Abstract
In mammals and plants, formation of heterochromatin is associated with hypermethylation of DNA at CpG sites and histone H3 methylation at lysine 9. Previous studies have revealed that maintenance of DNA methylation in Neurospora and Arabidopsis requires histone H3 methylation. A feedback loop from DNA methylation to histone methylation, however, is less understood. Its recent examination in Arabidopsis with a partial loss of function in DNA methyltransferase 1 (responsible for maintenance of CpG methylation) yielded conflicting results. Here we report that complete removal of CpG methylation in an Arabidopsis mutant null for DNA maintenance methyltransferase results in a clear loss of histone H3 methylation at lysine 9 in heterochromatin and also at heterochromatic loci that remain transcriptionally silent. Surprisingly, these dramatic alterations are not reflected in heterochromatin relaxation.
Maintenance of CpG methylation is essential for mammalian and plant development; it relies on proteins that are conserved between plants and mammals (1, 2). Failure to maintain this epigenetic mark in DNMT1 (DNA maintenance methyltransferase 1)-knockout mice results in embryonic lethality (3). The depletion of maintenance methyltransferase MET1 transcript in Arabidopsis by transgenic expression of its antisense RNA leads to a variety of pleiotropic phenotypes (4, 5). In transcriptionally silent heterochromatin, CpG methylation is associated with another epigenetic mark: namely, histone H3 dimethylated at lysine 9 (H3K9Me) (6). Recent results in Neurospora suggested that the status of histone methylation determines patterns of DNA methylation (7, 8), but in contrast to plants and mammals, DNA methylation in Neurospora is dispensable (9). In Arabidopsis mutants with the defective histone methyltransferase kryptonite (kyp) or a deficiency in a posttranscriptional gene-silencing component (argonaute4), DNA methylation is reduced only at plant-specific CpNpG or CpNpN sites (N = A, T, or C), not at CpGs (10, 11). Experiments addressing the question whether CpG methylation could direct H3K9Me formation yielded contradictory results. Cytological analyses and chromatin immunoprecipitation (ChIP) for histone H3K9Me performed in Arabidopsis mutants in the MET1 gene suggested that CpG methylation either does or does not influence histone methylation (12, 13). Reduction in H3K9Me at centromeres was detected cytologically (13), but ChIP for several target sequences, including centromeric repeats, revealed no changes in H3K9Me levels (12). It is likely that this discrepancy originated from a partial loss of function of the mutant allele (14) used in these studies. This partial loss of function, due to a point mutation that causes replacement of a nonconserved amino acid within the MET1 catalytic domain, results in an incomplete erasure of CpG methylation (up to 50% of CpG methylation is retained) (15, 16). Conclusions were complicated by the possibility that reduction of H3K9Me observed cytologically could be either secondary to transcriptional activation, as suggested (12, 17), or due to the dispersion of pericentromeric DNA out of chromocenters (13).
To readdress the relationship of methylated CpG to H3K9Me and possibly resolve previous inconsistencies, we examined a met1-null Arabidopsis strain, met1-3, that carries foreign DNA inserted into the sequence of the MET1 gene encoding the catalytic domain (18). We used a methodology directly comparable with the methodologies of both previous studies (12, 13). Importantly, as far as we can detect, CpG methylation is completely eliminated in our strain, homozygous for met1 mutation (18). We compared modifications of histones in the met1 strain to the corresponding WT strain by using immunocytological staining and ChIP directed toward transcribed and silent chromosomal loci. In the met1 strain we found a drastic reduction of H3K9Me levels at heterochromatic chromosomal regions, which could also occur without any detectable activation of transcription. Interestingly, the loss of both CpG methylation and H3K9Me at condensed, heterochromatic chromocenters had no significant effect on their structure; the DNA remained densely packed, characteristic of constitutive heterochromatin.
Materials and Methods
ChIP. The met1 strain contained a 7.1-kb portion of the bacterial Ti-plasmid (T-DNA) insert, disrupting the conserved motif region of MET1 (GenBank accession no. L10692). Leaves of 4-week-old WT Arabidopsis (Columbia ecotype) and met1 plants grown in soil were vacuum-infiltrated with formaldehyde cross-linking solution and ChIP was performed as described (12, 19). ChIP DNA was dissolved in 40 μl of TE (10 mM Tris/1 mM EDTA, pH 8) and was assayed and normalized for amplification of the Actin2/7 (20) gene as described (12). All PCRs were performed under conditions described (12). Amplification of single-copy sequences was performed for 40 cycles and for 180-bp repeats for 23 cycles. PCR products were scanned after electrophoretic separation with a Typhoon Scanner and quantified by using imagequant software (Amersham Biosciences). All the antibodies used are from Upstate Biotechnology (Lake Placid, NY).
RT-PCR. RNA was isolated from leaf tissues (as used for ChIP) by using the TRIzol reagent (GIBCO/BRL) according to the manufacturer's instructions. Aliquots of 5 μg of total RNA treated with RQ1 DNase (Promega) were reverse transcribed by using an oligo(dT)-anchored primer (Roche Pharmaceuticals, Nutley, NJ), and cDNA was PCR amplified for 30 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 30 s) with gene-specific primers. Amplification of Act2/7 RNA was used as an internal control. For At4g03870, additional reverse transcription reactions were performed with gene-specific primers located near the 3′ end of the coding region; cDNA was PCR amplified for 40 cycles as described above.
Immunostaining. Leaf protoplasts were isolated and fixed according to www.arabidopsis.org/cshl-course/7-gene_expression.html. After rehydration in PBS, slides were blocked in 2% BSA in PBS (30 min, 37°C) and incubated overnight at 4°C in 1% BSA in PBS containing antibodies (Upstate Biotechnology) specific to lysine-9-dimethylated H3 (1:100 dilution), lysine-4-dimethylated H3 (1:500 dilution), or tetraacetylated H4 (1:100 dilution). Detection was carried out with an FITC-coupled antibody to rabbit IgG (Molecular Probes; 1:100 dilution, 37°C, 40 min) in 0.5% BSA in PBS. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield (Vector Laboratories). Images were analyzed with a Deltavision deconvolution microscope (Applied Precision, Issaquah, WA). worx software (Applied Precision) was applied to deconvolute the images; single representative layers were chosen for Fig. 3. Fluorescent in situ hybridization with a 180-bp repeats probe and immunostaining analysis with a 5-methylcytosine antibody were performed as described (13, 21).
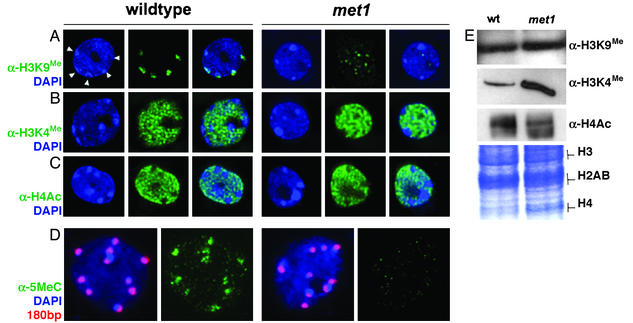
Fig. 3.
Immunolocalization of H3K9Me, H3K4Me, and tetraacetylated histone H4 in WT and met1 nuclei. (A) Detection of H3K9Me.(B) Detection of H3K4Me.(C) Detection of H4 acetylation. Shown are FITC immunostaining (green) and DAPI staining (blue). Chromocenters are visible as light blue, and densely DAPI-stained structures are marked by arrowheads in the leftmost A. (D) Visualization of WT and met1 chromocenters with fluorescent in situ hybridization using 180-bp centromeric repeats probe (red) and immunostaining using 5-methylcytosine antibody (α-5MeC, green). (E) Analysis of H3 and H4 methylation and acetylation, respectively, from WT and met1 plants by Western blot probed with antibody directed against H3K9Me (α-H3K9Me), H3K4Me (α-H3K4Me), or tetraacetylated H4 (α-H4Ac). (Bottom) Coomassie blue-stained proteins.
Western-Blot Analysis. Leaves of 4-week-old plants were homogenized in histone-extraction buffer (10 mM Tris·HCl, pH 7.5/2 mM EDTA/0.25 M HCl/5 mM 2-mercaptoethanol/0.2 mM PMSF). After centrifugation at 12,000 × g for 10 min, soluble proteins were precipitated with 25% trichloroacetic acid and collected by centrifugation at 17,000 × g for 30 min. The pellet was washed twice with acetone, resuspended in Laemmli buffer, separated electrophoretically, and Western blotted and detected by using standard procedures (22).
Results and Discussion
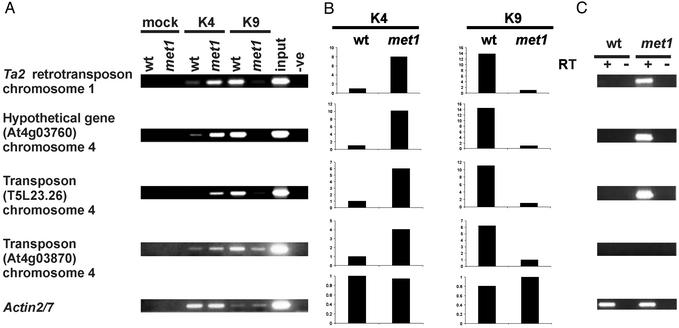
In our strain (named met1 throughout this article), we determined the methylation status of histone H3 at the nontranscribed Ta2 retrotransposon that resides in a pericentromeric region of chromosome 1. Ta2 has been shown to contain hypermethylated DNA that is packaged to nucleosomes with H3K9Me (12). We compared WT and met1 plants by using ChIP with antibodies recognizing H3K9Me or histone H3 dimethylated at lysine 4 (H3K4Me) (Fig. 1 A and B). H3K9Me, which is abundant at Ta2 in WT, was almost undetectable in the met1 mutant (Fig. 1 A and B). In met1, Ta2 consistently gained H3K4Me (Fig. 1 A and B), a marker for transcriptionally active genes (6). Indeed, in the met1 mutant, transcription of Ta2 was clearly activated (Fig. 1C). Depletion of CpG methylation at Ta2, therefore, likely changed the methylation status of histone H3. This change, however, could have been an indirect effect of transcriptional activation. To assess the generality of this observation and relate it to transcriptional reactivation, we examined levels of H3K9Me and H3K4Me in three additional target genes residing within the heterochromatic knob on chromosome 4 (At4g03760, T5L23.26, and At4g03870) (Fig. 1 A and B). Like Ta2, these loci are associated with H3K9Me in the WT (19). Four independent ChIP experiments were performed for each of the four target sequences and the Actin2/7 control (20); these ChIP experiments yielded equivalent results (Fig. 1 A and B). The abundant H3K9Me found in all of the targets in the WT was drastically reduced and replaced by H3K4Me in the met1 mutant (Fig. 1 A and B). When transcript levels originating from these loci in WT and met1 (Fig. 1C) were compared, transcripts were detected in the met1 mutant for Ta2, At4g03760, and T5L23.26 (Fig. 1C), but not for At4g03870 (Fig. 1C). This finding suggested that the loss of H3K9Me in the met1 mutant could occur also at silent templates; very low levels of transcription, however, cannot be excluded.
Fig. 1.
Histone 3 methylation analyzed by ChIP and transcript levels of selected heterochromatic loci. (A) ChIP with antibodies recognizing H3K4Me (K4) and H3K9Me (K9) of chromatin extracts from WT and the met1 mutant (met1). Controls: chromatin extract precipitated without antibody (mock), chromatin extract (input), and PCR without template (-ve). Analyzed target loci are listed to the left of the corresponding images. (B) Quantification of ChIP analysis corresponding to images presented in A. Ordinates are fold difference. (C) RT-PCR analysis of transcripts, with (+) and without (–) reverse transcriptase (RT).
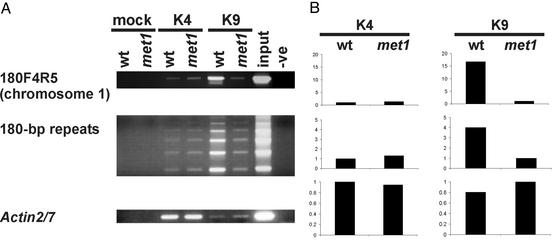
To substantiate this observation, we examined H3K9Me at 180-bp centromeric repeats considered transcriptionally inert in both WT and DNA methylation-deficient mutants (12). To examine changes in DNA methylation and histone modifications accurately and correlate them to the transcriptional status of repeats, we designed primers (180F4R5) specific to a single-copy sequence (GenBank accession no. AC083859, nucleotides 61536–62127) that flanks a particular set of 180-bp repeats residing in the pericentromeric region on chromosome 1. This primer eliminated interference by other 180-bp repeats distributed over all five chromosomes. As revealed by a bisulfite analysis assay, CpG methylation on 180F4R5 repeats is completely erased in met1, but reduced only to 57.6% and 73% at CpNpG and CpNpN sites, respectively (18). Analysis of other target sequences showed similar results (data not shown). The ChIP assays showed a strong enrichment in H3K9Me at these repeats in WT plants and a 17-fold depletion in the met1 mutant (Fig. 2 A and B). Importantly, multiple RT-PCR analyses did not detect RNA from this region in either WT or the met1 mutant (data not shown). To examine whether this finding was specific to this particular subset of 180-bp repeats or could be confirmed globally, we used unselective 180-bp repeat primers that generate a ladder because of multiple annealing sites throughout the genome (12). The met1 mutation also caused a strong reduction in H3K9Me at these combined targets (Fig. 2 A and B). In an RT-PCR assay with the same primers, however, we detected the appearance of traces of RNA in the met1 mutant (data not shown). This appearance of RNA traces is consistent with the earlier studies (12).
Fig. 2.
Histone 3 methylation analysis by ChIP of 180-bp centromeric repeats. (A) ChIP with antibodies recognizing H3K4Me (K4) and H3K9Me (K9) of chromatin extracts from WT and the met1 mutant (met1); controls as in Fig. 1. Amplification was performed using primers to a specific subset (180F4R5) or broader range of repeats, as described in the text. (B) Quantification of ChIP analysis corresponding to images presented in A. Ordinates are fold difference.
Because 180-bp repeats are an integral part of the centromeric region of all Arabidopsis chromosomes and histone methylation changes in the met1 mutant seem to be rather drastic, we attempted to visualize H3K9Me alterations by immunostaining WT and met1 nuclei by using an antibody that recognizes H3K9Me (α-H3K9). In the WT, the dense heterochromatic chromocenters, known to be enriched in H3K9Me, were clearly stained for H3K9Me (Fig. 3A). Similar staining performed previously with a point-mutant allele in the MET1 gene showed a reduction in H3K9Me, but part of the signal was still retained at the chromocenters (13). In met1 nuclei, in contrast, chromocenters detectable by DAPI staining remained unlabeled by the H3K9Me antibody (Fig. 3A). Thus in met1, the H3K9Me signal was reduced and completely redistributed away from chromocenters (Fig. 3A). Very similar images of met1 nuclei were obtained with antibodies directed against 5-methylcytosine (Fig. 3D), confirming a drastic, concerted change in DNA methylation and H3K9Me at the centromeric heterochromatin. Surprisingly, images obtained by fluorescent in situ hybridization with probes specific to centromeric 180-bp repeats appear to be very similar for the met1 and WT (Fig. 3D), which is in contrast to the ddm1 (decrease in DNA methylation 1) mutant, where centromeric DNA disperses (13, 23). The complete depletion of methylated CpGs and drastic reduction of H3K9Me in the met1 strain suggest that both modifications are dispensable for maintaining the compact, heterochromatic structures enclosing centromeric DNA. This finding is in accordance with a recent cytological study of the kyp mutant, indicating that H3K9Me is unessential for the maintenance of heterochromatic structure of Arabidopsis chromocenters (24).
In contrast to immunostaining for H3K9Me, α-H3K4Me showed exclusion of the signal from chromocenters (Fig. 3B) in both WT and met1. Thus, the cytological data were in agreement with ChIP analyses, demonstrating a severe reduction in H3K9Me at 180-bp repeats but retention of a constantly low level of H3K4Me in the met1 strain. Because cytological data suggested redistribution of H3K9Me away from chromocenters in the met1 mutant (rather than a general reduction in level) we compared by Western blotting the proportion of H3K9Me in enriched histone fractions. Indeed, similar levels of H3K9Me were detected in WT and met1, but there appeared to be an increase in abundance of H3K4Me, which may reflect the alleviation of silencing (Fig. 3E).
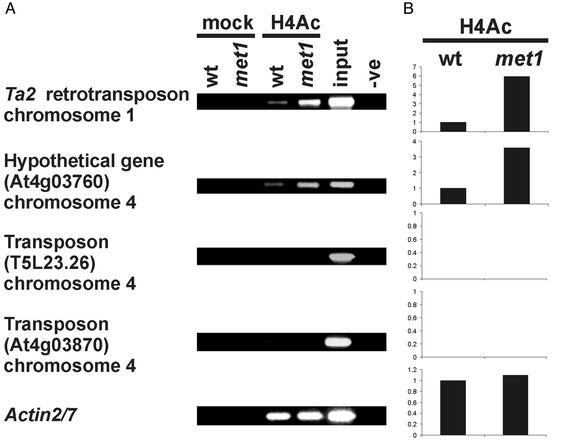
A tight link between CpG methylation and histone deacetylation and transcriptional repression has been postulated in mammalian cells (25–28). We investigated, therefore, whether depletion of CpG methylation in the met1 mutant affects levels of histone acetylation that could be related to changes in transcriptional activity. We performed ChIP with a tetraacetylated histone H4 antibody (α-H4Ac) by using all of the target sequences described above. According to the ChIP analyses, acetylated H4 (H4Ac) was enriched approximately 3- to 6-fold in the met1 mutant compared with the WT. However, this enrichment was restricted to targets that were transcriptionally reactivated in the met1 (Fig. 4). H4Ac was not detected at the At4g03870 locus or at nontranscribed 180-bp repeats (Fig. 4 and data not shown). Notably, H4Ac was almost undetectable at the T5L23.26 locus, reactivated in the met1 mutant (Figs. 1C and 4). Hyperacetylation, however, is not always associated with transcriptional reactivation of previously silent loci (29, 30) and the mechanism of T5L23.26 activation may be of this category. Immunostaining of WT and met1 nuclei with α-H4Ac antibody showed that chromocenters remained hypoacetylated in the mutant (Fig. 3C). Western blotting showed no change in histoneacetylation levels (Fig. 3E).
Fig. 4.
Histone 4 acetylation analyzed by ChIP of selected heterochromatic loci. (A) ChIP with antibodies recognizing histone H4 acetylated at lysines 5, 8, 12, and 16 (H4Ac) of chromatin extracts from WT and the met1 mutant; controls as in Fig. 1. Analyzed target loci are listed to the left of corresponding images. (B) Quantification of ChIP analysis corresponding to images presented in A. H4Ac was not detected at T5L23.26 and At4g03870 in either WT or met1. Ordinates are fold difference.
Our results clearly support the notion that methylation at H3K9 is directed by DNA methylation at CpG sites (13). They show, in addition, that this process probably occurs in a transcription-independent fashion. Obviously, very low transcriptional activity might have escaped detection. The recently discovered in vitro and in vivo interaction of human proteins recognizing methylated cytosine in CpG (MeCP2, MBD1) with histone methyltransferase also suggests such a link (31, 32). The effect of CpG methylation loss in the met1 is in contrast to methylation outside CpGs, which has no effect on H3K9Me when strongly reduced by cmt3 (chromomethylase 3) mutations (12, 33). Interestingly, in kyp mutants defective in histone H3 lysine 9 methyltransferase DNA methylation marks are affected only at CpNpG and CpNpN sites (not at CpGs sites), suggesting that non-CpG methylation is regulated by histone methylation (10). Recently, the putative chromatin-remodeling factor DDM1 (decrease in DNA methylation 1) was shown to be required for maintenance of H3K9Me (19). In ddm1 mutants, however, cytosine methylation in all sequence contexts including CpGs is affected, making it difficult to distinguish direct from indirect effects on histone methylation. Considering that CpG methylation determines the status of histone methylation, the loss of histone methylation in the ddm1 mutants could be due to the depletion of CpG methylation. Both met1 and ddm1 mutations also reduce levels of cytosine methylation in sequences outside CpGs. Although this reduction may contribute to the loss of histone methylation (12), given that DNA methylation outside CpGs acts downstream of histone methylation (10), changes in CpNpG and CpNpN observed in met1 and ddm1 are most likely secondary effects of these mutations on H3K9Me. It is notable that, in contrast to met1, chromocenters in ddm1 nuclei decompose (13, 21). It is conceivable, therefore, that structural alterations of heterochromatin seen in the ddm1 mutant are not linked directly to the methylated CpG/H3K9Me depletion but to another, unknown mechanism. Because cytological data suggested redistribution of H3K9Me in the met1 mutant, it would be of interest to map this redistribution in more detail.
Our results clearly support the existence in vivo of a self-reinforcing system that contributes to the formation of silent heterochromatin in which CpG methylation plays the role of a central scaffold directing histone methylation, possibly independent of transcription. This function of CpG methylation explains the severe and immediate phenotypes of met1 and the gametophytic effects (18) that are much less pronounced or absent in the weaker met1 mutant allele (16) used in all previous studies.
Acknowledgments
We thank Fang Lin Sun for helpful discussions regarding ChIP, Karin Afsar and Susanne Lienhardt for technical assistance, and Thomas Hohn, Witold Filipowicz, Barbara Hohn, Anne Blonstein, and Patrick King for helpful comments on the manuscript.
Abbreviations: ChIP, chromatin immunoprecipitation; H3K9Me, histone H3 dimethylated at lysine 9; H3K4Me, histone H3 dimethylated at lysine 4; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Finnegan, E. J. & Kovac, K. A. (2000) Plant Mol. Biol. 43, 189–201. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. (2002) Genes Dev. 16, 6–21. [DOI] [PubMed] [Google Scholar]
- 3.Li, E., Bestor, T. H. & Jaenisch, R. (1992) Cell 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 4.Finnegan, E. J., Peacock, W. J. & Dennis, E. S. (1996) Proc. Natl. Acad. Sci. USA 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ronemus, M. J., Galbiati, M., Ticknor, C., Chen, J. & Dellaporta, S. L. (1996) Science 273, 654–657. [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 7.Tamaru, H. & Selker, E. U. (2001) Nature 414, 277–283. [DOI] [PubMed] [Google Scholar]
- 8.Tamaru, H., Zhang, X., McMillen, D., Singh, P. B., Nakayama, J., Grewal, S. I., Allis, C. D., Cheng, X. & Selker, E. U. (2003) Nat. Genet. 34, 75–79. [DOI] [PubMed] [Google Scholar]
- 9.Kouzminova, E. & Selker, E. U. (2001) EMBO J. 20, 4309–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, J. P., Lindroth, A. M., Cao, X. & Jacobsen, S. E. (2002) Nature 416, 556–560. [DOI] [PubMed] [Google Scholar]
- 11.Zilberman, D., Cao, X. & Jacobsen, S. E. (2003) Science 299, 716–719. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, L., Cao, X. & Jacobsen, S. E. (2002) Curr. Biol. 12, 1360–1367. [DOI] [PubMed] [Google Scholar]
- 13.Soppe, W. J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M. S., Jacobsen, S. E., Schubert, I. & Fransz, P. F. (2002) EMBO J. 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartee, L. & Bender, J. (2001) Nucleic Acids Res. 29, 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vongs, A., Kakutani, T., Martienssen, R. A. & Richards, E. J. (1993) Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- 16.Kankel, M. W., Ramsey, D. E., Stokes, T. L., Flowers, S. K., Haag, J. R., Jeddeloh, J. A., Riddle, N. C., Verbsky, M. L. & Richards, E. J. (2003) Genetics 163, 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad, K. & Henikoff, S. (2002) Mol. Cell 9, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 18.Saze, H., Mittelsten Scheid, O. & Paszkowski, J. (2003) Nat. Genet. 34, 65–69. [DOI] [PubMed] [Google Scholar]
- 19.Gendrel, A. V., Lippman, Z., Yordan, C., Colot, V. & Martienssen, R. A. (2002) Science 297, 1871–1873. [DOI] [PubMed] [Google Scholar]
- 20.An, Y. Q., McDowell, J. M., Huang, S., McKinney, E. C., Chambliss, S. & Meagher, R. B. (1996) Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- 21.Mittelsten Scheid, O., Probst, A. V., Afsar, K. & Paszkowski, J. (2002) Proc. Natl. Acad. Sci. USA 99, 13659–13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 23.Probst, A. V., Fransz, P. F., Paszkowski, J. & Scheid, O. M. (2003) Plant J. 33, 743–749. [DOI] [PubMed] [Google Scholar]
- 24.Jasencakova, Z., Soppe, W. J., Meister, A., Gernand, D., Turner, B. M. & Schubert, I. (2003) Plant J. 33, 471–480. [DOI] [PubMed] [Google Scholar]
- 25.Ng, H. H., Zhang, Y., Hendrich, B., Johnson, C. A., Turner, B. M., Erdjument-Bromage, H., Tempst, P., Reinberg, D. & Bird, A. (1999) Nat. Genet. 23, 58–61. [DOI] [PubMed] [Google Scholar]
- 26.Wade, P. A., Gegonne, A., Jones, P. L., Ballestar, E., Aubry, F. & Wolffe, A. P. (1999) Nat. Genet. 23, 62–66. [DOI] [PubMed] [Google Scholar]
- 27.Jones, P. L., Veenstra, G. J., Wade, P. A., Vermaak, D., Kass, S. U., Landsberger, N., Strouboulis, J. & Wolffe, A. P. (1998) Nat. Genet. 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 28.Nan, X., Ng, H. H., Johnson, C. A., Laherty, C. D., Turner, B. M., Eisenman, R. N. & Bird, A. (1998) Nature 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 29.Lorincz, M. C., Schubeler, D., Goeke, S. C., Walters, M., Groudine, M. & Martin, D. I. (2000) Mol. Cell. Biol. 20, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamin, D. & Jost, J. P. (2001) Nucleic Acids Res. 29, 3603–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuks, F., Hurd, P. J., Wolf, D., Nan, X., Bird, A. P. & Kouzarides, T. (2003) J. Biol. Chem. 278, 4035–4040. [DOI] [PubMed] [Google Scholar]
- 32.Fujita, N., Watanabe, S., Ichimura, T., Tsuruzoe, S., Shinkai, Y., Tachibana, M., Chiba, T. & Nakao, M. (April 23, 2003) J. Biol. Chem., 10.1074/jbc.M302283200. [DOI] [PubMed]
- 33.Lindroth, A. M., Cao, X., Jackson, J. P., Zilberman, D., McCallum, C. M., Henikoff, S. & Jacobsen, S. E. (2001) Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]