Abstract
DNA sequence amplification is one of the most frequent manifestations of genomic instability in human tumors. We have shown previously that amplification of the dihydrofolate reductase (DHFR) gene in Chinese hamster cells is initiated by chromosome breaks, followed by bridge-breakage-fusion cycles that generate large intrachromosomal repeats; these are ultimately trimmed by an unknown process to smaller, more homogenous units manifested as homogenously staining chromosome regions (HSRs). However, in most human tumor cells, amplified DNA sequences are borne on unstable, extrachromosomal double minutes (DMs), which suggests the operation of a different amplification mechanism. In this study, we have isolated a large number of independent methotrexate-resistant human cell lines, all of which contained DHFR-bearing DMs. Surprisingly, all but one of these also had suffered partial or complete loss of one of the parental DHFR-bearing chromosomes. Cells in a few populations displayed what could be transient intermediates in the amplification process, including an initial HSR, its subsequent breakage, the appearance of DHFR-containing fragments, and, finally, DMs. Our studies suggest that HSRs and DMs both are initiated by chromosome breaks, but that cell types differ in how the extra sequences ultimately are processed and/or maintained.
Tumor cells arise from normal tissue by the accumulation of mutations in several different critical genes, each mutation imparting additional tumorigenic potential (reviewed in refs. 1–3). These genetic alterations include point mutations, deletions, inversions, translocations, and DNA sequence amplification and result in the activation or inactivation of protooncogenes or tumor suppressors, respectively. Amplification is extremely common in advanced tumors: the majority of mitotic chromosome spreads display either expanded, homogenously staining chromosome regions (HSRs) or small, acentric, autonomously replicating double minutes (DMs; reviewed in refs. 4–8). In many cases, these structures have been shown to contain cellular oncogenes (reviewed in refs. 9–11).
Amplification has been studied in in vitro model systems using a variety of competitive enzyme inhibitors to select drug-resistant amplificants (5–7, 12, 13). By far the most frequent mechanism for developing resistance in cultured cells is amplification of the relevant gene (14, 15). Luria–Delbruck fluctuation analyses suggest that amplification of drug-resistance markers occurs spontaneously at a frequency of ≈10−5-10−4 per cell generation (16, 17). Methotrexate-resistant murine cell lines usually maintain dihydrofolate reductase (DHFR) amplicons in DMs, whereas Chinese and Syrian hamster cells virtually always maintain DHFR (and all other) amplicons in HSRs (reviewed in refs. 5, 6, and 18). In human (19, 20) and rat (e.g., ref. 21) cell lines, DHFR amplicons can be manifested either as HSRs or DMs, although in human tumor samples, they most often appear as DMs (22).
Several mechanisms for initiating DNA sequence amplification in mammalian cells have been proposed, which fall either into the over-replication or nondisjunction camps (reviewed in refs. 6 and 23–26). To gain insight into possible mechanisms, we used fluorescence in situ hybridization (FISH) to analyze the chromosomal rearrangements that accompany the earliest detectable events in the amplification of the DHFR gene to form HSRs during the generation of methotrexate resistance in Chinese hamster ovary (CHO) cells (27, 28). These analyses provided compelling evidence that amplification is almost always initiated by a chromosomal break distal to the DHFR gene, followed by sister chromatid fusion. Thus, a dicentric chromosome containing a very large inverted duplication is formed, which leads to repeated bridge-breakage-fusion cycles. This general conclusion also was reached in studies on the CAD and AMPD2 loci in Syrian hamster and CHO cells, respectively (29–31). There is also some evidence that fragile sites are favored for the initiating breaks (32).
Amplification only occurs in tumor cells (15, 16, 33), probably because the majority of such cells are defective in the p53-mediated damage-sensing pathway (34–37). Because most human tumors bear amplified material on DMs (22), it was therefore possible that DM formation also might be initiated by chromosome breaks. In the present study, we have used FISH to analyze karyotypic rearrangements that accompany the earliest detectable stages of amplification of the DHFR gene in the human HeLa cell line. Our studies reveal a striking correlation between the appearance of DMs and the loss or fragmentation of one of the parental DHFR-bearing chromosomes, suggesting that DMs also arise from initiating chromosome breaks.
Materials and Methods
Cell Lines and Development of Independent Methotrexate-Resistant Variants.
The human tumor cell line, HeLa, was obtained from the American Type Culture Collection and was maintained in MEM supplemented with nonessential amino acids and 10% fetal clone I (HyClone) in an atmosphere of 5% CO2. The average population doubling time was ≈16 h. Methotrexate-resistant cell lines were developed by the strategy described by Tlsty et al. (16) and outlined in Fig. 1. Starting drug-sensitive cell lines either arose from a single cell (populations A6, C8, F5; Fig. 1) or were grown from 102 A6 cells to ≈107 (populations A6.1–A6.16). Methotrexate-resistant variants were selected from these cell lines by plating at various cell densities, with selection 8–16 h later on complete medium containing different methotrexate concentrations (see legend to Fig. 1).
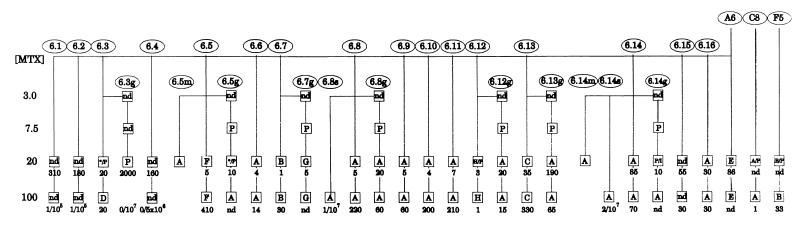
Figure 1.
Pedigree of drug-resistant cell lines selected at various methotrexate concentrations. Ellipses: drug-sensitive starting populations, and resistant derivatives designated A6.X, A6.Xg, A6.Xs, or A6.Xm (see text). Boxes: individual populations (number of clones per 106 cells plated surviving selection at that drug level is shown below). The letters A-H correspond to patterns with the same letters in Fig. 3. P designates the parental, nonrearranged karyotype; I designates an additional 5q isochromosome without DMs; * indicates a heterogenous mixture of patterns; nd indicates not determined.
Recombinant Cosmids and Chromosome-Specific Paints.
Cosmids c5–4, c5–92, c5–8, and c5–9 were isolated from a chromosome-5-specific cosmid library (Integrated Genetics) and map to 5p15, 5q12–13, 5q21, and 5q35, respectively (38). DHFR-specific clones (191C10.3, 83C2.7.2, 112A3.1, and 325J23) were identified by screening a second chromosome 5-specific cosmid library (39) or a bacterial artificial chromosome (BAC) library (Research Genetics, Huntsville, AL; ref. 40), using PCR and DHFR-specific primers (41). Labeling of cosmid or BAC DNA probes and the preparation of chromosome paints were as described (42).
FISH and Analysis.
The procedures for slide preparation and two-color FISH are exactly as described in ref. 42. At least 10 cells, and usually more than 20 cells, from each hybridization were analyzed by fluorescence microscopy (28, 42). The coordinates of chromosome termini, centromeres, and probes were determined from magnified photographic images projected onto a digitizing board (Summagraphics, Fairfield, CT) and entered into a computer for analysis (28).
Results
Strategy for Selecting Independent Drug-Resistant Variants.
The LD50 for methotrexate in three different drug-sensitive HeLa cell clones (A6, C8, and F5) was ≈3 nM, and 20 nM methotrexate (≈7× LD50) was the lowest drug level that selected for amplification of the DHFR gene (data not shown). To examine as many independent amplification events as possible and to determine the frequency of amplification in HeLa cells, we adopted the selection and analytical approach of Tlsty et al. (16).
We first determined that the baseline frequency of variants resistant to 20 nM methotrexate was ≈10−4 in each population (data not shown). One hundred A6 cells then were seeded into 16 individual dishes (designated A6.X, where X = 1–16); thus, none of these plates was likely to contain even a single resistant cell at the time of seeding. After ≈20 population doublings, ≈106 cells from each plate were replated, and 8–16 h later were subjected to 20 nM methotrexate. Resistant colonies (designated A6.X/20) ranged from 1 to 310 per plate, with a mean ± SD of 57 ± 90 and a variance/mean of 135 (Fig. 1). Such a large variance/mean value argues strongly that each resistant population was likely to have arisen spontaneously sometime during growth in drug-free medium and therefore must represent an independent mutational event (16). Luria–Delbruck fluctuation analysis indicated that the spontaneous rate of mutation to resistance at this drug level is ≈1.7 × 10-5 (calculations not shown). Additionally, 106 cells from the original clonal populations (A6, C8, and F5) were seeded into one plate each and were selected on 20 nM methotrexate for the equivalent of 20 cell generations. Each of the resulting 19 resistant populations (16 of the A6.X series plus the A6, C8, and F5 populations) then was subjected to a second selection step at 100 nM methotrexate.
Seven naive A6 populations also were selected at 3 nM, increasing to 7.5, 20, and 100 nM methotrexate, again starting with 100 cells per plate (this yielded the A6.Xg populations in Fig. 1). However, population-wide cell death was observed only at 20 nM methotrexate. Very few cells survived a single selection step directly to 100 nM (A6.8s and A6.14s in Fig. 1).
Chromosomal Locations of the DHFR Genes in Drug-Sensitive HeLa Cells.
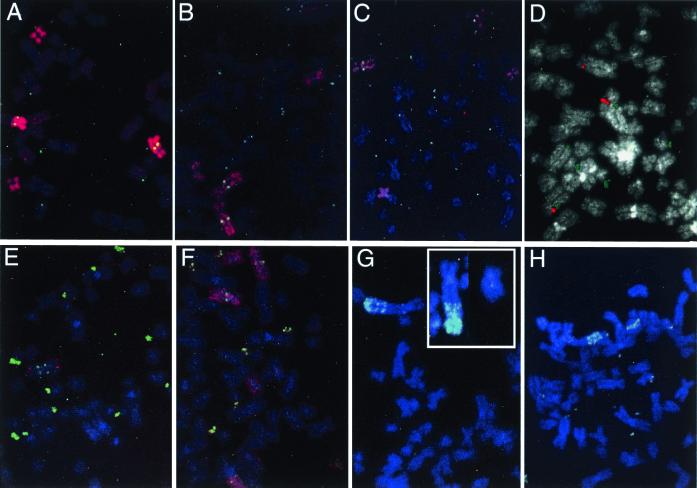
The HeLa cells used in this study were aneuploid, with a modal chromosome number of 69 and a range of 61 to 71 (n = 25). In all 19 of the starting drug-sensitive cell lines, the DHFR-specific cosmid (green) detected a single DHFR gene at 5q13 in each of the two normal homologues of chromosome 5 (Fig. 2A), as well as a third copy at band 5q13 on a long arm of 5 that has been translocated to the short arm of chromosome 3 [illuminated by red chromosome 3 and 5 paints in Fig. 2 A and B, respectively; designated der(3p5q)]. The chromosome 5 paint also labels two isochromosomes composed of the short arms of chromosome 5 [referred to as i(5p); Fig. 2A]. In addition to the der(3p5q), the chromosome 3 paint detects material on a karyotypically normal 3 homologue, two translocation products of chromosome 3 and unidentified chromosomes, and two acrocentric derivatives of chromosome 3 (Fig. 2B). Fluorescent images also are shown for the green DHFR probe in combination with red cosmids c5–92 (centromere-proximal; Fig. 2C), c5–8 (centromere-distal; Fig. 2D), or c5–9 (near the q-telomere; Fig. 2E), or with all three cosmids (Fig. 2F). Averaged and normalized distances among the relevant probes were determined as described (28) from more than 70 mitotic chromosome spreads of a representative drug-sensitive cell line (A6.8; Fig. 2G).
Figure 2.
Hybridization patterns of relevant probes to drug-sensitive HeLa cell lines. (A) The chromosome 5 paint (red) labels two normal 5s and the der(3p5q) translocation derivative, each of which carries a DHFR gene (detected with a DHFR-specific cosmid in green), as well as two i(5p) chromosomes. (B) The chromosome 3 paint detects a normal 3, the der(3p5q) translocation (which is DHFR-positive), two other translocation products, and two acrocentric derivatives. The two normal 5 homologues also are illuminated by the DHFR-specific cosmid. (C–E) FISH patterns of a mixture of the DHFR cosmid (green) and one of three other 5q marker cosmids (C5–92, C5–8, and C5–9, respectively; red). (F) A mixture of all four cosmids, with DHFR in green. (G) Consensus chromosome arrangements deduced as described in ref. 28.
Chromosome 5 Loss or Fragmentation Accompanies the Appearance of DHFR-Containing DMs in the Majority of Methotrexate-Resistant Variants.
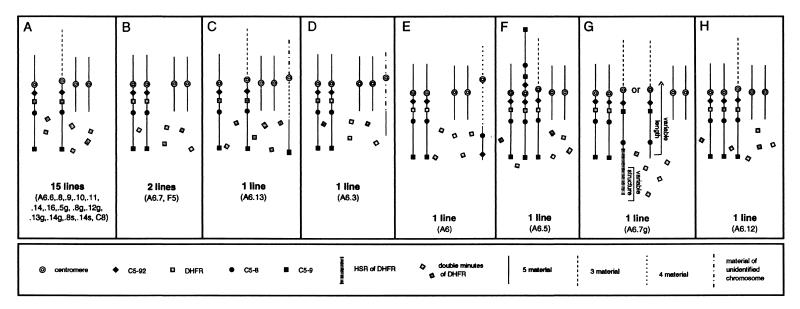
Only parental karyotypes were observed in populations grown in less than 20 nM methotrexate (Fig. 1, squares containing a P). With only a few exceptions (noted below), all of the populations selected on 20 nM or in a single step at 100 nM showed evidence of gene amplification. The karyotypes of cells resistant to 20 nM and 100 nM methotrexate within a cell line were not markedly different. A synopsis of the FISH analyses of 23 populations selected on 100 nM is diagrammed in Fig. 3 (the A6.1, A6.2, A6.4, and A6.15 populations were not further analyzed). These patterns fall into eight discernible groups, but 22 of 23 populations share two characteristic aberrations.
Figure 3.
The eight general karyotypic classes detected in 23 cell populations resistant to 100 nM methotrexate. Each panel corresponds to one of the eight karyotypes that were detected with a 5 paint/DHFR cosmid FISH combination. The number of DMs varied among cells and among populations from several to >100.
All 23 drug-resistant populations selected at 100 nM methotrexate and all but one of their predecessors selected at 20 nM contained DHFR-specific DMs (Figs. 1 and 3). The only exception at 20 nM was A6.14g/20, which carried five copies of DHFR [the three original ones and an additional two on a novel i(5q); not shown]. Relative to the 100 nM populations, most of the 20 nM populations displayed a lower average number of DMs, as well as a higher percentage of cells with no detectable DMs: in eight of the 20 nM populations, up to 50% of cells displayed the parental karyotype, indicating that they may not have acquired a stable genetic change to drug resistance and therefore were destined to die.
The most frequent and unexpected karyotypic alteration in addition to the appearance of DMs was the complete loss or fragmentation of one of the three DHFR-bearing chromosomes: 15 of the A6.X/100 populations lost one of the two normal chromosome 5 homologues and two populations lost the der(3p5q) (Fig. 3). For example, in a mitotic spread of the A6.8/100 population (Fig. 4A), single-copy DHFR signals (yellow dots) are visible on a normal 5 and on der(3p5q), the two i(5p)s are detected with the chromosome 5 paint, but the second normal 5 is absent. In the A6.7/100 cell line (Fig. 4B), both normal 5s and the two i(5p)s are detected with the 5 paint, but the der(3p5q) chromosome could not be detected in any of the spreads. In both cell lines, several DMs are highlighted with the DHFR-specific cosmid (green dots) and appear to contain only one or a few copies of the gene. However, these DMs are also visible with the 4′,6-diamidino-2-phenylindole stain (not shown), indicating that they contain DNA far in excess of the gene itself.
Figure 4.
FISH patterns demonstrating seven of the eight types of chromosome rearrangement detected. (A) The 5 paint/DHFR combination on A6.8/100, showing the loss of a normal 5 and the presence of several DMs. (B) The 5 paint/DHFR combination on A6.7/100, showing the loss of the der(3p5q) and the presence of numerous DMs. (C) The 5 paint/DHFR combination on A6.3/100, showing retention of two normal 5s, DMs, and a small fragment of der(3p5q) translocated to an unidentified chromosome (lower left). (D) The C5–92 cosmid (red) and DHFR (green) probe combinations on A6/100, showing the presence of two normal 5s, numerous DMs, and a third chromosome hybridizing to the C5–92 probe only. (E) A C5–9/DHFR combination on A6.5/100, showing the presence of only one normal 5, the der(3p5q) at the bottom, an i(5q), and numerous DMs. (F) A 5 paint/DHFR combination on A6.12/20 (the pattern is similar to A6.12/100), showing two normal 5s, a der(3p5q) on the right side, two i(5p)s, and numerous DMs. (G) The C5–92/DHFR combination on A6.7g, illustrating two normal chromosomes 5 and the HSR on the der(3p;inv5q); C5–92 is retained at its original position on all three chromosomes. (Inset) The C5–9/DHFR combination, showing the der(3p;inv5q) and a normal 5 from another mitotic cell, which best illustrates the inversion within the 5q arm. (H) An example from a second group of A6.7g cells that carries copies of the DHFR gene as DMs, in an HSR near the terminus of the der(3p;inv5q), and in large fragments of the HSR. The probe combination is C5–9/DHFR. Two normal 5s are present in each cell.
Five additional patterns indicative of chromosome 5 fragmentation events were displayed. In the A6.13/100 and A6.3/100 cell populations (Fig. 3 C and D), one of the parental DHFR-containing chromosomes is missing, but a fragment of chromosome 5 has been translocated to another unidentified chromosome. This pattern is indicated for the A6.3/100 variant in Fig. 4C: the 5-paint detects the two normal 5s, the two i(5p)s, and a small DHFR-negative chromosome 5 fragment translocated to a small chromosome (lower left), but fails to detect the characteristic der(3p5q) (compare with Fig. 2A). In both A6.13/100 and A6.3/100, the translocated fragments appear to have arisen from a region of chromosome 5 devoid of sequences represented by the DHFR, C5–92, and C5–8 cosmids (see Figs. 3 C and D and legends; additional FISH data not shown). In the A6/100 variant (Figs. 3E and 4D), the der(3p5q) is missing and a fragment of 5q has been translocated to the long arm of chromosome 4. In this case, the fragment lacks DHFR but retains the C5–92 and C5–8 markers that flank DHFR in the parental arrangement (Fig. 2G; additional FISH data not shown). Therefore, this fragment must have arisen by four separate breaks, two between the C5–92 and C5–8 positions, as well as two outside of this region. The A6.5/100 drug-resistant variant also displays a fragmented chromosome 5 (Figs. 3F and 4E), but in this case, the short arm of a normal 5 has been lost and an isochromosome of the long arm of chromosome 5 [i(5q)] appears. This i(5q) rearrangement is evident with the DHFR/C5–9 combination (Fig. 4E), as well as with combinations of DHFR and the other two cosmids or the 5-paint (not shown).
The A6.7g population (Figs. 3G and 4 G and H) displayed the widest range of chromosome rearrangements, falling into four general groups that could represent intermediates in the generation of DMs in this resistant population. In one group, there was no evidence of DMs, but a DHFR-positive HSR of variable length was detected at or near the terminus of the parental der(3p5q) chromosome (Fig. 4G and Inset; chromosomes identified with 3 and 5 paints; not shown). In each of these cells, the 5q arm of this chromosome also had suffered a paracentric inversion that juxtaposed the C5–9 marker to C5–92 and placed the DHFR gene at the chromosome terminus [hereafter termed the der(3p;inv5q) chromosome] (the C5–92 probe is only faintly visible in the insets of a normal and an HSR-containing der(3p;inv5q) in Fig. 4G, but was readily observed at the microscope in these and many other spreads). In a second group of cells, the HSR-bearing der(3p;inv5q) chromosome was present (although truncated in some cell lines) and was accompanied by variable numbers of DMs (Fig. 4H). A third group displayed DMs and truncated remnants of der(3p;inv5q) in various stages of fragmentation, with no remnants of the DHFR-containing HSR remaining on its terminus (not shown). The fourth group contained multiple DMs, but the der(3p;inv5q) chromosome was not detectable, even with the 3 paint (not shown).
The percentages of cells in each of the four prevalent groups among the A6.7g populations selected in increasing methotrexate levels showed an interesting trend. In cells selected in 20 nM MTX, the HSR-only karyotype prevailed (77%), whereas in cells propagated for 14 more generations at 100 nM, the DM-only karyotype prevailed (70%). Three additional observations are noteworthy. First, 31% of the A6.7g/100 population simultaneously contained DMs and either the HSR (13%) or fragmentation products of the HSR-bearing chromosome (18%). Secondly, the der(3p;inv5q) chromosome (both with and without the HSR) often was observed to be involved in bridge-breakage-fusion cycles, either with its sister chromatids joined at the terminus of the 5q arm or as part of a dicentric chromosome. Third, DHFR-specific fragments were observed in 10–15% of mitotic spreads, in many cases in the process of breaking away from the HSR (not shown). Collectively, these observations suggest that the HSR is giving rise to DMs in this cell line.
The A6.12-resistant population (Fig. 3H) was unique among the 23 cell lines examined. Multiple large DMs containing several closely spaced DHFR genes could be detected with the DHFR cosmid probe, but all three parental DHFR-bearing markers appeared to be intact (detectable in Fig. 4F with the 5 paint/DHFR combination) and retain all 5q markers (not shown).
Chromosome 5 Loss or Fragmentation and Appearance of DMs Occur Very Early in the Development of Drug Resistance.
Because the selection regimen that gave rise to the drug-resistant cell lines summarized in Fig. 2 was designed to ensure the generation of many independent amplificants, the nature of the selection regimen precluded analysis of resistant variants until 20–25 cell generations after application of methotrexate. To capture more telling intermediates, we used a method developed by Stark and coworkers (29) to isolate resistant variants as soon as possible after the acquisition of drug resistance. Thirty culture dishes were individually seeded with 2 × 106 cells from either the A6.5 or the A6.14 drug-sensitive clonal populations, and 20 nm methotrexate (7× the LD50) was added 12 h later. Samples enriched for mitotic (presumably drug-resistant) cells were isolated by shake-off from each set of 30 plates after 2.0, 5.2, 7.1, 11.0, 12.3, 14.3, and 15.6 cell generations (3, 8, 11, 17, 19, 22, and 24 days, respectively). Results for the two populations were similar and are combined in Table 1 (note that too few cells were recovered from the 2.0 generation sample to be analyzed).
Table 1.
Monitoring earliest events
| Generation post-MTX | Number of cells | % no change | % loss of 5 or der(3p5q)
|
|
|---|---|---|---|---|
| w/o DMs | with DMs | |||
| 5.2 | 84 | 80 | 20 | 0 |
| 7.1 | 34 | 24 | 58 | 18 |
| 11.0 | 47 | 13 | 2 | 85 |
| 12.3 | 52 | 4 | 4 | 92 |
| 14.3 | 105 | 0 | 1 | 99 |
| 15.6 | 101 | 0 | 0 | 100 |
MTX, methotrexate.
As the percentage of cells with a parental karyotype decreased with time, the percentage of cells that had lost a DHFR-bearing chromosome increased (assessed with the 5 paint and DHFR-specific cosmid, 191C10.3). Moreover, of the cells that had lost one of the three DHFR-bearing chromosomes, those with detectable DMs appeared much later than those with no DMs, a result predicted by a model in which DMs arise from a fragmented chromosome 5. Furthermore, of the DM-negative spreads detected in the first three time points, ≈72% contained DHFR-bearing chromosome 5 fragments, whereas the percentage of DM-positive cells containing DHFR-bearing fragments decreased from 50% to 0% between generations 7.1 and 12.3 (data not shown).
Discussion
FISH analysis of the resistant populations isolated at 20 and 100 nM in this study showed that all had amplified the DHFR gene via DMs (Fig. 4). Importantly, all but one of these had lost all or part of the parental DHFR-bearing chromosome, strongly suggesting that the two events are mechanistically linked. In the mitotic shake-off experiment designed to capture the earliest intermediates in the amplification process, populations that were likely to have arisen from at least two independent initiating events both displayed DMs, and each of these also had lost one of the chromosome 5 homologues. In 155 mitotic spreads examined in the starting drug-sensitive A6.12 and A6.8 cell populations, only 2.6% showed signs of breakage and/or translocation events involving these chromosomes, and none showed chromosome loss (B.J.T. and C.L.F., unpublished data).
These findings suggest that DMs arise from chromosome 5 or der(3p5q) fragmentation. One possible mode of amplification was exhibited in the drug-resistant A6.7g populations. At the 20 nM methotrexate resistance level, ≈80% of cells exhibited a DHFR-specific HSR on chromosome der(3p5q) and only 6% exhibited DMs (some of them in combination with an HSR). Ultimately, however, propagation at 100 nM methotrexate yielded populations that almost all contained DMs and very few contained the HSR. One other independent example of a DHFR-specific HSR was detected at the 2-nm resistance level, which gave way to DMs when the same cells were propagated on 100 nM methotrexate.
A possible scenario for the development of resistance in the A6.7g cell line is as follows. An initial chromosome break [as evidenced by the unbalanced der(3p5q) inversion] leaves the chromosome with a free end lacking a telomere. Bridge-breakage-fusion cycles lead to a terminal HSR with variably sized amplicons. To this point, the process is formally analogous to the generation of HSRs in Chinese hamster cells (27, 28). The HSR then is proposed to be unstable, giving rise to HSR fragments, which, in turn, give rise to DMs. In most cases, all traces of the parental der(3p5q) chromosome are lost. Space limitations preclude discussion of other equally plausible mechanisms.
Interestingly, recent studies on hamster cell lines have suggested that selective agents possessing clastogenic properties may induce bridge-breakage-fusion cycles that result in intrachromosomal amplification (32, 44), whereas extrachromosomal amplification may arise from a non-bridge-breakage-fusion mechanism (32, 45). Our results with the potentially clastogenic agent, methotrexate, suggest that the two processes are not mutually exclusive.
The results of our studies are compatible with an older series of experiments on HeLa cells in which methotrexate-resistant variants were shown to harbor the DHFR amplicons on extrachromosomal elements (ref. 46; note that this study did not specifically address the fate of the parental DHFR-bearing chromosomes in these variants). One of these cell lines, which contained a small number of visible DMs but a high copy number of genes, was shown to harbor most of these on submicroscopic, circular episomes (19). Episomes also have been detected during the generation of resistance to N-phosphonacetyl-l-aspartate in Chinese hamster cells that contained an integrated, tandem array of CAD genes (47). Additionally, in a study of DHFR gene amplification in a CHO hemizygote, the gene appeared to be amplified initially via 250- to 300-kb episomes, but the parental locus remained intact (26). Based on these observations, it was argued that episomes might be products of over-replication that mature into microscopically visible DMs, either by internal expansion or by coalescing with one another. In the case of CHO cells, it was further argued that episomes and/or DMs eventually integrate into chromosomes to form HSRs (26).
This proposal is difficult to reconcile with the data presented here. First of all, one parental DHFR-bearing chromosome was lost in virtually all of the resistant populations (Fig. 3A). In addition, in at least one population (A6.7g), the process appears to have proceeded in the opposite direction (i.e., an HSR giving rise to DMs). Some of the other drug-resistant populations developed in the present study also may have arisen via a very transient HSR that we were not able to detect.
Why do human and murine cells usually maintain amplicons in DMs, whereas CHO cells with rare exceptions (26, 47, 48) display HSRs? We have observed in both CHO and human cells that chromosomes carrying HSRs often are involved in bridge-breakage-fusion cycles, as evidenced by the presence of sister chromatid fusions, dicentrics, and chromosome fragments (this study; refs. 27 and 28). One possibility is that HeLa cells experience a greater anaphase delay than CHO cells when they contain broken chromosomes undergoing bridge-breakage-fusion cycles (49). Therefore, they may grow much more slowly than their counterparts that have lost the unhealed chromosome and retained extra copies of the DHFR gene in DMs. Another possibility is that CHO cells also may generate extrachromosomal copies of amplicons, but are incapable of maintaining them, so that only cells that manage to generate stable HSRs ultimately survive on methotrexate.
A third possible difference might be that HeLa cells are better than CHO cells at stabilizing extrachromosomal DNA fragments. Provided that the fragments thus formed can replicate efficiently, they should be maintained in the cell under selective conditions. Coupled with unequal segregation to daughter cells, even a single DM could quickly increase in copy number in cells that survive the ambient drug concentration.
Given that most of the gross chromosomal rearrangements that characterize tumor cells (i.e., translocations, inversions, deletions, insertions, and amplification) appear to be initiated by a chromosomal breakage event, an important question is what causes the initiating breaks in tumor cells that are not obviously exposed to clastogenic agents. In the absence of one or more intact repair pathways, which is the situation in many tumors, errors in normal DNA metabolic events and nucleotide metabolism may be the primary source of genetic instability (reviewed in ref. 43).
Acknowledgments
We thank Greg Landes (Genzyme) and Larry Deaven (Los Alamos National Laboratories), who provided cosmid clones and the human chromosome 5 library, respectively. We are very grateful to Kevin Cox and Carlton White, who provided expert technical assistance, and Mark Alexandrow for help with the figures. This project was supported by National Institutes of Health Grant RO1 CA52559 (to J.L.H. and B.J.T.). L.D.M. was supported by National Institutes of Health Grant T32CA 09109.
Abbreviations
- DHFR
dihydrofolate reductase
- HSR
homogenously staining chromosome region
- DM
double minute
- FISH
fluorescence in situ hybridization
- CHO
Chinese hamster ovary
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130194897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130194897
References
- 1.Bishop J M. Science. 1987;235:305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler K W. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 3.Knudson A G. J Can Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowell J K. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- 5.Schimke R T. J Biol Chem. 1988;263:5989–5992. [PubMed] [Google Scholar]
- 6.Hamlin J L, Leu T H, Vaughn J P, Ma C, Dijkwel P A. Prog Nucleic Acid Res Mol Biol. 1991;41:203–239. doi: 10.1016/s0079-6603(08)60010-0. [DOI] [PubMed] [Google Scholar]
- 7.Stark G R. Adv Cancer Res. 1993;61:87–113. doi: 10.1016/s0065-230x(08)60956-2. [DOI] [PubMed] [Google Scholar]
- 8.Schimke R T. Mutat Res. 1992;276:145–149. doi: 10.1016/0165-1110(92)90004-s. [DOI] [PubMed] [Google Scholar]
- 9.Anderson M W, Reynolds S H, You M, Maronpot R M. Environ Health Perspect. 1992;98:13–24. doi: 10.1289/ehp.929813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brison O. Biochim Biophys Acta. 1993;1155:25–41. doi: 10.1016/0304-419x(93)90020-d. [DOI] [PubMed] [Google Scholar]
- 11.Duffy M J. Clin Biochem. 1993;26:439–447. doi: 10.1016/0009-9120(93)80007-h. [DOI] [PubMed] [Google Scholar]
- 12.Knudson A G., Jr Annu Rev Genet. 1986;20:231–251. doi: 10.1146/annurev.ge.20.120186.001311. [DOI] [PubMed] [Google Scholar]
- 13.Smith K A, Agarwal M L, Chernov M V, Chernova O B, Deguchi Y, Ishizaka Y, Patterson T E, Poupon M F, Stark G R. Philos Trans R Soc London B. 1995;347:49–56. doi: 10.1098/rstb.1995.0008. [DOI] [PubMed] [Google Scholar]
- 14.Johnston R N, Beverley S M, Schimke R T. Proc Natl Acad Sci USA. 1983;80:3711–3715. doi: 10.1073/pnas.80.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tlsty T D. Proc Natl Acad Sci USA. 1990;87:3132–3136. doi: 10.1073/pnas.87.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tlsty T D, Margolin B H, Lum K. Proc Natl Acad Sci USA. 1989;86:9441–9445. doi: 10.1073/pnas.86.23.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zieg J, Clayton C E, Ardeshir F, Giulotto E, Swyryd E A, Stark G R. Mol Cell Biol. 1983;3:2089–2098. doi: 10.1128/mcb.3.11.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark G R, Debatisse M, Giulotto E, Wahl G M. Cell. 1989;57:901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- 19.Pauletti G, Lai E, Attardi G. Proc Natl Acad Sci USA. 1990;87:2955–2959. doi: 10.1073/pnas.87.8.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith K A, Chernova O B, Groves R P, Stark M B, Martinez J L, Davidson J N, Trent J M, Patterson T E, Agarwal A, Duncan P, et al. Proc Natl Acad Sci USA. 1997;94:1816–1821. doi: 10.1073/pnas.94.5.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fougere-Deschatrette C, Schimke R T, Weil D, Weiss M C. In: Gene Amplification. Schimke R T, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 29–32. [Google Scholar]
- 22.Benner S E, Wahl G M, Von Hoff D D. Anticancer Drugs. 1991;2:11–25. doi: 10.1097/00001813-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Schimke R T. Harvey Lect. 1980;76:1–25. [PubMed] [Google Scholar]
- 24.Stark G R, Wahl G M. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- 25.Wahl G M. Cancer Res. 1989;49:1333–1340. [PubMed] [Google Scholar]
- 26.Windle B, Draper B W, Yin Y X, O'Gorman S, Wahl G M. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 27.Trask B J, Hamlin J L. Genes Dev. 1989;3:1913–1925. doi: 10.1101/gad.3.12a.1913. [DOI] [PubMed] [Google Scholar]
- 28.Ma C, Martin S, Trask B J, Hamlin J L. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 29.Smith K A, Gorman P A, Stark M B, Groves R P, Stark G R. Cell. 1990;63:1219–1227. doi: 10.1016/0092-8674(90)90417-d. [DOI] [PubMed] [Google Scholar]
- 30.Toledo F, Le Roscouet D, Buttin G, Debatisse M. EMBO J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith K A, Stark M B, Gorman P A, Stark G R. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 33.Wright J A, Smith H S, Watt F M, Hancock M C, Hudson D L, Stark G R. Proc Natl Acad Sci USA. 1990;87:1791–1795. doi: 10.1073/pnas.87.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 35.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 36.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 37.Levine A J, Perry M E, Chang A, Silver A, Dittmer D, Wu M, Welsh D. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu-Kuo J M, Le Paslier D, Weissenbach J, Chumakov I, Cohen D, Ward D C. Human Mol Genet. 1994;3:99–106. doi: 10.1093/hmg/3.1.99. [DOI] [PubMed] [Google Scholar]
- 39.Deaven L L, McCormick M K, Grady D L, Robinson D L, Buckingham J M, Brown N C, Campbell E W, Campbell M L, Fawcett J J, Jewett P, et al. Proc. U. S. Dept. Energy Contractor-Grantee Workshop IV. 1994. p. 25. [Google Scholar]
- 40.Kim U-J, Birren S W, Slapak T, Mancino V, Boysen C, Kang H-L, Simon M I, Shizuya H. Genomics. 1996;34:213–216. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 41.Yang J K, Masters J N, Attardi G. J Mol Biol. 1984;176:169–187. doi: 10.1016/0022-2836(84)90419-4. [DOI] [PubMed] [Google Scholar]
- 42.Trask B J. In: Mapping Genomes, Genome Analysis: A Laboratory Manual Series. Birren B, Green E, Hietar P, Klaphole S, Myers R, Riethman H, Roskams J, editors. Vol. 4. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 303–413. [Google Scholar]
- 43.Almasan A, Linke S P, Paulson T G, Huang L C, Wahl G M. Cancer Metastasis Rev. 1995;14:59–73. doi: 10.1007/BF00690212. [DOI] [PubMed] [Google Scholar]
- 44.Poupon M F, Smith K A, Chernova O B, Gilbert C, Stark G R. Mol Biol Cell. 1996;7:345–354. doi: 10.1091/mbc.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toledo F, Buttin G, Debatisse M. Curr Biol. 1993;3:255–264. doi: 10.1016/0960-9822(93)90175-n. [DOI] [PubMed] [Google Scholar]
- 46.Maurer B J, Lai E, Hamkalo B A, Hood L, Attardi G. Nature (London) 1987;327:434–437. doi: 10.1038/327434a0. [DOI] [PubMed] [Google Scholar]
- 47.Carroll S M, DeRose M L, Gaudray P, Moore C M, Needham-Vandevanter D R, Von H D, Wahl G M. Mol Cell Biol. 1988;8:1525–1533. doi: 10.1128/mcb.8.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teeter L D, Atsumi S, Sen S, Kuo T. J Cell Biol. 1986;103:1159–1166. doi: 10.1083/jcb.103.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kung A L, Sherwood S W, Schimke R T. Proc Natl Acad Sci USA. 1990;87:9553–9557. doi: 10.1073/pnas.87.24.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]