Abstract
NY-ESO-1 is one of the most immunogenic proteins described in human cancers, based on its capacity to elicit simultaneous antibody and CD8+ T cell responses in vivo. Although HLA class II restricted epitopes from NY-ESO-1 have been identified, no broad survey has yet established the status of natural CD4+ T cell responses in cancer patients in relation to CD8+ and antibody responses. We used a recently developed general strategy for monitoring CD4+ responses that overcomes the need for prior knowledge of epitope or HLA restriction to analyze a series of 31 cancer patients and healthy donors for the presence of CD4+ T cells to NY-ESO-1, and related this response to NY-ESO-1 expression in tumor cells and serum antibodies to NY-ESO-1. None of the 18 patients that tested seronegative for NY-ESO-1 had detectable CD4+ T cell responses. On the contrary, 11 of 13 cancer patients with serum antibodies to NY-ESO-1 had polyclonal CD4+ T cell responses directed against various known and previously undescribed NY-ESO-1 epitopes. NY-ESO-1 peptide 80–109 was the most immunogenic, with 10 of 11 patients responding to this peptide. We show here that 12-mer determinants from NY-ESO-1 eliciting a CD4+ T cell response were peptide 87–98 with promiscuous HLA class II presentation, peptide 108–119 restricted by HLA-DP4, and peptides 121–132 and 145–156, both shorter epitopes from previously described HLA-DR4 peptides, also presented by HLA-DR7. This study represents the next step in compiling a comprehensive picture of the adaptive immune response to NY-ESO-1, and provides a general strategy for analyzing the CD4+ T cell response to other tumor antigens eliciting a humoral immune response.
NY-ESO-1 is a germ cell protein that is frequently expressed by cancer cells, but not normal somatic cells (1). It was discovered by screening the serum of an esophageal cancer patient for antibodies recognizing proteins expressed in autologous tumor cells. Humoral and CD8+ T cell responses to NY-ESO-1 are found in a high frequency of patients with NY-ESO-1+ tumors, making it one of the most immunogenic human tumor antigens (2).
Immune responses to NY-ESO-1 are generally restricted to patients with advanced cancer (3), and tend to disappear with tumor resection or therapy-induced regression (4). A possible reason for this dependence of NY-ESO-1 immunity on tumor size is that the tumor must reach a critical mass before sufficient antigen is released from necrotic areas of the tumor to prime antigen-presenting cells (APC) and initiate B cell and T cell responses. In monitoring a large series of cancer patients, we showed that humoral responses to NY-ESO-1 correlated with the presence of peripheral CD8+ T cells against NY-ESO-1 (5), suggesting the involvement of CD4+ helper T cells in coordinating this response. Furthermore, strong humoral responses to NY-ESO-1 are of IgG isotype (3), rather than IgM, which also predicts involvement of CD4+ T cells. Helper T cells are known to be critical for inducing class switching by B cells, and the major pathway for class switching occurs through surface interaction/stimulation of CD40 with CD40L (6, 7), with additional involvement of the more recently described molecule ICOS (8).
Independent of their helper role in humoral and CD8+ T cell responses, CD4+ T cells also provide a protective function by cytokine secretion and local inflammatory reactions, which has been shown to be independent of the CD40–CD40L interaction (6). It is thus interesting to determine whether CD4+ T cell responses to NY-ESO-1 correlate with the presence of antibody to NY-ESO-1, as do CD8+ T cell responses, or whether CD4+ T cell responses to NY-ESO-1 can be found more broadly in patients, as a first line of specific immune recognition preceding the other participants in the adaptive response.
CD4+ T cell responses to NY-ESO-1 have been recently described. Large epitopes restricted by HLA-DR4 (9–11) or promiscuous for several HLA-DR have been shown (12). HLA-DP4-restricted responses were also seen and were suggested to correlate with antibody status (13). Still, most approaches to study CD4+ T cells rely on the generation of T cell lines after multiple peptide stimulations, which are typically assessed for bulk cytokine secretion or proliferation. The lack of reliable and quantitative approaches for monitoring CD4+ T cell responses has hindered the analysis of larger series of patients.
We recently introduced a new methodology to analyze antigen-specific CD4+ T cells at the single cell level in enzyme-linked immunospot (ELISPOT) assays (14). Responses of purified CD4+ T cells to influenza nucleoprotein (NP) were assessed after a single round of in vitro antigen stimulation to recall memory responses only. Specific CD4+ T cells were detected not only against a peptide containing HLA-DR promiscuous epitope, but also against full-length NP expressed in APCs from recombinant fowlpox vectors. Through the use of innovative targets, namely phytohemagglutinin-expanded CD4+ T cells (T-APC), ELISPOT assays could be carried out with virtually no background, in a quantitative and reproducible manner.
We show here that the same approach can be adapted to monitor CD4+ T cell responses to NY-ESO-1 in cancer patients, as a counterpart to the general strategy that we previously developed to monitor CD8+ T cell responses to NY-ESO-1 in any patient (15). Because our approach to CD4+ T cell monitoring is not limited to known epitopes, it is applicable to any patient regardless of HLA restriction. In addition, it facilitates the discovery of new HLA class II restricted peptides from NY-ESO-1.
Materials and Methods
Peptides, Protein, and Viral Vectors. The following peptides were obtained from Bio-Synthesis (Lewisville, TX) with a purity of >90%: ESO80-109 (ARGPESRLLEFYLAMPFATPMEAELARRSL), ESO87-98 (LLEFYLAMPFAT), ESO108-119 (SLAQDAPPLPVP), ESO121-132 (VLLKEFTVSGNI), ESO143-154 (RQLQLSISSCLQ), ESO145-174 (LQLSISSCLQQLSLLMWITQCFLPVFLAQP), and NP206-229 (FWRGENGRKTRIAYERMCNILKGK). Peptide ESO79-109 (GARGPESRLLEFYLAMPFATPMEAELARRS) was obtained from Multiple Peptide Systems (San Diego) with a purity >80%. Overlapping 18-mer peptides from NY-ESO-1 were described (9). Adenoviral vector recombinant for NY-ESO-1 (AdESO) was obtained from Genzyme Corporation (Framingham, MA), and fowlpox vectors recombinant for NY-ESO-1 (FP-ESO) or for influenza nucleoprotein (FP-NP) were obtained from Therion Biologics (Cambridge, MA), and their construction was previously described (14, 15). The full-length recombinant NY-ESO-1 protein was expressed from Escherichia coli (3) and complexed with hydrophobized polysaccharide, pullulan (CHP) as described (16, 17).
In Vitro Sensitization with Peptides or Recombinant Adenovirus Constructs. Peripheral blood mononuclear cells (PBMC) were obtained with informed consent from cancer patients. HLA typing of donor PBMCs or derived cell lines was done by sequence specific oligonucleotide probing and sequence specific priming of genomic DNA by using standard procedures. CD4+ and CD8+ T lymphocytes were separated from PBMCs by using antibody-coated magnetic beads (Dynabeads, Dynal, Oslo) and seeded into round-bottomed 96-well plates (Corning) at a concentration of 5 × 105 cells per well in RPMI medium 1640 supplemented with 10% human AB serum (GemCell, Gemini BioProducts, Woodland, CA), l-glutamine (2 mM), penicillin (100 units/ml), streptomycin (100 μg/ml), and 1% nonessential amino acids (complete medium). As APC for antigen-specific presensitization, CD4–/CD8– PBMC depleted of CD4+ and CD8+ T cells were either pulsed with 10 μM peptide or infected with recombinant adenovirus at 1,000 infectious units per cell overnight at 37°C in 250 μl of serum-free medium (X-VIVO-15, Bio-Whittaker). Pulsed or infected CD4–/CD8– APC were then washed, irradiated, and added to plates containing CD4+ or CD8+ T cells at a concentration of 1 × 106 APC per well. After 8 h, IL-2 (10 units/ml, Roche Molecular Biochemicals) and IL-7 (20 ng/ml, R & D Systems) were added to culture wells, and this was repeated every 3–4 days thereafter.
Target Cells. A fraction of CD4+ T cells from the initial separation (see above) was seeded into 48-well plates (Corning) at a concentration of 1 × 106 cells per ml in complete medium supplemented with 10 μg/ml phytohemagglutinin (HA15, Murex Diagnostics, Dartford, U.K.). Cells were fed and expanded twice a week with complete medium containing IL-2 (10 units/ml) and IL-7 (20 ng/ml). The activated T cell APC (T-APC) were typically harvested and used as target cells after 20–30 days of culture.
Epstein–Barr virus (EBV)-transformed B lymphocytes (EBV-B cells) were cultured in RPMI medium 1640 supplemented with 10% FCS (Summit Biotechnology, Ft. Collins, CO), l-glutamine (2 mM), penicillin (100 units/ml), streptomycin (100 μg/ml), and 1% nonessential amino acids.
In all assays, target cell APC were washed twice in X-VIVO-15 medium to remove serum. Peptides (10 μM) or recombinant fowlpox virus (100 plaque forming units per cell) were then added and target cells were incubated overnight at 37°C. At the end of this incubation period, target cell APC were washed twice and resuspended in RPMI for testing.
ELISPOT Assays. Flat-bottomed, 96-well nitrocellulose plates (MultiScreen-HA, Millipore) were coated with IFN-γ mAb (2 μg/ml, 1-D1K, Mabtech, Stockholm) and incubated overnight at 4°C. After washing with RPMI, plates were blocked with 10% human AB type serum for 2 h at 37°C. Presensitized CD4+ T effector cells (5 × 104 or 1 × 104) and 1 × 105 target cells (T-APC or EBV-B cells) were added to each well and incubated for 20 h in RPMI medium 1640 without serum. For blocking experiments, 10 μg/ml of the indicated antibodies were added at culture initiation. Plates were then washed thoroughly with water containing 0.05% Tween 20 to remove cells, and IFN-γ mAb (0.2 μg/ml, 7-B6-1-biotin; Mabtech) was added to each well. After incubation for 2 h at 37°C, plates were washed and developed with streptavidin-alkaline phosphatase (1 μg/ml; Mabtech) for 1 h at room temperature. After washing, substrate (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium; Sigma) was added and incubated for 5 min. After final washes, plate membranes displayed dark-violet spots that were scanned and counted by using CTL ImmunoSpot analyzer and software (Cellular Technologies, Cleveland).
Results
Specificity of Presensitization with Peptides or Recombinant Viral Vectors. Melanoma patient NW-29 was previously shown to have a spontaneous CD4+ T cell response to NY-ESO-1 peptides 115-132, 121-138, and 139-156 (9), and was thus selected as a control for our monitoring approach. CD4+ T lymphocytes were stimulated with autologous CD4/CD8-depleted PBMC pulsed with either peptides from NY-ESO-1 or Influenza NP, or infected with viral vectors recombinant for full-length NY-ESO-1 or NP sequences. After 3 weeks of culture without additional restimulation, presensitized CD4 T+ cell effectors were tested by ELISPOT assay against autologous phytohemagglutinin-activated CD4+ T blasts (T-APC) pulsed with various peptides.
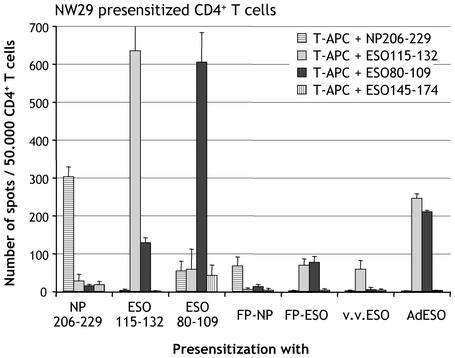
We confirmed that CD4+ T cells from patient NW-29 were reactive against influenza NP peptide 206-229, previously shown to be immunogenic in a majority of patients. A large number of IFN-γ producing CD4+ T cells specific for NP206-229 could be detected after presensitization with NP206-229, with low background against irrelevant NY-ESO-1 peptides (Fig. 1).
Fig. 1.
Specificity controls of CD4+ T cell presensitization from melanoma patient NW29. CD4+ T cells were presensitized with indicated peptides or recombinant vectors and tested by ELISPOT at d23 against autologous T-APC pulsed with peptides NP206-229, ESO115-132, ESO80-109, or ESO145-174. Results represent the mean number of spots per 50,000 effector CD4+ T cells in duplicate wells, with error bars indicating standard deviation.
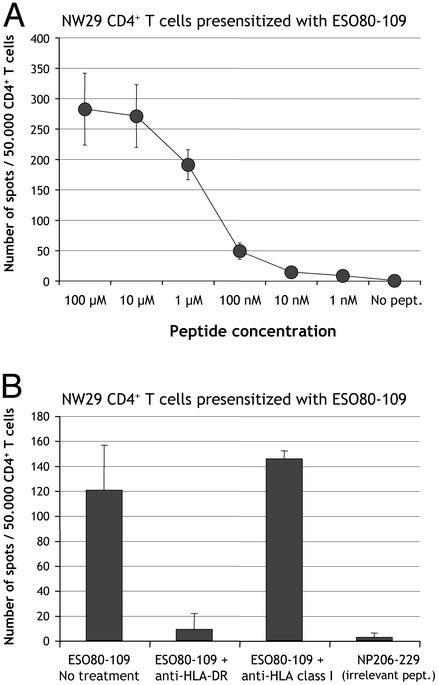
Next, we tested the immunogenicity of a known HLA class II NY-ESO-1 epitope and found that CD4+ T cells from patient NW-29 presensitized with NY-ESO-1 peptide 115-132 specifically produced IFN-γ in response to 115-132, but not to control peptides NY-ESO-1 145-174 nor NP 206-229 (Fig. 1). Unexpectedly, 115-132-presensitized cells also had significant reactivity against NY-ESO-1 polypeptide 80-109. To ask whether this reactivity was potentially related to a novel NY-ESO-1 specificity of CD4+ T cells from patient NW-29, CD4+ T cells were presensitized with peptide 80-109. The reactivity to 80-109 was very strong, comparable to that against 115-132 albeit with higher nonspecific background, thus revealing a new epitope spontaneously immunogenic in patient NW-29 (Fig. 1). Peptide 80-109 could be titrated down to 100 nM (Fig. 2A), and its recognition was blocked by antibodies against HLA-DR, but not against HLA class I (Fig. 2B).
Fig. 2.
(A) Titration of peptide ESO80-109 recognized by CD4+ T cells from patient NW29. CD4+ T cells were presensitized with peptide ESO80-109 and tested by ELISPOT against autologous T-APC pulsed with peptide ESO80-109 in concentrations ranging from 100 μM to 1 nM, or not pulsed. (B) Blocking of ESO80-109 recognition by anti-HLA-DR antibodies. The same effectors were tested by ELISPOT against targets pulsed with ESO80-109 or irrelevant control peptide NP206-229 in the presence or absence of anti-HLA-DR (10 μg/ml) or anti-HLA class I (10 μg/ml) monoclonal antibodies. Results represent the mean number of spots per 50,000 effector CD4+ T cells in duplicate wells, with error bars indicating standard deviation.
General Strategy for CD4+ T Cell Monitoring. To monitor patients CD4+ T cell responses regardless of specific HLA restriction or knowledge of defined epitopes, we adapted the general strategy developed for CD8+ T cell responses to NY-ESO-1 (15). This strategy was validated with a recombinant fowlpox vector used to introduce the full-length influenza NP sequence (FP-NP) in CD4/CD8-depleted cells, which in turn naturally process NP into class II epitope presented to CD4+ T cells. Although not as efficient as presensitization with peptide NP206-229, presensitization with FP-NP was able to significantly recall CD4+ T cells specific for NP206-229 from patient NW29 (Fig. 1).
Three different vectors coding for full-length NY-ESO-1 were then compared for CD4+ T cell presensitization: fowlpox virus (FP-ESO), vaccinia virus (v.v.ESO), and adenovirus (AdESO). Presensitization with FP-ESO was able to recall both specificities against ESO80-109 and ESO115-132, showing that the introduced NY-ESO-1 sequence had been naturally processed in endogenous pathways (Fig. 1). Recombinant vaccinia only recalled ESO115-132, thus exhibiting a differential pattern of processing or immunodominance specific to vaccinia vector, or simply lower general reactivity. Finally, presensitization with AdESO gave the highest responses to both immunogenic peptides, but not to control NP peptide or NY-ESO-1 peptide 145-174, which is not recognized in this patient (Fig. 1).
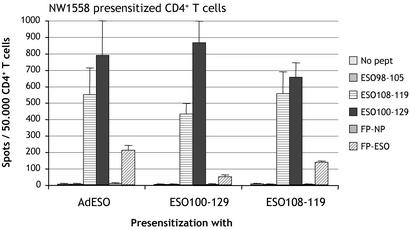
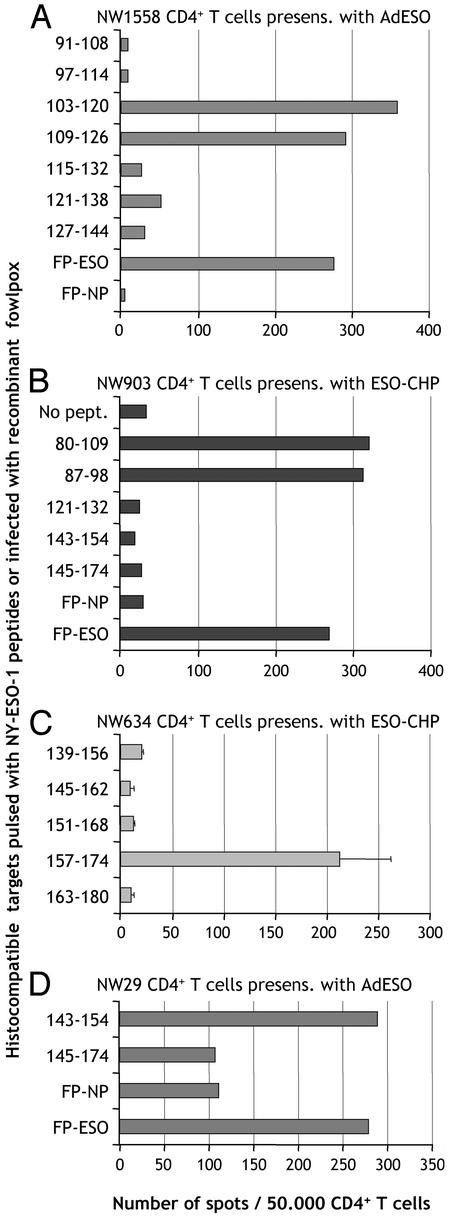
Different Routes of Class II Presentation and Characterization of 12-Mer Epitopes. This general strategy of monitoring CD4+ T cell responses to NY-ESO-1 regardless of the patient HLA haplotype or known epitopes was further pursued. Ovarian cancer patient NW1558 CD4+ T cells were presensitized with AdESO and tested by ELISPOT against autologous T-APC infected with FP-ESO or control FP-NP. Fowlpox was chosen over soluble protein to deliver NY-ESO-1 to T-APC, to avoid relying on the uptake capacity of target cells, and because it gave the most consistent results compared with other vectors such as vaccinia (not shown). From patient NW1558, a clear response specific for NY-ESO-1 was detected without the need to define HLA restriction usage (Fig. 3). To determine the epitope(s) responsible for CD4+ T cell recognition, we tested a 30-mer polypeptide from NY-ESO-1, 100-129, and found that it was very reactive with AdESO-presensitized CD4+ T cells (Fig. 3). The epitope was further mapped by using 18-mer overlapping peptides in the 100-129 region, and peptides 103-120 and 109-126 were still recognized (Fig. 4A). Finally, we searched for presence of HLA class II binding motif within these sequences by using a web-based predictive algorithm (18) and determined from potential anchor motifs that 12-mer peptide 108-119 would likely be long enough to contain the epitope and bind to HLA class II. Indeed, we confirmed that AdESO-presensitized CD4+ T cells recognized NY-ESO-1 peptide 108-119, and that both polypeptide 100-129 and 12-mer 108-119 were immunogenic when used to presensitize NW1558 CD4+ T cells (Fig. 3).
Fig. 3.
Presensitization of CD4+ T cells from ovarian cancer patient NW1558. CD4+ T cells were presensitized with AdESO or with peptides ESO100-129 or ESO108-119 and tested by ELISPOT at d19 against autologous T-APC pulsed with ESO98-105, ESO108-119, or ESO100-129, or infected with FP-NP or FP-ESO. Results represent the mean number of spots per 50,000 effector CD4+ T cells in duplicate wells, with error bars indicating standard deviation.
Fig. 4.
Mapping of epitopes recognized by CD4+ T cells from different cancer patients. (A) CD4+ T cells from patient NW1558 were presensitized with adenovirus recombinant for NY-ESO-1 (AdESO) and tested by ELISPOT against hsistocompatible EBV-B cells pulsed with indicated 18-mer NY-ESO-1 peptides, or infected with recombinant fowlpox. (B) CD4+ T cells from patient NW903 were presensitized with ESO-CHP and tested by ELISPOT against histocompatible EBV-B cells pulsed with indicated NY-ESO-1 peptides, or infected with recombinant fowlpox. (C) CD4+ T cells from patient NW634 were presensitized with ESO-CHP and tested by ELISPOT against histocompatible EBV-B cells pulsed with indicated NY-ESO-1 peptides. (D) CD4+ T cells from patient NW29 were presensitized with AdESO and tested by ELISPOT against histocompatible EBV-B cells pulsed with indicated NY-ESO-1 peptides, or infected with recombinant fowlpox.
As an alternative to infectious delivery of NY-ESO-1 for HLA class II presentation, we assessed soluble full-length NY-ESO-1 recombinant protein complexed with hydrophobized polysaccharide pullulan (ESO-CHP) for presensitizing patient CD4+ T cells (17). From melanoma patient NW903, we found that ESO-CHP could efficiently stimulate CD4+ T cells that recognized NY-ESO-1 recombinant fowlpox-infected targets, and we mapped the associated peptide epitope to polypeptide 80-109 (Fig. 4B). Within this 30-mer peptide, we found that 18-mer peptide 85-102 was still antigenic (not shown), and by using the predictive algorithms (18), determined that 87-98 was a 12-mer peptide still containing the epitope (Fig. 4B).
Similarly, we stimulated CD4+ T cells from melanoma patient NW634 with ESO-CHP and mapped one of the recognized epitopes to NY-ESO-1 peptide 157-174, which was previously shown to be presented by HLA-DP4 (Fig. 4C).
Finally, we found that, after presensitization with NY-ESO-1-recombinant adenovirus, peptides 115-132, 121-138, and 139-156 were antigenic in patient NW29 (Fig. 1 and data not shown), and mapped 12-mer epitopes still recognized within these peptides to 121-132 and 143-154 based on predictive algorithms (Fig. 4D and data not shown).
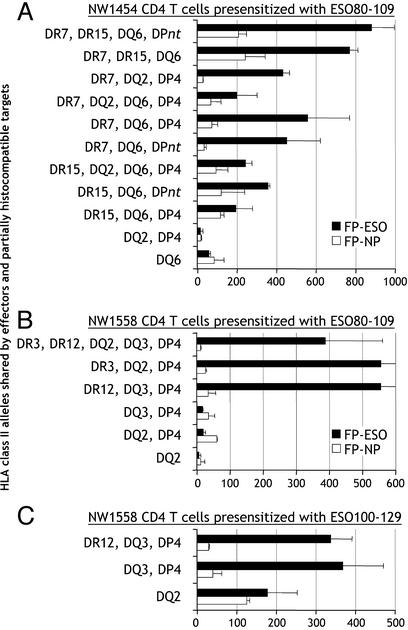
HLA Class II Restriction of NY-ESO-1 Epitopes. Recognition of NY-ESO-1 peptide 80-109 by CD4+ T cells from patient NW1454 was analyzed to determine the HLA class II allele used for presentation. Partially histocompatible EBV-B cells transduced with NY-ESO-1 recombinant fowlpox or control NP fowlpox were used to find matching HLA class II alleles able to present NY-ESO-1. We found that, in most cases, 80-109-specific CD4+ T cells recognized HLA-DRB1*07+ targets expressing NY-ESO-1, whereas other shared alleles did not significantly present the epitope (Fig. 5A). However, it could not be excluded that other alleles were responsible for the presentation of 80-109 in different patients, because NW1558 appeared to favor HLA-DRB1*03 or HLA-DRB1*12 usage for ESO80-109 presentation (Fig. 5B).
Fig. 5.
HLA restriction usage for presentation of NY-ESO-1 to CD4+ T cells specific for ESO80-109 and ESO100-129. (A) CD4+ T cells from patient NW1454 (HLA-DRB1*07,15; -DQB1*02,06; -DPB1*04,09) were presensitized with peptide ESO80-109 and tested by ELISPOT against partially histocompatible EBV-B cells infected with NY-ESO-1 or NP recombinant fowlpox vectors. HLA class II alleles shared between effectors and targets are indicated. *DPnt: targets not tested for HLA-DP compatibility with effectors. (B) Similar analysis with CD4+ T cells from patient NW1558 (HLA-DRB1*03,12; -DQB1*02,03; -DPB1*04,10) presensitized with peptide ESO80-109. (C) Similar analysis with CD4+ T cells from patient NW1558 presensitized with peptide ESO100-129. Results represent the mean number of spots per 50,000 effector CD4+ T cells in duplicate wells with error bars indicating standard deviation.
When similar approaches were used, responses to 121-132 and 143-154 were also found to be restricted by HLA-DRB1*07 in patient NW29 (not shown), and HLA-DPB1*04 appeared to be responsible for presentation of peptide 100-129, and thus likely for smaller epitope 108-119, in patient NW1558 (Fig. 5C).
Correlation of Antibody and CD4+ T Cell Responses to NY-ESO-1. To obtain a comprehensive picture of CD4+ T cell reactivity against NY-ESO-1, we analyzed a large series of cancer patients and healthy donors, selected according to their serological status for NY-ESO-1, and tested for their capacity to respond to NY-ESO-1 immunogenic polypeptide 80-109 or to full-length NY-ESO-1. After presensitization, CD4+ T cells were tested against autologous or histocompatible targets pulsed with polypeptide 80-109 or a negative control peptide, or infected with fowlpox recombinant for NY-ESO-1 or for control influenza NP. T-APC were used for peptide pulsed targets, because they usually evoked very low nonspecific background, and B-EBV were used for recombinant fowlpox infection, because responses were usually stronger than with infected T-APC, albeit with higher background too.
We found that there was a complete correlation between the presence of CD4+ T cell and antibody responses to NY-ESO-1 in the 31 patients tested (Table 1). No NY-ESO-1 seronegative patient or healthy donor had demonstrable CD4+ reactivity to NY-ESO-1. Among NY-ESO-1 antibody responders, 11 of 13 patients had CD4+ T cells reactive with polypeptide 80-109, either as an immunogen or as an antigen. Patient NW1557 could not be evaluated because of unexplained high background on autologous targets. Patient NW783 appeared negative despite a humoral response to NY-ESO-1. All patients responding to peptide 80-109 were capable of recognizing full-length NY-ESO-1 naturally processed from recombinant fowlpox (Table 1). In additional tests, patients NW29, NW903, NW1374, NW1454, and NW1558 all recognized shorter NY-ESO-1 peptides 85-102 or 87-98, contained within 80-109 (Fig. 4 and data not shown).
Table 1. Analysis of CD4+ T cell responses of cancer patients and healthy donors in response to NY-ESO-1 peptide or recombinant full-length antigen.
| Patient code | Cancer type | Antibody status | Presensitization with | No. of spots against T-APC* 80-109 (control) | No. of spots against EBV-B† FP-ESO (FP-NP) |
|---|---|---|---|---|---|
| NW29 | Melanoma | + | 80-109 | 684 (3) | 165 (8) |
| Ad-ESO | 207 (2) | 355 (11) | |||
| NW634 | Melanoma | + | 80-109 | 425 (0) | ND |
| ESO-CHP | 105 (2) | 163 (36) | |||
| NW783 | Melanoma | + | 80-109 | 0 (0) | ND |
| FP-ESO | 0 (0) | ND | |||
| NW903 | Melanoma | + | 80-109 | 21 (0) | ND |
| ESO-CHP | 314 (19) | 270 (30) | |||
| NW1289 | NSC Lung Ca. | + | 80-109 | 519 (19) | 171 (38) |
| Ad-ESO | 166 (4) | 237 (30) | |||
| NW1307 | Schwannoma | + | 80-109 | 185 (5) | ND |
| FP-ESO | 5 (1) | 111 (19) | |||
| NW1374 | NSC lung Ca. | + | 80-109 | 84 (7) | ND |
| FP-ESO | 1 (3) | ND | |||
| NW1454 | Melanoma | + | 80-109 | 334 (3) | 409 (28) |
| Ad-ESO | 30 (7) | 32 (21) | |||
| NW1557 | Esophageal Ca. | + | 80-109 | 132 (163) | 55 (51) |
| Ad-ESO | 39 (60) | 36 (26) | |||
| NW1558 | Ovarian Ca. | + | 80-109 | 457 (0) | 105 (4) |
| Ad-ESO | 370 (0) | 606 (18) | |||
| NW1567 | Ovarian Ca. | + | 80-109 | 78 (3) | 98 (64) |
| Ad-ESO | 0 (0) | 1 (5) | |||
| SS4 | Synovial sarcoma | + | 79-108 | 519 (3) | 61 (13) |
| Ad-ESO | 37 (0) | 229 (27) | |||
| LU-89 | NSC lung Ca. | + | 80-109 | 801 (12) | 54‡ (9) |
| NW208 | Melanoma | - | 80-109 | 6 (7) | 25 (15) |
| Ad-ESO | 4 (4) | 38 (48) | |||
| NW415 | Melanoma | - | 80-109 | 1 (0) | 115 (77) |
| Ad-ESO | 6 (7) | 33 (18) | |||
| NW681 | Melanoma | - | 80-109 | 7 (8) | 11 (33) |
| Ad-ESO | 9 (9) | 13 (17) | |||
| NW792 | Melanoma | - | 80-109 | 0 (0) | 20 (13) |
| Ad-ESO | 0 (0) | 26 (26) | |||
| NW886 | Melanoma | - | 80-109 | 5 (3) | 249 (269) |
| Ad-ESO | 0 (1) | 31 (31) | |||
| NW1028 | Melanoma | - | 80-109 | 2 (2) | 1 (5) |
| Ad-ESO | 2 (2) | 3 (7) | |||
| LU-82 | NSC lung Ca. | - | 80-109 | 0 (0) | ND |
| LU-86 | NSC lung Ca. | - | 80-109 | 4 (3) | ND |
| LU-279 | NSC lung Ca. | - | 80-109 | 3 (1) | ND |
| LU-281 | NSC lung Ca. | - | 80-109 | 2 (2) | ND |
| LU-346 | NSC lung Ca. | - | 80-109 | 0 (0) | ND |
| LU-395 | NSC lung Ca. | - | 80-109 | 0 (0) | ND |
| LU-400 | NSC lung Ca. | - | 80-109 | 0 (0) | ND |
| LU-449 | NSC lung Ca. | - | 80-109 | 0 (2) | ND |
| NC111 | Healthy donor | - | 80-109 | 0 (0) | ND |
| NC189 | Healthy donor | - | 80-109 | 3 (2) | ND |
| NC201 | Healthy donor | - | 80-109 | 3 (0) | ND |
| NC200 | Healthy donor | - | 80-109 | 0 (0) | ND |
CD4+ T cells were tested 20-30 days after presensitization. Boldface indicates significant difference compared to negative control (in parentheses). Results are representative of at least two separate reproducible ELISPOT assays. ND, not done; NSC lung Ca. non-small-cell lung cancer; Ca., cancer.
CD4+ T cells (50,000) presensitized with ESO80-109 or full-length NY-ESO-1 recombinant antigen were tested against T-APC pulsed with peptide 80-109 (or negative control peptide).
CD4+ T cells (10,000) presensitized with ESO80-109 or full-length NY-ESO-1 recombinant antigen were tested against EBV-B cells infected with NY-ESO-1 recombinant fowlpox FP-ESO (or control NP recombinant fowlfox FP-NP).
Tested on autologous T-APC infected with FP-ESO or FP-NP, not EBV-B.
Patient NW903 surprisingly failed to respond to 80-109 after peptide presensitization, but did so after ESO-CHP presensitization. Patients NW1454 and NW1567 failed to respond to AdESO presensitization despite specific responses after presensitization with NY-ESO-1 polypeptide 80-109. Patients NW1307 and SS4 appeared to preferentially respond to an epitope different from 80-109 after FP-ESO presensitization (we found that peptides 145-174 and 157-170 were respectively recognized in further tests not shown), even though ESO80-109 was also immunogenic on its own after peptide presensitization (Table 1).
Among responder patients, NW29, NW634, NW1307, NW1558, and SS4 were found to have polyclonal CD4 responses against at least two different NY-ESO-1 epitopes.
Discussion
Based on the remarkably high percentage of patients with advanced cancers developing NY-ESO-1 specific antibodies and CD8+ T cells in response to their own tumors (3, 5), NY-ESO-1 can be considered one of the most immunogenic human tumor antigens defined to date. The strength of NY-ESO-1 CD8+ responses is comparable to that of influenza memory effectors, as measured in vitro.
With the development of a new methodology to monitor CD4+ responses in an indirectly quantitative procedure similar to CD8+ analysis (14), we found that the comparison to influenza was still valid with regard to the extraordinary strength of NY-ESO-1 CD4+ T cell responses. Most importantly, CD4+ reactivity was tightly associated with the NY-ESO-1 antibody status of patients. Effectors analyzed in this study have been primed in vivo, and responses were not induced de novo from patients seronegative for NY-ESO-1. In contrast to other disease settings where CD4 responses are detected in the absence of antibodies or CD8+ T cells to a specific antigen (19), NY-ESO-1+ expression in tumors appears to induce an integrated immune response in vivo. These findings can be compared with high levels of antibodies, CD4+, and CD8+ T cells seen against HIV (20). In this context, successful viral and tumor protection in animal models is known to be associated with integrated immune responses (21, 22). CD4+ T cells are critical in mediating this protective immunity (23), and are essential for the development of functional CD8+ T cell memory (24). Recently, it was shown that CD4 help is required to induce CD8 responses through an indirect pathway of cross-presentation from tumor cells (25), thus favoring the hypothesis that local APCs take up the antigen to prime T cells rather than by direct presentation from tumor cells. The strong humoral response to NY-ESO-1 may greatly facilitate antigen uptake by APC with the resulting priming and cross-presentation leading to and maintaining a vigorous CD4+ and CD8+ T cell response to NY-ESO-1. This idea is supported by in vitro findings that NY-ESO-1 protein/antibody complexes are efficiently captured by dendritic cells for presentation to T cells (26).
Determining novel NY-ESO-1 epitopes proved relatively straightforward when our recently developed general strategy for CD4 monitoring in donors of any HLA haplotype was used (14). Many patients developed simultaneous responses to several epitopes, with no clear evidence of immunodominance. The central and N-terminal sequence of NY-ESO-1 appeared very rich in epitopes for CD4+ T cell recognition, as determined in this study with 12-mer immunogenic peptides: 87-98, 108-119, 121-132, and 143-154, which together with HLA-DP4 binding peptide 157-170 span this region of NY-ESO-1 in a nearly contiguous fashion (Fig. 6).
Fig. 6.
Epitope distribution along the NY-ESO-1 sequence. Above the sequence, defined HLA class I-restricted peptides; below the sequence, HLA class II-restricted peptides. Italics indicate peptides defined by other groups (10, 11, 13).
Notably, the most immunogenic peptide, NY-ESO-1 80-109, was found associated with multiple HLA class II restrictions. Its association with surface molecules other than MHC products should be explored to determine whether HLA class II promiscuity alone explains its high level of immunogenicity in antibody-positive patients.
The analysis of the class of Ig induced by NY-ESO-1 may also provide some clues about the in vivo function of CD4+ T cells. In diseases driven by type 1 immunity such as multiple sclerosis (27) or Lyme borreliosis (28), an association was found with specific Ig subclasses. From our preliminary data, a majority of patients develop Th1-related IgG1 isotype against NY-ESO-1, which appears in accordance with the presence of IFN-γ-producing CD4+ T cells specific for NY-ESO-1 derived from these patients.
The development of a general strategy to monitor CD4+ and CD8+ T cells against NY-ESO-1 in seropositive patients provides us with the methodology to now look for cellular responses to the large array of other serologically defined tumor antigens (www2.licr.org/CancerImmunomeDB/).
Acknowledgments
We thank K. Tuballes, S. J. Miranda, E. Ritter, and D. Santiago for excellent technical assistance, and we are grateful to the Cancer Research Institute for its support.
Abbreviations: APC, antigen-presenting cell; ELISPOT, enzyme-linked immunospot; NP, nucleoprotein; T-APC, target APC; CHP, hydrophobized polysaccharide pullulan; PBMC, peripheral blood mononuclear cells; EBV, Epstein–Barr virus; EBV-B, EBV-transformed B lymphocytes; AdESO, adenovirus recombinant for NY-ESO-1; FP-ESO, fowlpox virus recombinant for NY-ESO-1; FP-NP, fowlpox virus recombinant for influenza nucleoprotein.
References
- 1.Chen, Y.-T., Scanlan, M. J., Sahin, U., Türeci, Ö., Güre, A. O., Tsang, S., Williamson, B., Stockert, E., Pfreundschuh, M. & Old, L. J. (1997) Proc. Natl. Acad. Sci. USA 94, 1914–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jäger, E., Chen, Y.-T., Drijfhout, J. W., Karbach, J., Ringhoffer, M., Jäger, D., Arand, M., Wada, H., Noguchi, Y., Stockert, E., et al. (1998) J. Exp. Med. 187, 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockert, E., Jäger, E., Chen, Y.-T., Scanlan, M. J., Gout, I., Karbach, J., Arand, M., Knuth, A. & Old, L. J. (1998) J. Exp. Med. 187, 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jäger, E., Stockert, E., Zidianakis, Z., Chen, Y.-T., Karbach, J., Jäger, D., Arand, M., Ritter, G., Old, L. J. & Knuth, A. (1999) Int. J. Cancer 84, 506–510. [DOI] [PubMed] [Google Scholar]
- 5.Jäger, E., Nagata, Y., Gnjatic, S., Wada, H., Stockert, E., Karbach, J., Dunbar, P. R., Lee, S. Y., Jungbluth, A., Jäger, D., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4760–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oxenius, A., Campbell, K. A., Maliszewski, C. R., Kishimoto, T., Kikutani, H., Hengartner, H., Zinkernagel, R. M. & Bachmann, M. F. (1996) J. Exp. Med. 183, 2209–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dullforce, P., Sutton, D. C. & Heath, A. W. (1998) Nat. Med. 4, 88–91. [DOI] [PubMed] [Google Scholar]
- 8.McAdam, A. J., Greenwald, R. J., Levin, M. A., Chernova, T., Malenkovich, N., Ling, V., Freeman, G. J. & Sharpe, A. H. (2001) Nature 409, 102–105. [DOI] [PubMed] [Google Scholar]
- 9.Jäger, E., Jäger, D., Karbach, J., Chen, Y.-T., Ritter, G., Nagata, Y., Gnjatic, S., Stockert, E., Arand, M., Old, L. J. & Knuth, A. (2000) J. Exp. Med. 191, 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarour, H. M., Storkus, W. J., Brusic, V., Williams, E. & Kirkwood, J. M. (2000) Cancer Res. 60, 4946–4952. [PubMed] [Google Scholar]
- 11.Zeng, G., Touloukian, C. E., Wang, X., Restifo, N. P., Rosenberg, S. A. & Wang, R. F. (2000) J. Immunol. 165, 1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarour, H. M., Maillere, B., Brusic, V., Coval, K., Williams, E., Pouvelle-Moratille, S., Castelli, F., Land, S., Bennouna, J., Logan, T. & Kirkwood, J. M. (2002) Cancer Res. 62, 213–218. [PubMed] [Google Scholar]
- 13.Zeng, G., Wang, X., Robbins, P. F., Rosenberg, S. A. & Wang, R. F. (2001) Proc. Natl. Acad. Sci. USA 98, 3964–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atanackovic, D., Matsuo, M., Ritter, E., Mazzara, G., Ritter, G., Jäger, E., Knuth, A., Old, L. J. & Gnjatic, S. (2003) J. Immunol. Methods 278, in press. [DOI] [PubMed]
- 15.Gnjatic, S., Nagata, Y., Jäger, E., Stockert, E., Shankara, S., Roberts, B. L., Mazzara, G. P., Lee, S. Y., Dunbar, P. R., Dupont, B., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 10917–10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu, X. G., Schmitt, M., Hiasa, A., Nagata, Y., Ikeda, H., Sasaki, Y., Akiyoshi, K., Sunamoto, J., Nakamura, H., Kuribayashi, K. & Shiku, H. (1998) Cancer Res. 58, 3385–3390. [PubMed] [Google Scholar]
- 17.Ikuta, Y., Katayama, N., Wang, L., Okugawa, T., Takahashi, Y., Schmitt, M., Gu, X., Watanabe, M., Akiyoshi, K., Nakamura, H., et al. (2002) Blood 99, 3717–3724. [DOI] [PubMed] [Google Scholar]
- 18.Singh, H. & Raghava, G. P. (2001) Bioinformatics 17, 1236–1237. [DOI] [PubMed] [Google Scholar]
- 19.van der Burg, S. H., de Cock, K., Menon, A. G., Franken, K. L., Palmen, M., Redeker, A., Drijfhout, J., Kuppen, P. J., van de Velde, C., Erdile, L., et al. (2001) Eur. J. Immunol. 31, 146–155. [DOI] [PubMed] [Google Scholar]
- 20.Hogan, C. M. & Hammer, S. M. (2001) Ann. Intern. Med. 134, 761–776. [DOI] [PubMed] [Google Scholar]
- 21.Riberdy, J. M., Flynn, K. J., Stech, J., Webster, R. G., Altman, J. D. & Doherty, P. C. (1999) J. Virol. 73, 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reilly, R. T., Machiels, J. P., Emens, L. A., Ercolini, A. M., Okoye, F. I., Lei, R. Y., Weintraub, D. & Jaffee, E. M. (2001) Cancer Res. 61, 880–883. [PubMed] [Google Scholar]
- 23.Pardoll, D. M. & Topalian, S. L. (1998) Curr. Opin. Immunol. 10, 588–594. [DOI] [PubMed] [Google Scholar]
- 24.Kaech, S. M. & Ahmed, R. (2003) Science 300, 263–265. [DOI] [PubMed] [Google Scholar]
- 25.Yu, P., Spiotto, M. T., Lee, Y., Schreiber, H. & Fu, Y. X. (2003) J. Exp. Med. 197, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagata, Y., Ono, S., Matsuo, M., Gnjatic, S., Valmori, D., Ritter, G., Garrett, W., Old, L. J. & Mellman, I. (2002) Proc. Natl. Acad. Sci. USA 99, 10629–10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greve, B., Magnusson, C. G., Melms, A. & Weissert, R. (2001) J. Neuroimmunol. 121, 120–125. [DOI] [PubMed] [Google Scholar]
- 28.Widhe, M., Ekerfelt, C., Forsberg, P., Bergstrom, S. & Ernerudh, J. (1998) Scand. J. Immunol. 47, 575–581. [PubMed] [Google Scholar]