Abstract
Lactobacillus acidophilus is a probiotic organism that displays the ability to use prebiotic compounds such as fructooligosaccharides (FOS), which stimulate the growth of beneficial commensals in the gastrointestinal tract. However, little is known about the mechanisms and genes involved in FOS utilization by Lactobacillus species. Analysis of the L. acidophilus NCFM genome revealed an msm locus composed of a transcriptional regulator of the LacI family, a four-component ATP-binding cassette (ABC) transport system, a fructosidase, and a sucrose phosphorylase. Transcriptional analysis of this operon demonstrated that gene expression was induced by sucrose and FOS but not by glucose or fructose, suggesting some specificity for nonreadily fermentable sugars. Additionally, expression was repressed by glucose but not by fructose, suggesting catabolite repression via two cre-like sequences identified in the promoter–operator region. Insertional inactivation of the genes encoding the ABC transporter substrate-binding protein and the fructosidase reduced the ability of the mutants to grow on FOS. Comparative analysis of gene architecture within this cluster revealed a high degree of synteny with operons in Streptococcus mutans and Streptococcus pneumoniae. However, the association between a fructosidase and an ABC transporter is unusual and may be specific to L. acidophilus. This is a description of a previously undescribed gene locus involved in transport and catabolism of FOS compounds, which can promote competition of beneficial microorganisms in the human gastrointestinal tract.
The ability of select intestinal microbes to use substrates nondigested by the host may play an important role in their ability to successfully colonize the mammalian gastrointestinal (GI) tract. A diverse carbohydrate catabolic potential is associated with cariogenic activity of Streptococcus mutans in the oral cavity (1), adaptation of Lactobacillus plantarum to a variety of environmental niches (2), and residence of Bifidobacterium longum in the colon (3), illustrating the competitive benefits of complex sugar utilization. Prebiotics are nondigestible food ingredients that selectively stimulate the growth and/or activity of beneficial microbial strains residing in the host intestine (4). Among sugars that qualify as prebiotics, fructooligosaccharides (FOS) are a diverse family of fructose polymers used commercially in food products and nutritional supplements that vary in length and can be either derivatives of simple fructose polymers or fructose moieties attached to a sucrose molecule. The linkage and degree of polymerization can vary widely (usually between 2 and 60 moieties), and several names such as inulin, levan, oligofructose, and neosugars are used accordingly. The average daily intake of such compounds, originating mainly from wheat, onion, artichoke, banana, and asparagus (4, 5), is fairly significant, with ≈2.6 g of inulin and 2.5 g of oligofructose consumed in the average American diet (5). FOS are not digested in the upper GI tract and can be degraded by a variety of lactic acid bacteria (6–9), residing in the human lower GI tract (4, 10). FOS and other oligosaccharides have been shown in vivo to beneficially modulate the composition of the intestinal microbiota and specifically to increase bifidobacteria and lactobacilli (4, 10, 11). A variety of Lactobacillus acidophilus strains in particular have been shown to use several polysaccharides and oligosaccharides such as arabinogalactan, arabinoxylan, and FOS (6, 9). Despite the recent interest in FOS utilization, little information is available about the metabolic pathways and enzymes responsible for transport and catabolism of such complex sugars in lactobacilli.
In silico analysis of a particular locus within the L. acidophilus North Carolina Food Microbiology (NCFM) genome revealed the presence of a gene cluster encoding proteins potentially involved in prebiotic transport and hydrolysis. This specific cluster was analyzed computationally and functionally to reveal the genetic basis for FOS transport and catabolism by L. acidophilus NCFM.
Materials and Methods
Bacterial Strain and Media Used in This Study. The strain used in this study is L. acidophilus NCFM (12). Cultures were propagated at 37°C, aerobically in deMan, Rogosa, Sharpe broth (Difco). A semisynthetic medium consisted of: 1% bactopeptone (wt/vol) (Difco), 0.5% yeast extract (wt/vol) (Difco), 0.2% dipotassium phosphate (wt/vol) (Fisher), 0.5% sodium acetate (wt/vol) (Fisher), 0.2% ammonium citrate (wt/vol) (Sigma), 0.02% magnesium sulfate (wt/vol) (Fisher), 0.005% manganese sulfate (wt/vol) (Fisher), 0.1% Tween 80 (vol/vol) (Sigma), 0.003% bromocresol purple (vol/vol) (Fisher), and 1% sugar (wt/vol). The carbohydrates added were either glucose (dextrose) (Sigma), fructose (Sigma), sucrose (Sigma), or FOS. Two types of complex sugars were used as FOS: a GFn mix (manufactured by R. Hutkins, University of Nebraska), consisting of glucose monomers linked α-1,2 to two, three, or four fructosyl moieties linked β-2,1, to form kestose (GF2), nystose (GF3), and fructofuranosyl-nystose (GF4), respectively; and an Fn mix, Raftilose, derived from inulin hydrolysis (Orafti). Without carbohydrate supplementation, the semisynthetic medium was unable to sustain bacterial growth above OD600 nm ≈ 0.2.
Computational Analysis of the Putative Multiple Sugar Metabolism (msm) Operon. A 10-kbp DNA locus containing a putative msm operon was identified from the L. acidophilus NCFM genome sequence. ORF predictions were carried out by four computational programs: glimmer (13, 14), clone manager (Scientific and Educational Software, Durham, NC), the National Center for Biotechnology Information ORF finder (www.ncbi.nlm.nih.gov/gorf/gorf.html), and genomax (InforMax, Frederick, MD). glimmer was previously trained with a set of L. acidophilus genes available in public databases. The predicted ORFs were translated into putative proteins that were submitted to blastp analysis (15).
RNA Isolation and Analysis. Total RNA was isolated by using TRIzol (GIBCO/BRL), following the supplier's instructions. Cells in the exponential phase were harvested by centrifugation (2 min, 15,800 × g) and cooled on ice. Pellets were resuspended in TRIzol by vortexing and underwent five cycles of 1-min bead beating and 1 min on ice. Nucleic acids were subsequently purified by using three chloroform extractions and precipitated by using isopropanol and centrifugation for 10 min at 11,600 × g. The RNA pellet was washed with 70% ethanol and resuspended into diethyl pyrocarbonate-treated water. RNA samples were treated with DNAse I according to the supplier's instructions (Boehringer Mannheim). First-strand cDNA was synthesized by using the Invitrogen RT-PCR kit according to the supplier's instructions. cDNA products were subsequently amplified by using PCR with primers internal to genes of interest. For RNA slot blots, RNA samples were transferred to nitrocellulose membranes (Bio-Rad) using a slot-blot apparatus (Bio-Dot SF, Bio-Rad), and the RNAs were UV crosslinked to the membranes. Blots were probed with DNA fragments generated by PCR that had been purified from agarose gels (GeneClean III kit, Midwest Scientific, St. Louis). Probes were labeled with α-32P with the Amersham Pharmacia Multiprime Kit and consisted of 700- and 750-bp fragments internal to the msmE and bfrA genes, respectively. Hybridization and washes were carried out according to the supplier's instructions (Bio-Dot Microfiltration Apparatus, Bio-Rad), and radioactive signals were detected by using a Kodak Biomax film. Primers are listed in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Comparative Genomic Analysis. A gene cluster bearing a fructosidase gene was selected after computational data-mining of the L. acidophilus NCFM genome. Additionally, microbial clusters containing fructosidase EC 3.2.1.26 orthologs or bearing an ATP-binding cassette (ABC) transport system associated with an α-galactosidase EC 3.2.1.22 were selected from public databases (National Center for Biotechnology Information, The Institute for Genomic Research). The sucrose operon is a widely distributed cluster consisting of either three or four elements, namely: a regulator, a sucrose phosphotransferase (PTS) transporter, a sucrose hydrolase, and occasionally a fructokinase. Two gene cluster alignments were generated: (i) a PTS alignment representing similarities over the sucrose operon, bearing a PTS transport system associated with a sucrose hydrolase; and (ii) an ABC alignment representing similarities over the multiple sugar metabolism cluster, bearing an ABC transport system usually associated with a galactosidase. Sequence information is available in Table 3, which is published as supporting information on the PNAS web site.
Phylogenetic Trees. Nucleotide and protein sequences were aligned computationally by using the clustalw algorithm (16). The multiple alignment outputs were used for generating unrooted neighbor-joining phylogenetic trees by using mega2 (17). In addition to a phylogenetic tree derived from 16S rRNA genes, trees were generated for ABC transporters, PTS transporters, transcription regulators, fructosidases, and fructokinases.
Gene Inactivation. Gene inactivation was conducted by site-specific plasmid integration into the L. acidophilus chromosome via homologous recombination (18). Internal fragments of the msmE and bfrA genes were cloned into pORI28 by using Escherichia coli as a host (19), and the constructs were subsequently purified and transformed into L. acidophilus NCFM. The ability of the mutant strains to grow on a variety of carbohydrate substrates was investigated by using growth curves. Strains were grown on semisynthetic medium supplemented with 0.5% wt/vol carbohydrate.
Results
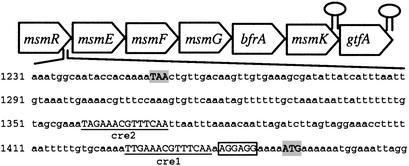
Computational Analysis of the msm Operon. Analysis of the msm locus using four ORF-calling programs revealed the presence of seven putative ORFs. Because most of the encoded proteins were homologous to those of the msm operon present in S. mutans (20), a similar gene nomenclature was used. The analysis of the predicted ORFs suggested the presence of a transcriptional regulator of the LacI repressor family, MsmR; a four-component transport system of the ABC family, MsmEFGK; and two enzymes involved in carbohydrate metabolism, namely a fructosidase EC 3.2.1.26, BfrA; and a sucrose phosphorylase EC 2.4.1.7, GtfA. A putative Shine–Dalgarno sequence 5′AGGAGG3′ was found within 10 bp upstream of the msmE start codon. A dyad symmetry analysis revealed the presence of two stem–loop structures that could act as putative Rho-independent transcriptional terminators: one between msmK and gtfA (between base pairs 6,986 and 7,014), free energy –13.6 kcal·mol–1, and one 20 bp downstream of the last gene of the putative operon (between base pairs 8,500 and 8,538), free energy –16.5 kcal·mol–1. The operon structure is shown in Fig. 1.
Fig. 1.
Operon layout. The start and stop codons are shaded, the putative ribosome binding site is boxed, and the cre-like elements are underlined. Terminators are indicated by hairpin structures.
The regulator contained two distinct domains: a DNA-binding domain at the N terminus with a predicted helix-turn-helix motif (pfam00354), and a sugar-binding domain at the C terminus (pfam00532). The transporter elements consisted of a periplasmic solute-binding protein (pfam01547), two membrane-spanning permeases (pfam00528), and a cytoplasmic nucleotide-binding protein (pfam 00005), characteristic of the different subunits of a typical ABC transport system (21). A putative anchoring motif LSLTG was present at the N terminus of the substrate-binding protein. Each permease contained five trans-membrane regions predicted computationally (22). Analyses of ABC transporters in recently sequenced microbial genomes have defined four characteristic sequence motifs (23, 24). The predicted MsmK protein included all four ABC conserved motifs, namely: Walker A: GPSGCGKST (consensus GxxGxGKST or [AG]xxxxGK[ST]); Walker B: IFLMDEPLSNLD (consensus hhhhDEPT or DExxxxxD); ABC signature sequence: LSGG; and Linton and Higgins motif: IAKLHQ (consensus hhhhH±, with h, hydrophobic and ±, charged residues). The putative fructosidase showed high similarity to glycosyl hydrolases (pfam 00251). The putative sucrose phosphorylase shared 63% residue identity with that of S. mutans.
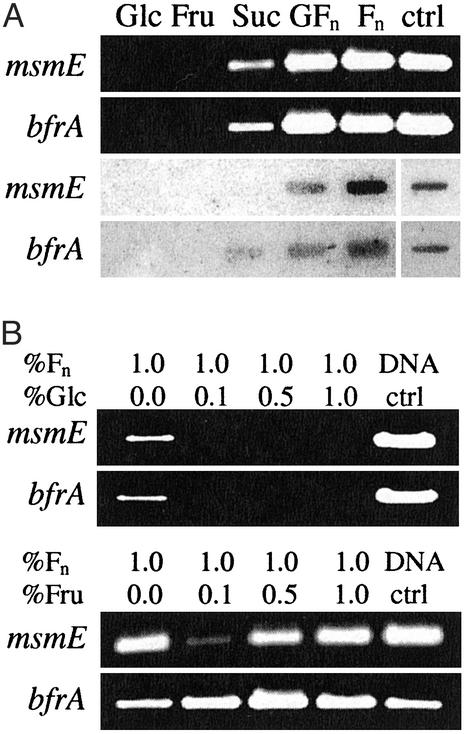
Sugar Induction and Coexpression of Contiguous Genes. Transcriptional analysis of the msm operon by using RT-PCR and RNA slot blots showed that sucrose and both types of oligofructose (GFn and Fn) were able to induce expression of msmE and bfrA (Fig. 2A). In contrast, glucose and fructose did not induce transcription of those genes, suggesting specificity for nonreadily fermentable sugars and the presence of a regulation system based on carbohydrate availability. In the presence of both FOS and readily fermentable sugars, glucose repressed expression of msmE, even if present at a lower concentration, whereas fructose did not (Fig. 2B). Analysis of the transcripts induced by oligofructose indicated that all genes within the operon are coexpressed (Fig. 6, which is published as supporting information on the PNAS web site) in a manner consistent with the S. mutans msm operon (25).
Fig. 2.
Sugar induction and repression. (A) Transcriptional induction of the msmE and bfrA genes, monitored by RT-PCR (Upper) and RNA slot blots (Lower). Cells were grown on glucose (Glc), fructose (Fru), sucrose (Suc), FOS GFn, and FOS Fn. Chromosomal DNA was used as a positive control for the probe. (B) Transcriptional repression analysis of msmE and bfrA by variable levels of Glc and Fru: 0.1% (5.5 mM), 0.5% (28 mM), and 1.0% (55 mM), in the presence of 1% Fn. Cells were grown in the presence of Fn until OD600 approximated 0.5–0.6, glucose was added, and cells were propagated for an additional 30 min.
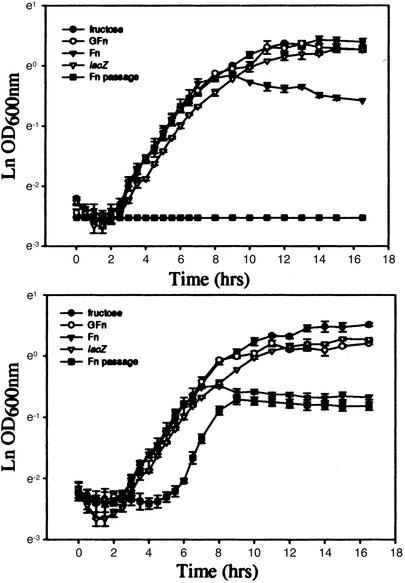
Mutant Phenotype Analysis. The ability of the bfrA (fructosidase) and msmE (ABC transporter) mutant strains to grow on a variety of carbohydrates was monitored by both optical density at 600 nm and colony-forming units. The mutants retained the ability to grow on glucose, fructose, sucrose, galactose, lactose, and FOS-GFn, in a manner similar to that of the control strain (Fig. 7, which is published as supporting information on the PNAS web site), a lacZ mutant of the L. acidophilus parental strain also generated by plasmid integration (18). This strain was chosen because it also bears a copy of the plasmid used for gene inactivation integrated in the genome. In contrast, both the bfrA and msmE mutants halted growth on FOS-Fn prematurely (Fig. 3), likely on exhaustion of simple carbohydrate from the semisynthetic medium. After one passage, the msmE mutant displayed slower growth on FOS-Fn, whereas the bfrA mutant could not grow (Fig. 3). Additionally, terminal cell counts from overnight cultures grown on FOS-Fn were significantly lower for the mutants, especially after one passage (Fig. 7).
Fig. 3.
Growth curves. The two mutants, bfrA (Upper) and msmE (Lower), were grown on semisynthetic medium supplemented with 0.5% wt/vol carbohydrate: fructose (•), GFn (○), Fn (▾), after one passage on Fn (▪). The lacZ mutant grown on Fn was used as control (▿).
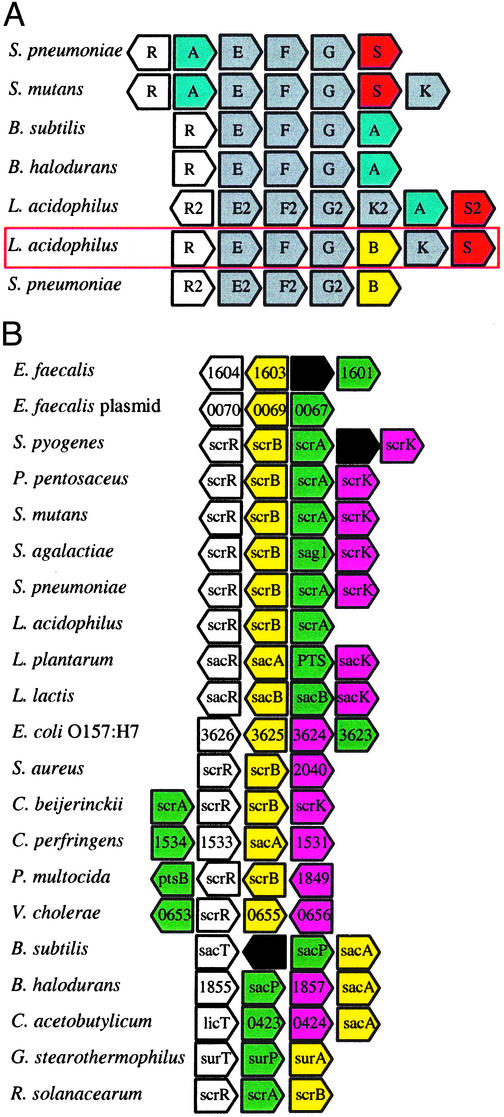
Comparative Genomic Analyses and Locus Alignments. Comparative genomic analysis of gene architecture between L. acidophilus, S. mutans, Streptococcus pneumoniae, Bacillus subtilis, and Bacillus halodurans revealed a high degree of synteny within the msm cluster, except for the core sugar hydrolase (Fig. 4A). In contrast, gene content was consistent, whereas gene order was not well conserved for the sucrose operon (Fig. 4B). The lactic acid bacteria exhibit a divergent sucrose operon, where the regulator and hydrolase are transcribed opposite the transporter and the fructokinase. In contrast, gene architecture was variable among the proteobacteria.
Fig. 4.
Operon architecture analysis. (A) Alignment of the msm locus from selected bacteria. Regulators, white; α-galactosidases, blue; ABC transporters, gray; fructosidases, yellow; sucrose phosphorylases, red. (B) Alignment of the sucrose locus from selected microbes. Regulators, white; fructosidases, yellow; PTS transporters, green; fructokinases, purple; putative proteins, black.
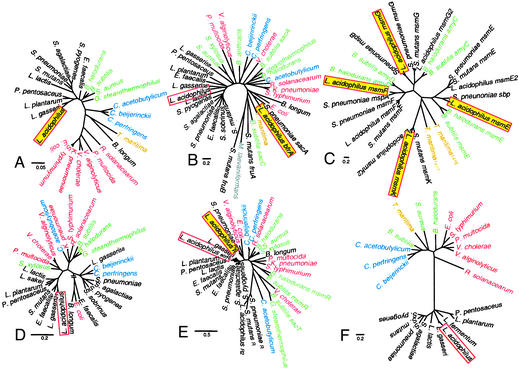
Phylogenetic Trees. Phylogenetic trees were generated to investigate whether there was a correlation between protein similarity, gene architecture, and the phylogenic relationships of the selected microorganisms. The phylogenetic relationships were obtained from 16S ribosomal DNA alignment. All proteobacteria appeared distant from the lactic acid bacteria, and the Clostridium species formed a well defined cluster between Thermotoga maritima and the bacillales (Fig. 5A).
Fig. 5.
Neighbor-joining phylogenetic trees. Lactobacillales, black; bacillales, green; clostridia, blue; thermotogae, yellow; proteobacteria, red. (A) 16S; (B) fructosidase; (C) ABC; (D) PTS; (E) regulators; (F) fructokinase. L. acidophilus proteins are boxed and shaded when encoded by the msm locus. Bars indicate scales for computed pairwise distances.
For the fructosidases, all enzymes obtained from the LAB sucrose operons clustered extremely well together at the left end of the tree, whereas there was apparent shuffling of the other three groups (Fig. 5B). The paralogs of those fructosidases in S. mutans, S. pneumoniae, and L. acidophilus clustered at the opposite end of the tree. Interestingly, the L. acidophilus fructosidase was distant from the LAB sucrose hydrolases cluster and showed strong homology to enzymes experimentally associated with oligosaccharide hydrolysis, in organisms such as T. maritima, Microbacterium laevaniformans, and B. subtilis.
Each component of the ABC transport system clustered together (Fig. 5C), namely MsmE, MsmF, MsmG, and MsmK for substrate-binding membrane-spanning proteins and nucleotide-binding unit, respectively. For MsmE, MsmF, and MsmG, three consistent subclusters were obtained: (i) the two Bacillus species; (ii) L. acidophilus, S. mutans, and S. pneumoniae from the operons bearing a galactosidase; and (iii) L. acidophilus and S. pneumoniae from the operons bearing a fructosidase.
For the PTS transporters, the clustering did not proceed according to phylogeny, especially for lactic acid bacteria, which formed two separate clusters (Fig. 5D). The two distant transporters at the bottom of the tree are non-PTS sucrose transporters of the major facilitator family of transporters, as suggested by their initial annotation.
All regulators were repressors, with the exception of those regulators of L. acidophilus, S. pneumoniae, and S. mutans clustering at the bottom of the tree (Fig. 5E), which activate transcription of operons bearing an ABC transport system associated with a galactosidase (20). In contrast, the msm regulators for both S. pneumoniae and L. acidophilus seemed to be repressors similar to that of the sucrose operon (5E). The helix-turn-helix DNA-binding motif of the regulator was very well conserved among selected regulators of the LacI family (Fig. 8A, which is published as supporting information on the PNAS web site), as shown previously (26). In contrast, the seven regulators at the bottom of the tree did not contain this conserved motif.
The fructokinase clustering was the most similar to that of the 16S phylogenetic tree, with distinct clustering of lactobacillales, bacillales, clostridia, and proteobacteria (Fig. 5F). The lack of correlation between phylogeny, gene architecture, and protein similarity may be due to extensive gene transfer among bacteria and independent sequence divergence.
Catabolite Response Element (cre) Analysis. Analysis of the promoter–operator region upstream of the msmE gene revealed the presence of two 17-bp palindromes separated by 30 nucleotides, showing high similarity to a consensus sequence for the cis-acting sites controlling catabolite repression in Gram-positive bacteria, notably B. subtilis (27–29). Several cre-like sequences highly similar to those found in B. subtilis and S. mutans (27–30) were also retrieved from the promoter–operator region of the L. acidophilus NCFM sucrose operon as well as that of the other msm locus (Table 1). Interestingly, sequences nearly identical to the cre-like elements found in the L. acidophilus msm operon were found in the promoter–operator region of the msm locus in S. pneumoniae (Table 1).
Table 1. cre sequences.
| Bacterium | Sequence* | Origin |
|---|---|---|
| B. subtilis | WTGNAANCGNWNNCW | Ref. 28, search sequence |
| B. subtilis | WWTGNAARCGNWWWCAWW | Ref. 28, new consensus |
| B. subtilis | TGWAANCGNTNWCA | Ref. 29, consensus |
| B. subtilis | TGTAAGCGCTTACA | Ref. 29, optimal operator |
| B. subtilis | TGTAAACGTTATCA | Ref. 27 |
| L. acidophilus cre1 | ATTG-AAACGTTT-CAA | Upstream of msmE |
| L. acidophilus cre2 | ATAG-AAACGTTT-CAA | Upstream of msmE |
| S. pneumoniae cre1 | AATG-AAACGTTT-CAA | Upstream of msmE2 |
| S. pneumoniae cre2 | AATG-AAACGTTT-CAA | Upstream of msmE2 |
| L. acidophilus scr | AATAAAAGCGTTTACAT | Upstream of scrB |
| L. acidophilus cre3 | TATGAAAGCGCTTAAAA | Upstream of msmE2 |
| S. mutans creW | AGATAGCGATTTGG | Ref. 30 |
| S. mutans creS | AGATAGCGCTTACA | Ref. 30 |
N, any; W, A or T; R, G or A; bold nucleotides were specifically conserved and consistent with consensus sequences.
Discussion
The L. acidophilus NCFM msm operon encodes an ABC transporter associated with a fructosidase, both of which are induced in the presence of FOS. Sucrose and both types of oligofructose induced expression of the operon, whereas glucose and fructose did not. Additionally, glucose repressed expression of the operon, suggesting the presence of a regulation mechanism of preferred carbohydrate utilization based on availability. Specific induction by FOS and sucrose and repression by glucose indicated transcriptional regulation, likely through cre present in the operator–promoter region, similar to those found in B. subtilis (28) and S. mutans (30). Catabolite repression is a mechanism widely distributed among Gram-positive bacteria, usually mediated in cis by cre sequences and in trans by repressors of the LacI family responsible for transcriptional repression of genes encoding catabolic enzymes in the presence of readily fermentable sugars (29, 31, 32).
A variety of enzymes have been associated with microbial utilization of FOS, namely: fructosidase EC 3.2.1.26 (33, 34), inulinase EC 3.2.1.7 (35–37), levanase EC 3.2.1.65 (38), fructofuranosidase EC 3.2.1.26 (39–41), fructanase EC 3.2.1.80 (7), and levan biohydrolase EC 3.2.1.64 (42, 43). Despite the semantic diversity, these enzymes are functionally related and should be considered as members of the same β-fructosidase superfamily that incorporates members of both glycosyl family 32 and 68 (44). All those enzymes share the conserved motif H-x (2)-P-x (4)-[LIVM]-N-D-P-N-G and all are involved in the hydrolysis of β-d-fructosidic linkages to release fructose. Generally, fructosidases across genera share ≈25–30% identity and 35–50% similarity (30), with several regions widely conserved across the glycosyl hydrolase 32 family (44). The two residues shown to be involved in the enzymatic activity of fructan-hydrolases, namely Asp-47 and Cys-230 (33, 45), as well as motifs highly conserved in the β-fructosidase superfamily, such as the NDPNG, FRDP, and ECP motifs (33, 44), were extremely well conserved among all fructosidase sequences (Fig. 8B).
Because the L. acidophilus fructosidase was similar to that of T. maritima and S. mutans' FruA (see Fig. 5B), two enzymes that have been associated experimentally with oligofructose hydrolysis (33, 34), we initially hypothesized that BfrA is responsible for FOS hydrolysis. Induction and gene inactivation data confirmed the correlation between the msm locus and FOS utilization. The L. acidophilus BfrA fructosidase was most similar to that of T. maritima, which has the ability to release fructose from sucrose, raffinose, levan (β2,6), and inulin (β2,1) in an exo-type manner (33). It was also very similar to other enzymes that have been characterized experimentally and associated with hydrolysis of FOS compounds by S. mutans (30) and M. laevaniformans (43). Analysis of FOS degradation by S. mutans showed that FruA is involved in hydrolysis of levan, inulin, sucrose, and raffinose (7, 20, 30, 34). Additionally, it was shown that expression of this gene was regulated by cre sequences (30, 32), and that fruA transcription was induced by levan, inulin, and sucrose, while repressed by readily metabolizable hexoses (30, 34).
In S. mutans, FruA was shown to be an extracellular enzyme that is anchored to the cell wall by a LPxTG motif (46), which catalyzes the degradation of available complex carbohydrates outside of the cell. Additionally, microbial fructosidases associated with FOS hydrolysis such as M. laevaniformans LevM (43) and Streptomyces exfoliatus levanbiohydrolase (42) have been reported as extracellular enzymes as well. In contrast, the L. acidophilus NCFM fructosidase does not contain an anchoring signal, thus is likely a cytoplasmic enzyme requiring transport of its substrate(s) through the cell membrane. No additional secreted levanase or inulinase was found in the L. acidophilus genome sequence. Because transporter genes are often coexpressed with genes involved in the metabolism of the transported compounds (47), in silico analysis of the msm operon indicates that the substrate of the fructosidase is transported by an ABC transport system. This is rather unusual, because when the fructosidase is not extracellular, the fructosidase gene is commonly associated with a sucrose PTS transporter (Fig. 4), notably in lactococci, streptococci, and bacilli (48, 49), or a sucrose permease of the major facilitator family, as in B. longum (3). Those fructosidases usually associated with PTS transporters are generally sucrose-6-phosphate hydrolases that do not have FOS as cognate substrate. Therefore, L. acidophilus NCFM may have combined the ABC transport system usually associated with an α-galactosidase with a fructosidase in the msm locus. The genetic makeup of NCFM is seemingly distinct and exclusively similar to that of S. pneumoniae. Additionally, recent evidence in Lactobacillus paracasei suggested that an ABC transport system might be involved in FOS utilization (50), which further supports the hypothesis that FOS is transported by an ABC transporter in L. acidophilus.
Lateral gene transfer (LGT) has increasingly been shown to account for a significant number of genes in bacterial genomes (51) and may account for a large proportion of the strain-specific genes found in microbes, as shown in Helicobacter pylori (52), Campilobacter jejuni (53), S. pneumoniae (54), and T. maritima (55). Notably, in T. maritima, genes involved in sugar transport and polysaccharide degradation represent a large proportion of variable genes, with ABC transporters having the highest horizontal gene transfer frequency (55). In addition, it was recently suggested that the oligosaccharide catabolic capabilities of B. longum have been expanded through horizontal transfer, as part of its adaptation to the human GI tract (3), and that the large set of sugar uptake and utilization genes in L. plantarum was acquired through LGT (2).
Intestinal microbes would benefit greatly from acquisition of gene clusters involved in transport and catabolism of undigested sugars, especially if they conferred a competitive edge toward successful colonization of the host GI tract. It is possible that L. acidophilus acquired the ability to use FOS through genetic exchange, because ABC transporters and polysaccharide degradation enzymes have a high horizontal gene transfer frequency (55). The two fructosidase paralogs seemed fairly distant from one another, sharing 28% identity and 44% similarity, suggesting those genes might have arisen from LGT rather than gene duplication. Also, because no neighboring genes or sequences are common to those two genes, a duplication event seems unlikely. Given the lack of consistency between phylogeny, gene architecture, and protein similarity, it is possible both the msm and sucrose operons underwent gene rearrangements. However, there was no evidence the msm cluster was obtained through LGT, because the GC content was very similar to that of the genome, and there was no discrepancy in the genetic code usage.
On the basis of these observations, we conclude that L. acidophilus has combined the ABC transport system derived from the raffinose operon with a β-fructosidase to form a distinct gene cluster involved in transport and catabolism of prebiotic compounds including FOS, suggesting a possible adaptation of the sugar catabolism system toward different complex sugars. The catabolic properties of this operon might differ from those of the raffinose and sucrose operons (Fig. 9, which is published as supporting information on the PNAS web site). In light of the theory that environmental factors and ecology might be dominant over phylogeny for variable genes (55), we may hypothesize that L. acidophilus has acquired FOS utilization capabilities through LGT or rearranged its genetic make-up to build a competitive edge toward colonization of the human GI tract by using prebiotic compounds, ultimately contributing to a more beneficial microbiota.
Supplementary Material
Acknowledgments
We thank Joseph Sturino for insightful discussions and for reviewing this manuscript, and Tri Duong for technical help. We also acknowledge the Streptococcus mutans Genome Sequencing Project, The Institute for Genome Research, and the Joint Genome Institute for making sequencing data available to the public before publication. This work was sponsored by Dairy Management, Incorporated; The Southeast Dairy Foods Research Center; and Rhodia, Incorporated (Madison, WI).
Abbreviations: ABC, ATP-binding cassette; cre, catabolite response element; FOS, fructooligosaccharides; MSM, multiple sugar metabolism; PTS, phosphotransferase system; LGT, lateral gene transfer; GI, gastrointestinal; NCFM, North Carolina Food Microbiology.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY172019, AY172020, and AY177419).
References
- 1.Ajdic, D., McShan, W. M., McLaughlin, R. E., Savic, G., Chang, J., Carson, M. B., Primeaux, C., Tian, R., Kenton, S., Jia, H., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleerebezem, M., Boekhorst, J., van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., Tarchini, R., Peters, S. A., Sandbrink, H. M., Fiers, M. W., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1990–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M. C., Desiere, F., Bork, P., Delley, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson, G. R. & Roberfroid, M. B. (1995) J. Nutr. 125, 1401–1412. [DOI] [PubMed] [Google Scholar]
- 5.Moshfegh, A. J., Friday, J. E., Goldman, J. P. & Ahuja, J. K. C. (1999) J. Nutr. 129, 1407s–1411s. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan, H. & Hutkins, R. W. (2000) Appl. Environ. Microbiol. 66, 2682–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartemink, R., Quataert, M. C. J., Vanlaere, K. M. J., Nout, M. J. R. & Rombouts, F. M. (1995) J. Appl. Bacteriol. 79, 551–557. [DOI] [PubMed] [Google Scholar]
- 8.Hartemink, R., VanLaere, K. M. J. & Rombouts, F. M. (1997) J. Appl. Microbiol. 83, 367–374. [DOI] [PubMed] [Google Scholar]
- 9.Van Laere, K. M., Hartemink, R., Bosveld, M., Schols, H. A. & Voragen, A. G. (2000) J. Agric. Food Chem. 48, 1644–1652. [DOI] [PubMed] [Google Scholar]
- 10.Orrhage, K., Sjostedt, S. & Nord, C. E. (2000) J. Antimicrob. Chemother. 46, 603–612. [DOI] [PubMed] [Google Scholar]
- 11.Rycroft, C. E., Jones, M. R., Gibson, G. R. & Rastall, R. A. (2001) J. Appl. Microbiol. 91, 878–887. [DOI] [PubMed] [Google Scholar]
- 12.Barefoot, S. F. & Klaenhammer, T. R. (1983) Appl. Environ. Microbiol. 45, 1808–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salzberg, S. L., Delcher, A. L., Kasif, S. & White, O. (1998) Nucleic Acids Res. 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 18.Russell, W. M. & Klaenhammer, T. R. (2001) Appl. Environ. Microbiol. 67, 4361–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law, J., Buist, G., Haandrikman, A., Kok, J., Venema, G. & Leenhouts, K. (1995) J. Bacteriol. 177, 7011–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell, R. R. B., Aduseopoku, J., Sutcliffe, I. C., Tao, L. & Ferretti, J. J. (1992) J. Biol. Chem. 267, 4631–4637. [PubMed] [Google Scholar]
- 21.Quentin, Y., Fichant, G. & Denizot, F. (1999) J. Mol. Biol. 287, 467–484. [DOI] [PubMed] [Google Scholar]
- 22.Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001) J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 23.Braibant, M., Gilot, P. & Content, J. (2000) FEMS Microbiol. Rev. 24, 449–467. [DOI] [PubMed] [Google Scholar]
- 24.Linton, K. J. & Higgins, C. F. (1998) Mol. Microbiol. 28, 5–13. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin, R. E. & Ferretti, J. J. (1996) FEMS Microbiol. Lett. 140, 261–264. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, C. C. & Saier, M. H., Jr. (1995) FEBS Lett. 377, 98–102. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto, H., Serizawa, M., Thompson, J. & Sekiguchi, J. (2001) J. Bacteriol. 183, 5110–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miwa, Y., Nakata, A., Ogiwara, A., Yamamoto, M. & Fujita, Y. (2000) Nucleic Acids Res. 28, 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weickert, M. J. & Chambliss, G. H. (1990) Proc. Natl. Acad. Sci. USA 87, 6238–6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burne, R. A., Wen, Z. T., Chen, Y. Y. M. & Penders, J. E. C. (1999) J. Bacteriol. 181, 2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hueck, C. J., Hillen, W. & Saier, M. H., Jr. (1994) Res. Microbiol. 145, 503–518. [DOI] [PubMed] [Google Scholar]
- 32.Wen, Z. T. & Burne, R. A. (2002) J. Bacteriol. 184, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liebl, W., Brem, D. & Gotschlich, A. (1998) Appl. Microbiol. Biotechnol. 50, 55–64. [DOI] [PubMed] [Google Scholar]
- 34.Burne, R. A., Schilling, K., Bowen, W. H. & Yasbin, R. E. (1987) J. Bacteriol. 169, 4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onodera, S. & Shiomi, N. (1988) Agric. Biol. Chem. 52, 2569–2576. [Google Scholar]
- 36.Xiao, R., Tanida, M. & Takao, S. (1989) J. Ferment. Bioeng. 67, 331–334. [Google Scholar]
- 37.Mckellar, R. C. & Modler, H. W. (1989) Appl. Microbiol. Biotechnol. 31, 537–541. [Google Scholar]
- 38.Menendez, C., Hernandez, L., Selman, G., Mendoza, M. F., Hevia, P., Sotolongo, M. & Arrieta, J. G. (2002) Curr. Microbiol. 45, 5–12. [DOI] [PubMed] [Google Scholar]
- 39.Oda, Y. & Ito, M. (2000) Curr. Microbiol. 41, 392–395. [DOI] [PubMed] [Google Scholar]
- 40.Perrin, S., Grill, J. P. & Schneider, F. (2000) J. Appl. Microbiol. 88, 968–974. [DOI] [PubMed] [Google Scholar]
- 41.Muramatsu, K., Onodera, S., Kikuchi, M. & Shiomi, N. (1992) Biosci. Biotech. Biochem. 56, 1451–1454. [Google Scholar]
- 42.Saito, K., Kondo, K., Kojima, I., Yokota, A. & Tomita, F. (2000) Appl. Environ. Microbiol. 66, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song, E. K., Kim, H., Sung, H. K. & Cha, J. (2002) Gene 291, 45–55. [DOI] [PubMed] [Google Scholar]
- 44.Naumoff, D. G. (2001) Proteins 42, 66–76. [DOI] [PubMed] [Google Scholar]
- 45.Reddy, V. A. & Maley, F. (1990) J. Biol. Chem. 265, 10817–10820. [PubMed] [Google Scholar]
- 46.Burne, R. A. & Penders, J. E. (1992) Infect. Immun. 60, 4621–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert, A., Osteras, M., Mandon, K., Poggi, M. C. & Le Rudulier, D. (2001) J. Bacteriol. 183, 4709–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiratsuka, K., Wang, B., Sato, Y. & Kuramitsu, H. (1998) Infect. Immun. 66, 3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luesink, E. J., Marugg, J. D., Kuipers, O. P. & de Vos, W. M. (1999) J. Bacteriol. 181, 1924–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan, H., and Hutkins, R. W. (2003) Appl. Environ. Microbiol. 69, 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koonin, E. V., Makarova, K. S. & Aravind, L. (2001) Annu. Rev. Microbiol. 55, 709–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salama, N., Guillemin, K., McDaniel, T. K., Sherlock, G., Tompkins, L. & Falkow, S. (2000) Proc. Natl. Acad. Sci. USA 97, 14668–14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dorrell, N., Mangan, J. A., Laing, K. G., Hinds, J., Linton, D., Al-Ghusein, H., Barrell, B. G., Parkhill, J., Stoker, N. G., Karlyshev, A. V., et al. (2001) Genome Res. 11, 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hakenbeck, R., Balmelle, N., Weber, B., Gardes, C., Keck, W. & de Saizieu, A. (2001) Infect. Immun. 69, 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nesbo, C. L., Nelson, K. E. & Doolittle, W. F. (2002) J. Bacteriol. 184, 4475–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.