluorescence (relative units) was plotted versus the initial G1E input cDNA concentration. The plot illustrates the linearity and range of signals used to measure target cDNA. (E) Quantitative real-time RT-PCR analysis of GATA-2 mRNA expression in untreated and tamoxifen-treated (48 h) G1E-ER-GATA cells. Primers amplified transcripts transcribed from the 1S promoter, the 1G promoter, and from both promoters (exon 3/4). β-Globin expression was measured as a control. Relative expression levels were normalized by GAPDH expression (mean ± SEM; five independent experiments). (F) Western blot analysis of GATA-2 expression in untreated and tamoxifen-treated (24 h) G1E-ER-GATA cells. The asterisk denotes a broadly expressed crossreactive band.
Repression of GATA-2 occurs as hematopoiesis proceeds (38). Expression of GATA-1 (31, 39) and the lympho-myeloid-specific factor PU.1 (40) correlates with GATA-2 repression, but a mechanistic link has not been established. It was hypothesized that PU.1 binding to GATA-2 disrupts positive autoregulation of GATA-2 transcription (40). Despite this intriguing hypothesis, evidence did not exist for GATA-2 binding to the GATA-2 domain or for autoregulation. GATA-1 activates and represses genes via interactions with Friend of GATA-1 (FOG-1) (41–44). GATA-2 also binds FOG-1, but a GATA-2 mutant defective in FOG-1 binding rescues hematopoiesis (45).
GATA-2 regulates hematopoietic stem cell function (5, 7, 20, 22), and therefore the control of GATA-2 expression has major implications for hematopoiesis. As severalfold differences in the concentration of PU.1 regulate the decision for progenitors to differentiate into lymphoid or myeloid cells (46), changes in concentrations of cell-type-specific factors can be crucial determinants of cell fate. To address how GATA-2 is regulated, we analyzed the native nucleoprotein structure of the active and inactive GATA-2 domain. This analysis revealed a mechanism in which GATA-1 represses GATA-2 transcription through disruption of positive autoregulation and via broad chromatin modification.
Materials and Methods
Cell Culture and Transfection. G1E (47) and G1E-ER-GATA-1 (39, 48) cells were maintained and transfected as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Quantitative Chromatin Immunoprecipitation (ChIP) Assay. Real-time PCR quantitative ChIP analysis was conducted as described (49–51). Chromatin fragments averaged ≈400 bp. Before tamoxifen treatment, cells were grown for at least 12 h in medium containing 15% charcoal-stripped FBS to eliminate steroids. Cells were grown in medium containing 15% FBS with or without 1 μM tamoxifen (Sigma) for 48 h. In Fig. 4, cells were grown in medium containing 7.5% FBS/7.5% charcoal-stripped FBS with or without 1 μM tamoxifen for various times. Immunoprecipitated DNA was analyzed by real-time PCR (ABI Prism 7000, PE Applied Biosystems). Primers were designed by primer express 1.0 (PE Applied Biosystems) to amplify 50- to 150-bp amplicons and were based on GenBank accession no. AB009272 and sequences in Ensembl (www.ensembl.org/Mus_musculus/geneview?gene=ENSMUSG00000015053). Samples from three or more immunoprecipitations were analyzed. Product was measured by SYBR green fluorescence in 25-μl reactions. The amount of product was determined relative to a standard curve of input chromatin. Dissociation curves showed that PCRs yielded single products. Primers and antibodies are described in Supporting Materials and Methods.
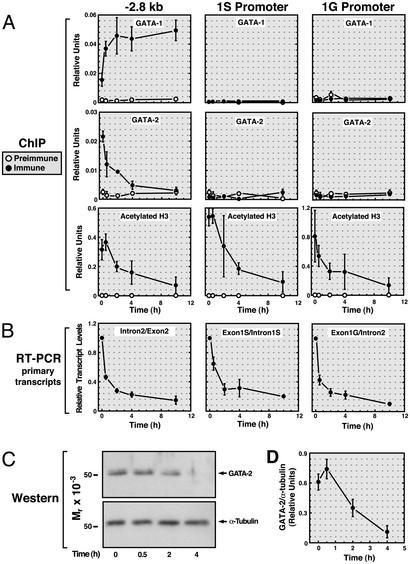
Fig. 4.
Rapid GATA-1-dependent displacement of GATA-2 from the –2.8-kb site. G1E-ER-GATA-1 cells were incubated for 0, 0.5, 2, 4, or 10 h with 1 μM tamoxifen. (A) Quantitative ChIP analysis was used to measure the binding of GATA-1 and -2, as well as H3 acetylation at the –2.8-kb site and at the 1S and 1G promoters. Samples were analyzed quantitatively relative to a standard curve generated from input chromatin. The plots depict relative levels of binding and acetylation (means from four independent experiments). (B) RT-PCR was used to measure primary GATA-2 transcripts arising from use of the 1S promoter (Exon1S/Intron1S), the 1G promoter (Exon1G/Intron2), or both promoters (Intron2/Exon2). GAPDH mRNA was measured as a control. Relative GATA-2 primary transcript levels were normalized by the levels of GAPDH transcripts. The plots depict the mean GATA-2/GAPDH ratios from four independent experiments. (C) Time course for GATA-1-dependent reduction in GATA-2 protein levels. Whole-cell lysates isolated from the same samples as those analyzed by ChIP and RT-PCR were subjected to Western blot analysis. A representative Western blot of GATA-2 and α-tubulin is shown. (D) GATA-2 and α-tubulin levels were quantitated by means of densitometric analysis. The plot depicts GATA-2/α-tubulin ratios at various times after tamoxifen treatment. Note that GATA-2 protein levels are not reduced upon 30 min of tamoxifen treatment, despite GATA-1 binding, GATA-2 displacement, and reduced primary transcripts.
RNA Isolation and Quantitative RT-PCR. RNA was prepared from the same cultures used for ChIP. Total RNA was purified with TRIzol (GIBCO/BRL) and analyzed as described in Supporting Materials and Methods.
Protein Analysis. Protein analysis was conducted as described in Supporting Materials and Methods.
Results and Discussion
GATA-1-Dependent Repression of GATA-2 Transcription Correlates With Highly Selective Occupancy of GATA Sites Within the GATA-2 Domain. GATA-1 expression in GATA-1-null G1E cells induces erythroid differentiation and represses GATA-2 (42). It is unknown whether GATA-1 regulates the hematopoietic-specific 1S promoter and the 1G promoter, which is active in diverse cells. Real-time RT-PCR was used to measure GATA-2 mRNA transcripts generated via the usage of 1S or 1G promoters and total GATA-2 transcripts in untreated and tamoxifen-treated G1E cells expressing estrogen receptor hormone-binding domain fused to GATA-1 (G1E-ER-GATA-1). ER-GATA-1 expression was lower than endogenous mouse erythroleukemia (MEL) cell GATA-1 (Fig. 1C). Tamoxifen-mediated activation of ER-GATA-1 for 48 h repressed both promoters (67%, 85%, and 79% decrease for 1S, 1G, both promoters, respectively; Fig. 1 D and E) and abrogated GATA-2 protein expression (Fig. 1F).
Because activation of ER-GATA-1 induces erythroid differentiation of G1E cells (39), GATA-1 might directly repress GATA-2, or repression might be a consequence of differentiation. Through usage of FOG-1, GATA-1 mediates activation and repression (42, 52, 53). Direct repression would require GATA-1 binding to the GATA-2 domain, which contains >80 GATA consensus sites (Fig. 2A). Based on the simplicity of this site, functional sites cannot be predicted. Analogous to our analysis of GATA-1 binding to the murine β-globin domain (48), quantitative ChIP assays were conducted with an anti-GATA-1 antibody to assess whether ER-GATA-1 occupies sites within the GATA-2 domain (Fig. 2A). Very-low-level ER-GATA-1 binding was detected at the 1G promoter, and no binding was evident at the 1S promoter and at other sites. Strong binding was detected 2.8 kb upstream of the 1S exon. Primers flanking the amplicon containing the –2.8-kb region by 1.4 and 1.0 kb (5′ and 3′, respectively) revealed much less binding, thus defining an ≈2.4-kb restricted region bound by GATA-1. Located centrally within the amplicon are tandem, inverted GATA sites separated by a 14-bp spacer containing a CCAAT box; two additional GATA sites are nearby (Fig. 2C). The double GATA-CCAAT motif differs from mouse to man by a single base in the CCAAT box.
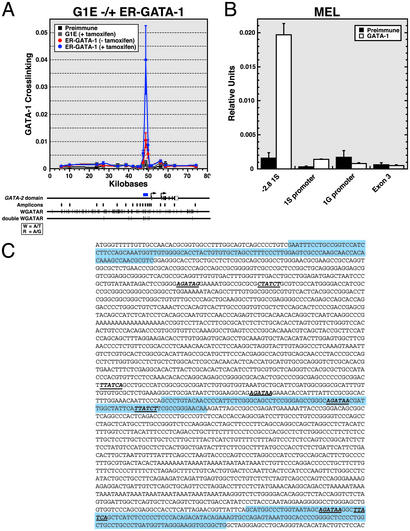
Fig. 2.
Highly selective discrimination by GATA-1 among GATA sites of the GATA-2 domain. (A) Quantitative ChIP analysis of G1E-ER-GATA-1 binding within the GATA-2 domain in tamoxifen-treated G1E and untreated and tamoxifen-treated ER-GATA cells. The graph depicts data from at least three independent experiments (mean ± SEM). The diagram below the graph shows the murine GATA-2 domain. Vertical lines below the locus indicate the position of the amplicons analyzed. The distribution of individual and double (two sites within 25 bp) consensus GATA-1 sites is also indicated. The consensus motif is defined on the left. (B) ChIP analysis of ER-GATA-1 binding to the GATA-2 domain of MEL cells treated with 1.5% DMSO for 4 days. Note that the strong binding 2.8 kb upstream of the 1S exon (–2.8 1S) corresponds to the peak of binding in A. (mean ± SEM; four independent experiments). (C) GATA-2 domain sequence containing the region bound by GATA-1. The blue shaded sequences represent three amplicons analyzed in A. The middle sequence represents the amplicon in which high-level GATA-1 binding was detected. GATA consensus sites are indicated in bold and italics and are underlined. Note that a double GATA site resides within the amplicon.
To determine whether the pattern of GATA-1 occupancy is unique to G1E cells, endogenous GATA-1 binding was analyzed in DMSO-induced MEL cells. Induced MEL cells express GATA-1 but only very low levels of GATA-2 (data not shown). ER-GATA-1 and GATA-1 occupy the β-globin locus similarly in G1E-ER-GATA-1 and MEL cells, respectively (48). Strong GATA-1 crosslinking was detected at the –2.8-kb region, and very low or no binding was detected at the 1S promoter, the 1G promoter, and exon 3 (Fig. 2B). Thus, the analysis identified a highly restricted region of ER-GATA-1 and GATA-1 binding, indicating that GATA-1 discriminates exquisitely among the many GATA sites. Although one cannot state unequivocally which GATA site mediates binding, the –2.8-kb region contains only the double GATA-CCAAT motif, two consensus GATA sites, and one nonconsensus site. Sequences ≈800 bp to either side of the –2.8-kb amplicon lack GATA sites. Because our ChIP assay can measure major differences in protein–DNA interactions between amplicons that are considerably less than 800 bp apart (51, 54), it is highly likely that one or more of the five GATA sites mediate binding. Palindromic GATA sites mediate particularly high-affinity GATA-1 binding in vitro (55).
GATA-1-Dependent Remodeling of the Histone Modification Pattern of the GATA-2 Domain. Given that GATA-1 binds the GATA-2 domain in vivo (Fig. 2) and represses both GATA-2 promoters that are separated by ≈5 kb, GATA-1 might instigate broad chromatin remodeling to generate repressive chromatin. FOG-1 has been implicated in GATA-1-dependent repression (42–44). To investigate how GATA-1 represses GATA-2, ChIP was used to define the histone modification state of the active and inactive GATA-2 domains. ChIP analysis was conducted in G1E cells with or without inactive or activated ER-GATA-1 by using antibodies against diacetylated histone H3, multiacetylated H4, and histone H3 methylated at lysine 4 (H3-meK4). H4 acetylation can be distributed broadly throughout domains (51, 56–58). H3 acetylation has a more restricted distribution, being highest at promoters and at active genes, although low-level H3 acetylation can reside throughout domains (51, 56, 57, 59). H3-meK4 is enriched at active domains (60, 61), but the enrichment can be highly variable (51). Although the patterns of these modifications can be similar, establishment and maintenance of individual modifications are uniquely regulated (49–51, 54).
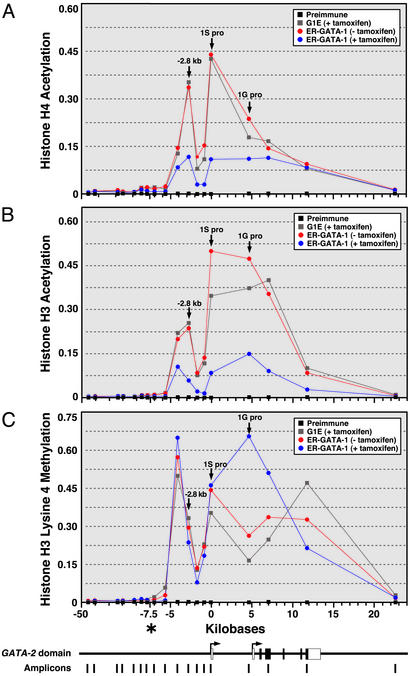
The GATA-2 domain in G1E cells was characterized by broad, nonuniform H4 (Fig. 3A) and H3 (Fig. 3B) acetylation and H3-meK4 (Fig. 3C) from ≈4.2 kb upstream of the 1S exon to the 3′ end of GATA-2. Activation of ER-GATA-1 decreased acetylation domain-wide. H3-meK4 was not affected by GATA-1, with the exception of the 1G promoter and a 3′ site, in which H3-meK4 increased upon ER-GATA-1 activation. The lack of a role for GATA-1 in establishing H3-meK4 resembles the finding that GATA-1 deficiency in G1E cells does not affect H3-meK4 at the βmajor promoter (51). However, GATA-1 differentially regulates histone acetylation at the βmajor promoter and GATA-2, inducing H3 and H4 acetylation at the βmajor promoter (51) and reducing acetylation at GATA-2 (Fig. 3).
Fig. 3.
GATA-1-dependent remodeling of the histone modification pattern of the GATA-2 domain. Quantitative ChIP analysis of H4 (A) and H3 (B) acetylation and H3-meK4 (C) at the murine GATA-2 domain. The relative levels of acetylated H3 and H4 and H3-meK4 were plotted as a function of the position within the locus (mean ± SEM; at least three independent experiments). SEM values did not exceed 25% of the corresponding means. Positions of the –2.8-kb region and the 1S and 1G promoters are indicated by arrows. The diagram at the bottom depicts the murine GATA-2 domain and the amplicons analyzed by ChIP. *, the position of the x axis in which the scale changes.
How does reduced acetylation affect GATA-2 regulation? A ≈3-fold decrease in acetylation can greatly enhance higher-order chromatin folding (62), and factor access to cis-elements is increased by acetylation (63, 64). The large reduction in acetylation induced by GATA-1 might decrease accessibility of the GATA-2 promoters reducing RNA polymerase II (pol II) recruitment. ChIP analysis was conducted to assess pol II binding to the promoters, to exons 3 and 6, and to a site 9.1 kb downstream of GATA-2 (+9.1 kb). ER-GATA-1 activation strongly decreased pol II recruitment to both promoters and lowered pol II levels at the exons (Fig. 7A, which is published as supporting information on the PNAS web site). No pol II was detected at the +9.1 kb site. Thus, GATA-1-mediated repression involves a domain-wide reduction in acetylation and decreased pol II recruitment. We examined acetylation and pol II binding to the constitutive Pol II large subunit (RPII215) (Fig. 7B) and brain-specific necdin promoters (Fig. 7C). Acetylated H3 and H4 and H3-meK4 were high at the RPII215 promoter and were not affected by ER-GATA-1. Acetylated H3 and H4 and H3-meK4 were very low at the necdin promoter, and ER-GATA-1 did not affect these modifications, nor did it affect pol II binding to necdin and RPII215 promoters. Thus, GATA-1-dependent reductions in acetylation and pol II recruitment to the GATA-2 domain are highly specific actions.
Rapid Displacement of GATA-2 by GATA-1 from the –2.8-kb Region. GATA-1 and GATA-2 bind the GATA consensus motif (WGATAR) similarly (65, 66). Because GATA-1 binds the –2.8-kb region of the GATA-2 domain, which contains multiple WGATAR motifs, and GATA-2 is expressed in G1E cells, we asked whether GATA-2 binds this region. In G1E-ER-GATA-1 cells, GATA-2 was crosslinked to the –2.8-kb region but not to the 1S and 1G promoters (Fig. 4A). A similar pattern of GATA-2 occupancy was seen in FOG-1-null erythroid precursors (data not shown), which express GATA-2 (67).
The binding of GATA-2 to the –2.8-kb region suggested that GATA-1-dependent repression of GATA-2 requires displacement of GATA-2. To test this model, a time course was conducted in which ER-GATA-1 binding, GATA-2 binding, histone acetylation, primary GATA-2 transcripts, and GATA-2 protein levels were measured after tamoxifen treatment of G1E-ER-GATA-1 cells (Fig. 4). ER-GATA-1 binding to the –2.8-kb region was maximal between 30 min and 2 h of tamoxifen treatment. GATA-2 binding to the –2.8-kb region was highest in untreated cells and decreased maximally by 2–4 h. H3 acetylation was highest in untreated cells and decreased maximally by 10 h. Primary GATA-2 transcripts (total transcripts, 1S promoter-derived transcripts, and 1G promoter-derived transcripts) were quantitated to measure transcription (Fig. 4B). Primary transcripts decreased maximally by 2–4 h concomitant with GATA-2 displacement. GATA-2 protein levels were unchanged by 30 min and decreased maximally by 4 h (Fig.4 C and D). Thus, ER-GATA-1 binding and GATA-2 displacement are rapid events tightly coupled to repression. The decrease in acetylation was slower than the GATA switch, suggesting that reduced acetylation is a late event in repression.
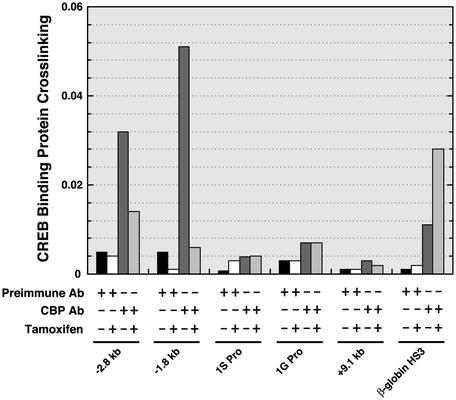
How Does the GATA Switch Repress GATA-2 Transcription? How can two highly related GATA factors induce opposite transcriptional responses? Both GATA-1 and GATA-2 interact with the histone acetylase CBP (68) and FOG-1 (41). We used ChIP to test whether CBP is recruited to the –2.8-kb region (Fig. 5). CBP binding to the –2.8-kb region was detected, with little or no binding at the promoters and the +9.1 kb site. Binding was also detected between the –2.8-kb site and the 1S promoter (–1.8 kb). Activation of ER-GATA-1 for 20 h induced CBP displacement from both sites. Thus, CBP recruitment correlates with GATA-2 occupancy of the –2.8-kb site. ER-GATA-1 displaces GATA-2 and abrogates CBP recruitment. Because GATA-1, ER-GATA-1, and GATA-2 bind CBP (68), the differential activities of GATA-1 and GATA-2 to recruit CBP were unexpected.
Fig. 5.
GATA-1-dependent displacement of CBP from the –2.8-kb region and from a downstream site. G1E-ER-GATA-1 cells were treated with 1 μM tamoxifen for 20 h. CBP binding to selected sites of the GATA-2 locus and to HS3 of the β-globin locus control region was measured by quantitative ChIP analysis (mean ± SEM; two independent experiments).
ChIP analyses strongly implicated the –2.8-kb region in transcriptional control, which is supported by the finding that a DNaseI HS resides in this region. Comparison of mouse, rat, and human sequences revealed that this region is highly conserved (Fig. 8, which is published as supporting information on the PNAS web site). We tested whether a 389-bp sequence located at the –2.8-kb region can activate an SV40 promoter (–2.8/SV40 pro) in transient transfection assays in G1E and 3T3 cells. The activity of –2.8/SV40 pro was compared with the SV40 promoter alone or with the SV40 enhancer. The –2.8-kb sequence increased promoter activity 3.4-fold in G1E cells but had no activity in 3T3 cells (Fig. 9, which is published as supporting information on the PNAS web site). The SV40 enhancer strongly activated the SV40 promoter in 3T3 cells. Thus, the –2.8-kb sequence exhibited weak cell-type-specific enhancer activity. HS3 from the β-globin locus control region represents another example of a tissue-specific regulatory element with only weak enhancer activity in transients (69). Because multiple regulatory sequences often interact to control transcription within chromatin domains, it would not be surprising if the –2.8-kb region cooperates with other conserved elements of the GATA-2 domain.
Establishment of the Inactive GATA-2 Domain. GATA-2 repression correlates with transcriptional activation of lineage-restricted factors such as GATA-1 (31) and PU.1 (40, 70). However, mechanistic links had not been established. Because ER-GATA-1 and GATA-1 occupy the –2.8-kb region, GATA-2 repression involves binding of GATA-1 to the GATA-2 domain. These results support a mechanism in which GATA-1 directly represses GATA-2. Because GATA-2 occupies the –2.8-kb region when GATA-2 is transcriptionally active, repression involves displacement of GATA-2 by GATA-1. Because the GATA switch at the –2.8-kb region was rapid and tightly coupled to repression, it is likely that GATA-2 displacement instigates repression. The GATA switch occurred before decreased GATA-2 protein levels, inconsistent with reduced GATA-2 occupancy resulting from down-regulation of GATA-2 protein.
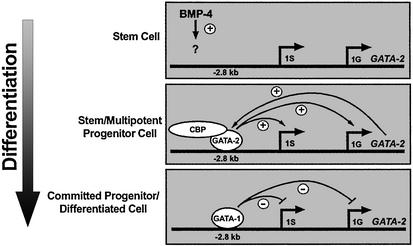
Based on the role of BMP-4 signaling in activating GATA-2 transcription early in hematopoiesis (33, 35) and work described herein, we propose that BMP-4 induces levels of GATA-2 sufficient to establish positive autoregulation (Fig. 6). Autoregulation might sustain GATA-2 transcription in hematopoietic stem cells and progenitors. Truncation of GATA-2 upstream sequences in transgenes, which removes the –2.8-kb region, abrogates GATA-2 expression in hematopoietic cells (71), consistent with this model. Elevated GATA-1 levels in erythromyeloid progenitors displace GATA-2 from the –2.8-kb region, abolishing positive autoregulation. Importantly, the GATA switch does not only occur with ER-GATA-1, because endogenous GATA-1 occupies the –2.8-kb region in MEL cells (Fig. 2B), and ER-GATA-1 was expressed at a lower level than endogenous MEL cell GATA-1 (Fig. 1C). Analogous to GATA-1-dependent repression, GATA-2 repression by PU.1 during macrophage differentiation has been postulated to involve disruption of autoregulation (40). However, no evidence existed for occupancy of the GATA-2 domain by GATA-1 or GATA-2.
Fig. 6.
Regulation of the GATA-2 domain. The models depict the regulation of GATA-2 transcription during hematopoiesis. BMP-4 signals activate GATA-2 at the earliest stages of hematopoiesis (33, 35). The analysis described herein shows that GATA-2 binds the –2.8-kb site when the locus is transcriptionally active. BMP-4-dependent activation of GATA-2 would yield a sufficient concentration of GATA-2 to bind the –2.8-kb site and to positively autoregulate GATA-2 expression. As GATA-1 levels increase, GATA-1 would competitively displace GATA-2, disrupting positive autoregulation. GATA-2 displacement is accompanied by loss of CBP from the –2.8-kb site and from a downstream –1.8-kb site. GATA-1 also establishes a repressive chromatin structure by means of broad chromatin modification, but GATA-1-dependent hypoacetylation is delayed temporally relative to repression. It is hypothesized that hypoacetylation locks the locus in an inaccessible state once repression has been instigated via the GATA switch.
Our model assumes that GATA-1 and -2 confer repression and activation, respectively, when bound to the –2.8-kb region. Accordingly, the concentrations of GATA-1 and -2 and factors that modulate their DNA binding would be critical determinants of GATA-2 transcription. Transgene rescue experiments showed that GATA-1, -2, and -3 rescued embryonic lethality resulting from mutation of GATA-1 (72). Only GATA-1 rescued anemia in adult mice, indicating that GATA factors have qualitatively different activities in vivo. GATA-1, but not GATA-2, activates the granule major basic protein promoter in transient transfection assays in Jurkat T cells (73).
How do highly related GATA factors function differentially? Although both GATA factors interact with FOG-1 (41), no coactivators have been shown to interact exclusively with GATA-1 or GATA-2. GATA-1 and GATA-2 also interact with the histone acetylase CBP/p300 (68, 74). GATA-2 interacts with histone deacetylase 3 and 5 (HDAC) (75). Given the requirement of the GATA-2 zinc fingers for HDAC binding and the high conservation of the zinc fingers of GATA-1 and -2, it is likely that GATA-1 also interacts with HDACs. The differential activities might reflect the fact that GATA-1 opposes CBP recruitment, whereas GATA-2 occupancy is associated with CBP recruitment.
In summary, ChIP analysis revealed GATA-1 binding to an unexpected –2.8-kb region of the GATA-2 domain, providing a direct link between chromatin occupancy by GATA-1 and GATA-2 repression. GATA-1 rapidly displaces GATA-2 and CBP from the –2.8-kb region, induces domain-wide remodeling of histone acetylation, and represses GATA-2. We propose that GATA-1 and GATA-2 elicit opposing functions through the –2.8-kb region, and GATA-1 uniquely confers repression through this region. The exquisite control of GATA-2 levels by means of the bimodal mechanism of disrupting positive autoregulation and establishing a broad domain of repressive chromatin would be predicted to be critical for regulating hematopoiesis.
Supplementary Material
Acknowledgments
We thank Kirby D. Johnson and Carol M. Kiekhaefer for critical reviews of the manuscript, and Dr. Gerd Blobel for advice on CBP ChIP methodology. This work was funded by National Institutes of Health Grant DK50107 (to E.H.B.). E.H.B. is a Romnes Scholar and a Shaw Scientist.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BMP-4, bone morphogenetic protein 4; CBP, CREB-binding protein; pol II, RNA polymerase II; ChIP, chromatin immunoprecipitation; ER-GATA-1, estrogen receptor hormone-binding domain fused to GATA-1; FOG-1, Friend of GATA-1; HDAC, histone deacetylase; H3-meK4, histone H3 methylated at lysine 4; HS, hypersensitive site; MEL, mouse erythroleukemia.
References
- 1.Weiss, M. J. & Orkin, S. H. (1995) Exp. Hematol. 23, 99–107. [PubMed] [Google Scholar]
- 2.Molkentin, J. D. (2000) J. Biol. Chem. 275, 38949–38952. [DOI] [PubMed] [Google Scholar]
- 3.Patient, R. K. & McGhee, J. D. (2002) Curr. Opin. Genet. Dev. 12, 416–422. [DOI] [PubMed] [Google Scholar]
- 4.Tsai, S. F., Martin, D. I., Zon, L. I., D'Andrea, A. D., Wong, G. G. & Orkin, S. H. (1989) Nature 339, 446–451. [DOI] [PubMed] [Google Scholar]
- 5.Tsai, F. Y., Keller, G., Kuo, F. C., Weiss, M., Chen, J., Rosenblatt, M., Alt, F. W. & Orkin, S. H. (1994) Nature 371, 221–226. [DOI] [PubMed] [Google Scholar]
- 6.Orlic, D., Anderson, S., Biesecker, L. G., Sorrentino, B. P. & Bodine, D. M. (1995) Proc. Natl. Acad. Sci. USA 92, 4601–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai, F.-Y. & Orkin, S. H. (1997) Blood 89, 3636–3643. [PubMed] [Google Scholar]
- 8.Minegishi, N., Ohta, J., Suwabe, N., Nakauchi, H., Ishihara, H., Hayashi, N. & Yamamoto, M. (1998) J. Biol. Chem. 273, 3625–3634. [DOI] [PubMed] [Google Scholar]
- 9.Dasen, J. S., O'Connell, S. M., Flynn, S. E., Treier, M., Gleiberman, A. S., Szeto, D. P., Hooshmand, F., Aggarwal, A. K. & Rosenfeld, M. G. (1999) Cell 97, 587–598. [DOI] [PubMed] [Google Scholar]
- 10.Nardelli, J., Thiesson, D., Fujiwara, Y., Tsai, F.-Y. & Orkin, S. H. (1999) Dev. Biol. 210, 305–321. [DOI] [PubMed] [Google Scholar]
- 11.Zhou, Y., Yamamoto, M. & Engel, J. D. (2000) Development (Cambridge, U.K.) 127, 3829–3838. [DOI] [PubMed] [Google Scholar]
- 12.Pevny, L., Simon, M. C., Robertson, E., Klein, W. H., Tsai, S. F., D'Agati, V., Orkin, S. H. & Costantini, F. (1991) Nature 349, 257–260. [DOI] [PubMed] [Google Scholar]
- 13.Simon, M. C., Pevny, L., Wiles, M. V., Keller, G., Costantini, F. & Orkin, S. H. (1992) Nat. Genet. 1, 92–98. [DOI] [PubMed] [Google Scholar]
- 14.Pevny, L., Lin, C. S., D'Agati, V., Simon, M. C., Orkin, S. H. & Costantini, F. (1995) Development (Cambridge, U.K.) 121, 163–172. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi, S., Onodera, K., Motohashi, H., Suwabe, N., Hayashi, N., Yanai, N., Nabesima, Y. & Yamamoto, M. (1997) J. Biol. Chem. 272, 12611–12615. [DOI] [PubMed] [Google Scholar]
- 16.Tsang, A. P., Fujiwara, Y., Hom, D. B. & Orkin, S. H. (1998) Genes Dev. 12, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu, C., Cantor, A. B., Yang, H., Browne, C., Wells, R. A., Fukiwara, Y. & Orkin, S. H. (2002) J. Exp. Med. 195, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirawawa, R., Shimuzu, R., Takahashi, S., Osawa, M., Takayanagi, S., Kato, Y., Onodera, M., Minegishi, N., Yamamoto, M., Fukao, K., et al. (2002) J. Exp. Med. 195, 1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griegel, K., Lim, K. C., Plank, C., Beug, H., Engel, J. D. & Zenke, M. (1993) Genes Dev. 7, 1097–1109. [DOI] [PubMed] [Google Scholar]
- 20.Heyworth, C., Gale, K., Dexter, M., May, G. & Enver, T. (1999) Genes Dev. 13, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persons, D. A., Allay, J. A., Allay, E. R., Ashmun, R. A., Orlic, D., Jane, S. M., Cunningham, J. M. & Nienhuis, A. W. (1999) Blood 93, 488–499. [PubMed] [Google Scholar]
- 22.Kitajima, K., Masuhara, M., Era, T., Enver, T. & Nakano, T. (2002) EMBO J. 17, 3060–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai, S. F., Strauss, E. & Orkin, S. H. (1991) Genes Dev. 5, 919–931. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, M., Nishikawa, K. & Yamamoto, M. (2001) Development (Cambridge, U.K.) 128, 2341–2350. [DOI] [PubMed] [Google Scholar]
- 25.McDevitt, M. A., Fujiwara, Y., Shivdasani, R. A. & Orkin, S. H. (1997) Proc. Natl. Acad. Sci. USA 94, 7976–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onodera, K., Takahashi, S., Nishimura, S., Ohta, J., Motohashi, H., Yomogida, K., Hayashi, N., Engel, J. D. & Yamamoto, M. (1997) Proc. Natl. Acad. Sci. USA 94, 4487–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vyas, P., McDevitt, M. A., Cantor, A. B., Katz, S. G., Fujiwara, Y. & Orkin, S. H. (1999) Development (Cambridge, U.K.) 126, 2799–2811. [DOI] [PubMed] [Google Scholar]
- 28.Wadman, I. A., Osada, H., Grutz, G. G., Agulnick, A. D., Westphal, H., Forster, A. & Rabbitts, T. H. (1997) EMBO J. 16, 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shivdasani, R. A., Fujiwara, Y., McDevitt, M. A. & Orkin, S. H. (1997) EMBO J. 16, 3965–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto, M., Ko, L. J., Leonard, M. W., Beug, H., Orkin, S. H. & Engel, J. D. (1990) Genes Dev. 4, 1650–1662. [DOI] [PubMed] [Google Scholar]
- 31.Weiss, M. J., Keller, G. & Orkin, S. H. (1994) Genes Dev. 8, 1184–1197. [DOI] [PubMed] [Google Scholar]
- 32.Cheng, T., Shen, H., Giokas, D., Gere, J., Tenen, D. G. & Scadden, D. T. (1996) Proc. Natl. Acad. Sci. USA 93, 13158–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeno, M., Mead, P. E., Kelley, C., Xu, R. H., Kung, H. F., Suzuki, A., Ueno, N. & Zon, L. I. (1996) Blood 88, 1966–1972. [PubMed] [Google Scholar]
- 34.Ghatpande, S., Ghatpande, A., Sher, J., Zile, M. H. & Evans, T. (2002) Blood 99, 2379–2386. [DOI] [PubMed] [Google Scholar]
- 35.Friedle, H. & Knochel, W. (2002) J. Biol. Chem. 277, 23871–23881. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Y., Musci, T. & Dernyck, R. (1997) Curr. Biol. 7, 270–276. [DOI] [PubMed] [Google Scholar]
- 37.Nony, P., Hannon, R., Gould, H. & Felsenfeld, G. (1998) J. Biol. Chem. 273, 32910–32919. [DOI] [PubMed] [Google Scholar]
- 38.Cantor, A. B. & Orkin, S. H. (2002) Oncogene 21, 3368–3376. [DOI] [PubMed] [Google Scholar]
- 39.Gregory, T., Yu, C., Ma, A., Orkin, S. H., Blobel, G. A. & Weiss, M. J. (1999) Blood 94, 87–96. [PubMed] [Google Scholar]
- 40.Walsh, J. C., DeKoter, R. P., Lee, H.-J., Smith, E. D., Lancki, D. W., Gurish, M. F., Friend, D. S., Stevens, R. L., Anastasi, J. & Singh, H. (2002) Immunity 17, 665–676. [DOI] [PubMed] [Google Scholar]
- 41.Tsang, A. P., Visvader, J. E., Turner, C. A., Fujiwara, Y., Yu, C., Weiss, M. J., Crossley, M. & Orkin, S. H. (1997) Cell 90, 109–119. [DOI] [PubMed] [Google Scholar]
- 42.Crispino, J. D., Lodish, M. B., MacKay, J. P. & Orkin, S. H. (1999) Mol. Cell 3, 219–228. [DOI] [PubMed] [Google Scholar]
- 43.Deconinck, A. E., Mead, P. E., Tevosian, S. G., Crispino, J. D., Katz, S. G., Zon, L. I. & Orkin, S. H. (2000) Development (Cambridge, U.K.) 127, 2031–2040. [DOI] [PubMed] [Google Scholar]
- 44.Fox, A. H., Liew, C., Holmes, M., Kowalski, K., MacKay, J. P. & Crossley, M. (1999) EMBO J. 18, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang, A. N., Cantor, A. B., Fujiwara, Y., Lodish, M. B., Droho, S., Crispino, J. D. & Orkin, S. H. (2002) Proc. Natl. Acad. Sci. USA 99, 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeKoter, R. P. & Singh, H. (2000) Science 288, 1439–1441. [DOI] [PubMed] [Google Scholar]
- 47.Weiss, M. J., Yu, C. & Orkin, S. H. (1997) Mol. Cell. Biol. 17, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson, K. D., Grass, J. A., Boyer, M. E., Kiekhaefer, C. M., Blobel, G. A., Weiss, M. J. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 11760–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson, K. D., Christensen, H. M., Zhao, B. & Bresnick, E. H. (2001) Mol. Cell 8, 465–471. [DOI] [PubMed] [Google Scholar]
- 50.Johnson, K. D. & Bresnick, E. H. (2002) Methods 26, 27–36. [DOI] [PubMed] [Google Scholar]
- 51.Kiekhaefer, C. M., Grass, J. A., Johnson, K. D., Boyer, M. E. & Bresnick, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 14309–14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katz, S. G., Cantor, A. B. & Orkin, S. H. (2002) Mol. Cell. Biol. 22, 3121–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, X., Crispino, J. D., Letting, D. L., Nakazawa, M., Poncz, M. & Blobel, G. A. (2002) EMBO J. 21, 5225–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Im, H., Grass, J. A., Christensen, H. M., Perkins, A. & Bresnick, E. H. (2002) Biochemistry 41, 15152–15160. [DOI] [PubMed] [Google Scholar]
- 55.Trainor, C. D., Omichinski, J. G., Vandergon, T. L., Gronenborn, A. M., Clore, G. M. & Felsenfeld, G. (1996) Mol. Cell. Biol. 16, 2238–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forsberg, E. C., Downs, K. M., Christensen, H. M., Im, H., Nuzzi, P. A. & Bresnick, E. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elefant, F., Su, Y., Liebhaber, S. A. & Cooke, N. E. (2000) EMBO J. 19, 6814–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forsberg, E. C. & Bresnick, E. H. (2001) BioEssays 23, 820–830. [DOI] [PubMed] [Google Scholar]
- 59.Schubeler, D., Francastel, C., Cimbora, D. M., Reik, A., Martin, D. I. & Groudine, M. (2000) Genes Dev. 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- 60.Litt, M. D., Simpson, M., Gaszner, M., Allis, C. D. & Felsenfeld, G. (2001) Science 293, 2453–2455. [DOI] [PubMed] [Google Scholar]
- 61.Noma, K., Allis, C. D. & Grewal, S. I. (2001) Science 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 62.Tse, C., Sera, T., Wolffe, A. P. & Hansen, J. C. (1998) Mol. Cell. Biol. 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee, D. Y., Hayes, J. J., Pruss, D. & Wolffe, A. P. (1993) Cell 72, 73–84. [DOI] [PubMed] [Google Scholar]
- 64.Vettese-Dadey, M., Grant, P. A., Hebbes, T. R., Crane-Robinson, C., Allis, C. D. & Workman, J. L. (1996) EMBO J. 15, 2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 65.Ko, L. J. & Engel, J. D. (1993) Mol. Cell. Biol. 13, 4011–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merika, M. & Orkin, S. H. (1993) Mol. Cell. Biol. 13, 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cantor, A. B., Katz, S. G. & Orkin, S. H. (2002) Mol. Cell. Biol. 22, 4268–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blobel, G. A., Nakajima, T., Eckner, R., Montminy, M. & Orkin, S. H. (1998) Proc. Natl. Acad. Sci. USA 95, 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellis, J., Tan-Un, K. C., Harper, A., Michalovich, D., Yannoutsos, N., Philipsen, S. & Grosveld, F. (1996) EMBO J. 15, 562–568. [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, P., Behre, G., Pan, J., Iwama, A., Wara-aswapati, N., Radomska, H. S., Auron, P. E., Tenen, D. G. & Sun, Z. (1999) Proc. Natl. Acad. Sci. USA 96, 8705–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minegishi, N., Ohta, J., Yamagiwa, H., Suzuki, N., Kawauchi, S., Zhou, Y., Takahashi, S., Hayashi, N., Engel, J. D. & Yamamoto, M. (1999) Blood 93, 4196–4207. [PubMed] [Google Scholar]
- 72.Takahashi, S., Shimuzu, R., Suwabe, N., Kuroha, T., Yoh, K., Ohta, J., Nishimura, S., Lim, K. C., Engel, J. D. & Yamamoto, K. R. (2000) Blood 96, 910–916. [PubMed] [Google Scholar]
- 73.Yamaguchi, Y., Ackerman, S. J., Minegishi, N., Takiguchi, M., Yamamoto, M. & Suda, T. (1998) Blood 91, 3447–3458. [PubMed] [Google Scholar]
- 74.Hung, H. L., Lau, J., Kim, A. Y., Weiss, M. J. & Blobel, G. A. (1999) Mol. Cell. Biol. 19, 3496–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ozawa, Y., Towatari, M., Tsuzuki, S., Hayakawa, F., Maeda, T., Miyata, Y., Tanimoto, M. & Saito, H. (2001) Blood 98, 2116–2123. [DOI] [PubMed] [Google Scholar]
- 76.Schwartz, S., Zhang, Z., Frazer, K. A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R. & Miller, W. (2000) Genome Res. 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.