Abstract
While an arbitrary level of complexity may be included in simulations of spatial epidemics, computational intensity and analytical intractability mean that such models often lack transparency into the determinants of epidemiological dynamics. Although numerous approaches attempt to resolve this complexity–tractability trade-off, moment closure methods arguably offer the most promising and robust frameworks for capturing the role of the locality of contact processes on global disease dynamics. While a close analogy may be made between full stochastic spatial transmission models and dynamic network models, we consider here the special case where the dynamics of the network topology change on time-scales much longer than the epidemiological processes imposed on them; in such cases, the use of static network models are justified. We show that in such cases, static network models may provide excellent approximations to the underlying spatial contact process through an appropriate choice of the effective neighbourhood size. We also demonstrate the robustness of this mapping by examining the equivalence of deterministic approximations to the full spatial and network models derived under third-order moment closure assumptions. For systems where deviation from homogeneous mixing is limited, we show that pair equations developed for network models are at least as good an approximation to the underlying stochastic spatial model as more complex spatial moment equations, with both classes of approximation becoming less accurate only for highly localized kernels.
Keywords: epidemic modelling, spatial models, contact networks, spatial kernels, moment closure methods, foot and mouth disease
1. Introduction
While assuming homogeneous contact processes often provides a sufficient basis for the simple modelling of many physical and biological systems, the mean-field approach becomes less accurate when contact processes are spatially localized. Spatial effects may have profound impacts on both population and epidemiological dynamics (Hastings 1990; Kareiva 1994; Bolker & Grenfell 1995; Lloyd & May 1996; Tilman & Kareiva 1997; Keeling 1999a,b; Dieckmann et al. 2000; Hagenaars et al. 2004), yet few analytical tools exist to even qualitatively understand such effects. Approaches beyond the mean-field approximation include integro-partial differential equations (PDEs) and other PDEs based on the reaction–diffusion equation, patch or metapopulation models, coupled map lattices and interacting particle systems and these have been reviewed in some detail elsewhere (Keeling 1999a). Durrett & Levin (1994) have contrasted the various approaches to spatial modelling in more depth, together with a more detailed consideration of the limits under which the methods coincide. The often complex nature of models explicitly incorporating space, however, usually results in simulation or numerical solution as the only way forward.
A more promising approach to simulation for the analysis of systems with local interactions is moment closure methods. These characterize the local and global dynamics of systems, thus allowing insight into the establishment of local correlations, by modelling the dynamics of low-order moments of the system. For processes continuous in space and time, spatial moment equations summarize the ensemble behaviour of the underlying stochastic model (Bolker & Pacala 1997, 1999; Bolker et al. 2000; Bolker 1999, 2003), while analogous methods (Keeling et al. 1997; Keeling 1999a; Rand 1999; Bauch & Rand 2000; Ferguson & Garnett 2000) have been developed for modelling disease spread on contact networks by capturing how the number of pairs of connected individuals vary over time.
Space may be included in epidemic models within continuous or discrete space frameworks and the choice of model depends on both epidemiological and modelling considerations. Infectious diseases may be transmitted via airborne agents over a wide range of spatial scales (e.g. the foot and mouth disease virus has been shown to travel very long distances over sea (Hugh-Jones & Wright 1970) and land (Gloster et al. 1981), together with more frequent local transmissions resulting from the imposition of movement restrictions and biosecurity measures) and many other viruses may also have sufficiently long survival times to remain a threat by this means (Wallinga et al. 1999). Such systems may lend themselves more naturally to continuous space modelling due to the wide range of scales over which transmission may occur. Discrete space frameworks on the other hand may represent potentially more convenient frameworks for spatial processes occurring on much shorter spatial scales and with a limited number of local contacts. Note, however, that spatial models may offer a less useful basis for modelling systems with low host population densities and that in such cases, individual-based models often provide a more realistic alternative. In this research, we exclusively consider models developed in continuous space to account for transmission over a range of spatial scales (and a sufficiently high-population density to justify such an approach), although note that the techniques and approximations developed are by no means restricted to this case: similar models may be readily developed within discrete space frameworks and an analogous methodology developed.
While network models more generally provide an intuitive basis for modelling biological interactions, such approaches typically lack an explicit means of capturing geographical space. Network approaches may be particularly appropriate to modelling systems in which the probability of disease transmission is not directly related to geographical space (e.g. diseases spread by extreme close contact only), but is more dependent and intuitively related to contact space. Many childhood diseases, sexually transmitted diseases and other airborne agents such as influenza and tuberculosis are highly dependent on contact networks of individuals and the change in movement patterns over time. Modelling the spread of such infectious diseases on contact networks, either defined by the modeller or more realistically constructed from contact tracing, therefore provides an intuitive mathematical framework in which disease spread may be investigated.
In this paper, we consider the extent to which the ensemble behaviour of stochastic spatial epidemic models may be captured by modelling disease processes as occurring on networks derived from the underlying spatial model and therefore the degree to which the two approaches provide comparable frameworks for epidemic modelling. We consider here a mapping of spatial epidemic models parameterized by a spatial contact kernel onto network models, an exact representation of spatial systems in the limiting case where the dynamics of the network are much faster than the time-scale of disease dynamics. Freezing the network results in a static network model, and we discuss the conditions under which this provides a good approximation to the original spatial process. We then demonstrate that third-order ODE-based moment closure approximations to spatial and static network models are comparable, thus demonstrating the equivalence of network and spatial models in the limit where mixing is close to mass-action.
2. A general mapping
A simple means of modelling spatially localized transmission processes is to introduce a function U(r) to capture the decreasing probability of disease transmission with increasing distance between individuals. U(r) is commonly referred to as a contact kernel and is normalized so that for any one individual in a population of size N
| (2.1) |
where Ω is the area within which transmission occurs and U(r) is averaged over all other individuals in this area. For simplicity, consider a population uniformly distributed in Ω with unit density. We define the infection hazard posed by an infectious individual i at location yi to a susceptible host j at location xj to be βU(xj−yi), where β is the contact rate between infectious and susceptible individuals. Additionally, defining the hazard that an infected individual recovers from infection to be γ defines a simple, spatially localized, closed epidemic process.
A contact network can also be defined using the same kernel U(r). We link all pairs of individuals (i, j) with probability , resulting in a network where the number of connections of any given node is Bernoulli distributed with a mean degree or neighbourhood size n. The spatial locality captured by the kernel of the original process is encoded in this network by the higher probability of links existing between nearby compared with distant individuals. There is a degree of arbitrariness however: how do we choose n such that contact processes occurring on the network resemble those occurring in the original spatially explicit model? Clearly, as n→N−1, all spatial information is lost and the model reverts to assuming homogeneous mixing.
One way of overcoming this arbitrariness is to consider a defining characteristic of local contact processes, namely the saturation in the number of potential contacts. When contacts are mostly local, the probability that two contacts drawn at random from the population will be with the same individual is much higher than the 1/(N−1) expected with homogeneous mixing. For a network model, the probability p2 that two contacts will be with the same individual is simply p2=1/n. For the spatial contact process, the probability of finding an individual in the infinitesimal area dr around an arbitrary location r is just dr given our assumption of unit density. The probability of contacting an individual in the area dr if contacts are picked randomly from the population using the kernel U(r) is then U(r)dr and the probability of picking that same individual twice is just U(r)2dr. Integrating over all space gives p2=1/A where 1/A is defined as
| (2.2) |
Demanding that p2 is equal for the spatial and network contact processes gives
| (2.3) |
and generalizing to non-unit density ρ merely involves multiplying the RHS of (2.3) by ρ. This expression for the effective neighbourhood size A is identical to that derived heuristically by Bolker (1999), yet we motivate its use here not only dimensionally, but by demonstrating that this definition of the neighbourhood size quantifies the degree of contact saturation generated by a particular kernel.
The clustering or connectivity coefficient ϕ is often additionally defined for contact networks and captures the average local density of connections of any given individual. In the sociology literature, ϕ is commonly referred to as the network transitivity, reflecting the likelihood that two contacts of a given individual are themselves contacts of each other. If we therefore define ϕ as the proportion of all triples that form triangles (Keeling et al. 1997), it is shown in the electronic supplementary material, section 2, that for a contact network defined from a spatial kernel U(r)
| (2.4) |
where and 0≤ϕ≤1 by definition.
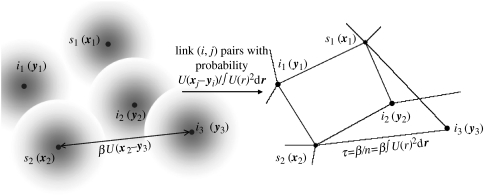
So far, we have only considered general contact processes occurring either spatially or on the network. If we consider the spatial transmission process defined above, the hazard of infection between two individuals was defined to be βU(xj−yi). Writing this as (where q(i, j) is the probability of (i, j) being connected in the network), it is clear that the equivalent network transmission process is defined by a constant hazard of infection between connected individuals of τ=(β/n). The resultant mapping from a spatial transmission model to a network representation is shown schematically in figure 1.
Figure 1.
Infectious individuals i in the spatial model pose an infection hazard βU(xj−yi) to susceptible neighbours j, with the dependence on distance being determined by the kernel U(r) (here schematically represented by the greyscale shading around each individual). Linking all (i, j) pairs according to this spatial kernel allows one to define an equivalent contact network with a fixed infection hazard between pairs of τ=(β/n), where the mean neighbourhood size and the network connectivity , so that the corresponding network may be parameterized directly from knowledge of U(r) (see §2).
The question then arises as to the conditions under which the dynamics of the network transmission process thus defined are equivalent to the original spatial model. If we consider the contact network to be static during the entire epidemic process occurring on the network, it is clear that the dynamics are not necessarily equivalent. For the spatial model, the basic reproduction number R0 (the mean number of secondary infections generated by a single primary infection in an infinite population) is shown in the electronic supplementary material, section 3, to be given by
| (2.5) |
while for the network model, . (Note, however, that in highly heterogeneous neighbourhood distributions, the effective number of contacts, defined as the mean number of contacts plus the variance-to-mean ratio of this distribution, is the more epidemiologically relevant quantity (Anderson & May 1992). A more general receipe for calculating the basic reproduction number in heterogeneous populations defines R0 as the dominant eigenvalue of the linear next-generation operator, so that iteratively applying this operator gives the expected number of infected individuals in successive generations (Diekmann & Heesterbeek 2000)). Expanding to second order in β/γ gives
| (2.6) |
for both the network and spatial models and this expression converges to the mass-action result R0=β/γ as n→∞ as required.
The two models diverge, however, as β→∞ since R0→n for the network model, while R0→∞ for the spatial model. This reflects a fundamental difference between the two approaches: in the spatial model, each contact is picked at random from the entire population (meaning that an infinite number of individuals can in principle be contacted), while for the static network model, a finite set of possible contacts is picked for each individual prior to the start of the epidemic. The only network representation, which is therefore formally equivalent to a spatial model is a dynamic network model in which new links are made (and old ones destroyed) at a rate much faster than the epidemic process, meaning in essence that each new contact is picked from the entire population. It should be stressed however that this problem as β→∞ is characteristic of the approach we adopt here for mapping the two models. Alternative approaches, for example based on the finite number of secondary infections caused per generation (quantified by R0), may not suffer this problem, representing an interesting problem for future research. The mapping we propose here is likely to represent only one of a number of possible mappings between the two approaches, although we argue that the basis of the mapping here represents one of the simplest possible translations based on an important distinction between the two models.
Equation (2.6) gives us some insight into when we might expect the static network model to be a good approximation to the spatial epidemic process, namely when , or when there is only limited saturation of susceptibles in the local contact neighbourhood of an infected individual. When saturation is limited, the finite maximum on the number of individuals connected to any one infected individual in the network model no longer has a significant dynamical effect, meaning in essence that prior selection of a finite number of contacts is expected to be equivalent to dynamical selection of a new contact prior to each transmission event.
While simulation represents one means of examining the degree of correspondence between network and spatial models (and indeed considerations based on R0 another), we focus here on using the mapping described above to motivate the derivation of an analytical framework based on moment closure methods for modelling spatial epidemic processes. As a demonstration of the robustness of the above mapping, we derive moment closure approximations to spatial and network models separately before demonstrating their dynamical equivalence.
3. Spatial contact models and moment methods in continuous space and time
Although the development of spatial contact models for ecological and epidemiological systems is well established (Mollison 1972, 1977), convolutions of the kernel often lead to analytical intractability. While such integro-differential equations may be approximated by diffusion models, the need to obtain insight into a wider range of spatial models has led to the recent development of spatial moment methods, predominantly applied to the study of plant disease epidemiology (Bolker & Pacala 1997, 1999; Bolker et al. 2000; Bolker 1999, 2003). As well as implicitly accounting for stochasticity and the discreteness of individuals, moment equations are, in general, easily parameterizable with field data, provide a more accessible and transparent analytical framework for investigating disease spread and allow for heterogeneous mixing via the inclusion of a contact kernel (or dispersal kernel in plant systems). Contact kernels, in general, reflect the decreasing probability of disease transmission with increasing distance from the source and a variety of functional forms may be considered.
The essence of spatial moment equations for systems continuous in space and time is to derive differential equations for the first m spatial moments of the underlying spatial distribution of individuals. In this paper, we adopt a second-order approach and derive equations for the mean number density , the average expected number of individuals of type X per unit area, and covariance density , capturing the degree of association or correlation between hosts of type X and Y separated by a distance r. The covariance is used as a quantitative measure of the spatial component of the system and the autocovariance (when X=Y) and cross-covariance (X≠Y) measure the degree of aggregation within and between species respectively. Positive, negative and zero covariances, respectively, correspond to clustered, evenly spaced and randomly distributed individuals.
While moment equations provide a deterministic approximation to ensemble behaviour by averaging the underlying model over all stochastic realizations, a key drawback lies in the inability to obtain a closed set of equations since the evolution of mth-order moments depends on moments of order m+1. Defining a model by mth-order moments therefore requires a suitable moment closure assumption to define higher-order moments in terms of those of interest (of order m), and this is often determined either by desirable or intuitive physical or mathematical properties of the closure or from best-fit to simulation (Dieckmann & Law 2000). We adopt here the so-called power-1 closure, formally derived by setting third-order central spatial moments to zero, and moment equations under this closure have been shown to match simulation extremely well for spatial scales other than the very short (Bolker 1999; Filipe et al. 2004). Filipe et al. (2004) have recently shown that threshold conditions exist on the initial density of infectives below which power-1 closure breaks down and demonstrate that power-3 closure does not suffer this drawback, accurately matching simulation for all initial conditions and a wider range of spatial scales. However, we focus here on power-1 closure since our results are independent of the closure we adopt and provide a simpler set of equations to illustrate the methodology.
While power-1 closure originates from setting third-order moments to zero, the symmetric power-2 closure attaches equal importance to all pairs of individuals in triples and can be justified on probabilistic grounds (Dieckmann & Law 2000). The asymmetric power-2 closure, on the other hand, may be more appropriate in low clustering regimes, since it assumes that one of the pairs in a triple contributes far less to the dynamics than the other two pairs. Combinations of asymmetric power-2 closures may also be developed (with different weightings on each pair), and have been applied to a number of ecological problems (Murrell & Law 2000; Law et al. 2003). Power-3 closure has been applied extensively in the statistical mechanics literature in the context of disorder in spatially homogeneous systems (Ziman 1979), and is often known as the Kirkwood superposition approximation (Kirkwood 1935).
Consider the simple SIR epidemic model proposed in §2 and let S(x) and I(y) be the probability that patches centred at x and y contain a susceptible and infected individual respectively at time t, with associated mean densities and . For a fixed contact rate β, average infectious period 1/γ and a normalized spatial transmission kernel U(r), let
and
Taking the expectation over all stochastic realizations and defining spatial averages (denoted for a function f(x, y)) to be weighted across all non-uniform spatial probability distributions p(x, y) so that
| (3.1) |
(where p(x, y)=U(x−y) here), the moment equations under power-1 closure are shown in Bolker (1999) to be given by
| (3.2) |
where we assume a spatially homogeneous and isotropic landscape, (A*B)(r) denotes a convolution between functions A and B and is simply
| (3.3) |
Note that the equivalent moment equations under power-2 and power-3 closures simply consist of the linear covariance terms in (3.2) plus higher-order covariances dependent on the exact form of the closure.
Since we are looking to map the spatially averaged behaviour of (3.2) onto a network representation, taking ∂/∂t of (3.3) and substituting in (3.2) eliminates the explicit spatial dependence of the covariances by averaging each term over all space. While this reduces the covariances to their weighted averages and the weighted average of the kernel itself is simply related to the effective neighbourhood size A (see equation (2.3)), averaging the convolution terms presents a problem since we cannot express the result in terms of the state variables of interest. In fact, (3.2) can only be reduced to a set of ordinary differential equations (ODEs) for all U(r) if we approximate the convolution such that
| (3.4) |
where k is a kernel-dependent factor related to the moments of U(r) and 0≤k≤1 by definition (where the extreme case of U(r)=δ(r) exactly corresponds to k=1 so that increasing kernel locality corresponds to increasing k). Recall that in the full stochastic model, (U*I) is proportional to the probability of new S–I pairs forming, so that (U*cXY) may therefore be interpreted as proportional to the probability that transmission occurs between S–X–Y triples where at least one of X and Y must be of type I for this probability to be non-zero. With this interpretation of (U*cXY) and the fact that cXY in some sense captures the likelihood of X–Y pair formations, simple rearrangement of (3.4) leads to an interpretation of k as the conditional probability that transmission occurs between a susceptible individual at x and an X–Y pair given that the X–Y pair is formed. k is therefore a connectivity parameter capturing the spatial clustering associated with a given U(r) (equivalently the degree to which pairs make up triples), as well as implicitly capturing the relative importance of local versus global connections in the network. k is dependent on a number of factors including the initial conditions, the moment closure adopted and the degree of locality in the kernel.
While best-fit k may be calculated numerically, an analytical approach is ideally required. To this end, let us first propose that the spatial covariances are separable so that
| (3.5) |
implying that the spatial and temporal epidemic components evolve independently. Numerical investigation has shown this to become unreliable only at very small r, as it is only in the very early stages of the epidemic that these components are strongly coupled. It is shown in the electronic supplementary material, section 4, that this allows k to be estimated by
| (3.6) |
where . Note that the assumption of separability eliminates the true time-dependence of k to a constant for a given kernel (corresponding to a spatially averaged network connectivity) and that (3.6) is independent of the kernel scale parameter. Since several expressions for k may also be derived (electronic supplementary material, section 5), table 1 compares these with the average best-fit values for three moment closure assumptions and contact kernels.
Table 1.
Comparison of the best-fit values of k, obtained by fitting the moment equations (3.2) with and without the convolution approximation (3.4), with values obtained analytically from (3.6), and eqns (8) and (16) in the electronic supplementary material, for the spatial model under power-1 (P1), symmetric power-2 (P2) and power-3 (P3) closures. We compare the normalized offset power-law (with exponent 3), exponential and Gaussian kernels for the case where γ=0, β=1, A∼25, and we assume a randomly distributed host population at t=0 of whom 5% are infectious.
| best-fit k | analytical estimate | |||||
|---|---|---|---|---|---|---|
| contact kernel | P1 | P2 | P3 | (3.6) | (8) | (16) |
| offset power-law | 0.56 | 0.64 | 0.66 | 0.48 | 0.55 | 0.61 |
| exponential | 0.68 | 0.77 | 0.79 | 0.57 | 0.59 | 0.65 |
| Gaussian | 0.74 | 0.81 | 0.82 | 0.67 | 0.71 | 0.77 |
Having defined the effective uniform neighbourhood size A and substituted the convolution approximation (3.4), we can reduce (3.2) to
| (3.7) |
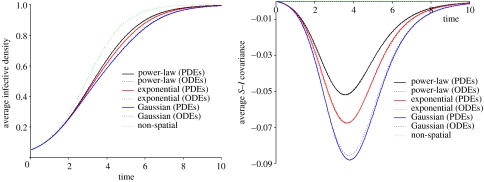
and figure 2 compares the epidemic curves and spatial correlations predicted by (3.2) and (3.7) for three different kernels. We find in general that the ODEs match the moment equations extremely well throughout the epidemic for all three kernels, accurately capturing the dynamics in parameter regimes in which the moment equations themselves capture the ensemble behaviour of the underlying stochastic model. A similar result is obtained for moment equations under power-2 and power-3 closures.
Figure 2.
Comparing the moment equations (3.2) and ODEs (3.7) for an offset power-law, exponential and Gaussian kernel for the mean infective densities and average S–I covariance over time. The ODEs are plotted for best-fit k and with all other parameters as per table 1.
4. Network models and correlation equations
The study of complex networks has become a recent focus across a wide range of scientific disciplines and the mathematical modelling of physical and biological systems on networks has received increasing attention (Albert & Barabási 2002; Dorogovtsev & Mendes 2002; Newman 2003). More recent research has also considered the applicability of contact networks to epidemiological modelling (Anderson et al. 1990; Kretzschmar & Morris 1996; May & Lloyd 2001; Newman 2002; Read & Keeling 2003; Keeling & Eames 2005).
Networks consist simply of a set of nodes (or vertices) and the links (or edges) between them and a wide variety of network architectures may be constructed. Examples include a static versus growing population of nodes, weighted-edge networks (whether or not we attach more importance to certain links over others), directed and undirected networks (whether links can exist both ways or in one direction only), static and dynamically linked networks, and so on. Such a diverse set of networks are typically characterized by a number of attributes including the degree distribution (the probability that a randomly selected node has a given number of links), average path length (quantifying the average number of links required to go between two nodes) and the network clustering (capturing the cliquishness between contacts).
Early work focused on random graph models (Erdös & Rényi 1959) and regular lattices and much research followed on the existence of percolation phenomena in such systems. More recent research has focused on intermediate regimes between randomness and order (Watts & Strogatz 1998) and the dynamics on so-called small-world and scale-free topologies have been widely investigated. This has highlighted for example the role of intervention strategies based on network topologies (e.g. targeting control efforts at core groups in the face of limited resources), the stability of biological networks and the existence of epidemic threshold phenomena. More recent work has also examined weighted-edge networks in more detail and their topological properties in relation to air transportation and scientific collaboration networks (Barrat et al. 2004), as well as aiming to develop analytical tools to investigate the properties of such networks. The highly complex nature of many real networks has led to much dependence on computational insights and more recent research has developed analytic tools to complement such approaches: the work by Newman et al. (2001), for example, considers how probability generating functions may be used to obtain insight into in-degree and out-degree distributions and epidemic thresholds for complex epidemiological models run on directed networks.
While spatial moment equations provide a means of capturing the ensemble behaviour of stochastic spatial models, correlation equations provide an analogous tool for capturing the role of local correlations in network models of disease spread (Keeling et al. 1997; Keeling 1999a; Rand 1999). For the simple SIR model considered in §2 for a population of N individuals, an analogous set of equations may be derived governing disease evolution on a static, regular and fixed-edge contact network (so that links between individuals are constant in time, occur with a fixed transmissibility between nodes τ and all individuals have the same average number of neighbours n). If we let [X]=[X](t) be the expected number of individuals of type X, [XY]=[XY](t) the number of X–Y pairs and [XYZ]=[XYZ](t) the number of X–Y–Z triples, it is readily shown in Rand (1999) that
| (4.1) |
where 1/g is the average infectious period. Adopting a second-order approach explicitly considers only the number of singles and pairs of each type as dynamical variables. By employing a moment closure assumption to close this system of equations, such pair approximations make simple assumptions about the way triples are formed from singles and pairs.
One such assumption is to assume that the number of neighbours of a given individual is Bernoulli distributed and this leads to the simple approximation that
| (4.2) |
This is equivalent in spirit to the asymmetric power-2 spatial moment closure discussed in §3 by considering only the [XY] and [YZ] pairs as comprising the triple and neglecting [XZ] pairs by comparison: this has been shown elsewhere to provide a good approximation to a wide range of static network models (Rand 1999). However, this assumption evidently becomes less accurate for spatially clustered systems in which transitivity becomes important. In such cases, a common assumption is to assume that a certain fraction ϕ of triples form triangles and this leads to the assumption that
| (4.3) |
(Keeling et al. 1997), where the network connectivity ϕ plays the role of the clustering coefficient in standard complex network analysis. With respect to the spatial moment closures in §3, (4.3) represents the combination of an asymmetric power-2 and power-3 closure.
Note that correlations CXY=CXY(t) between individuals of type X and Y may be captured by defining
| (4.4) |
(Keeling et al. 1997), where CXY=1 represents mass-action mixing, CXY>1 positive correlations and CXY<1 avoidance or segregation within or between species.
5. Mapping under moment closure
Although §2 considers a more general mapping, we focus here on demonstrating a correspondence between spatial and network models under moment closure approximations to both and investigate the accuracy to which pair equations capture the dynamics of corresponding spatial models.
To map the first moment in the spatial approach onto the number of singles in the network model, note first that the derivation of (3.2) averages single probability densities over all stochastic realizations and then over all space to obtain the expected number of individuals per unit area. Together with dimensional arguments, we suggest a priori that an intuitive mapping of the first moments is provided by
| (5.1) |
since A also plays the role of the effective neighbourhood area or the uniform transmission neighbourhood corresponding to a non-uniform transmission kernel, and furthermore that
| (5.2) |
follows naturally and maps the second moments. Recall that the overbar notation denotes a spatial average with respect to a non-uniform spatial probability distribution (here the transmission kernel) and it is the effective neighbourhood size A of this kernel that plays the crucial role in this mapping: this fact is useful when models become more complex, for example introducing multiple spatial kernels, although we do not consider this further here.
Moreover, substituting (5.1) and (5.2) into the network equations (4.1) demonstrates that both
| (5.3) |
and
| (5.4) |
follow and are necessary for dimensional and model consistency; the former is precisely the heuristic mapping derived from first principles in §2 and the latter provides a trivial mapping between the SIR models. In addition, adopting the moment closure approximation (4.3) and substituting (5.1)–(5.4) into (4.1) allows one to demonstrate that the pair equations map identically onto the spatially derived ODEs (3.7) to first order in the covariances providing that the further mapping
| (5.5) |
is made (electronic supplementary material, section 6). While ϕ captures the degree of local clustering in a network, k plays an analogous role in the spatial models here by reflecting the degree of locality in U(r) and hence the relative importance of local versus global connections in affecting the dynamics; in this sense, (5.5) is unsurprising. Note additionally that the earlier expressions for ϕ and k (equations (2.4) and (3.6), respectively) for a given U(r) are identical, although other expressions for k may also be derived.
First-order covariances dominate the dynamics whenever departure from mass-action mixing is minimized and spatial correlations are fairly weak: it is in these regimes that we therefore expect pair correlation equations to accurately capture the essence of their underlying spatial models. Moreover, the fact that these terms arise independent of the moment closure we adopt implies that this result is true more generally whenever departure from homogeneous mixing is not significant.
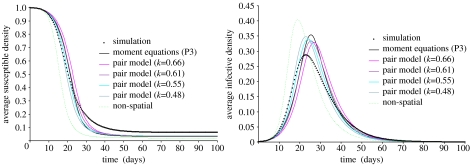
These moment and parameter mappings allow us to investigate the extent to which pair correlation equations approximate spatial dynamics. Figure 3 illustrates a simple example and compares simulation of the full spatial model with the spatial moment equations and pair correlation equations derived from the mappings above for the simple SIR model. The full model is simulated using a discrete-time approximation to the underlying stochastic spatial process run with a time-step of 0.1 days for a population size of 4 million. Figure 3 represents the average of 100 stochastic realizations of the full model. Note that important computational issues arise in comparing simulations of such models with deterministic approximations: subtle variations in model architecture may lead to significant effects on aspects of temporal dynamics such as R0 and the epidemic generation time, although the importance of such effects is intimately related to the connectivity of the contact space considered (Green et al. 2005). We assume that such effects are unimportant for the reasonably high effective neighbourhood size illustrated here.
Figure 3.
Comparing the SIR dynamics resulting from the average of 100 stochastic realizations of the full spatial model, the spatial model under power-3 closure and the pair equations derived from the mappings for an offset power-law kernel. Parameters as per figure 2 except , the basic reproduction number R0=4 and the infectious period 1/γ=7 days. The pair equations are plotted for the four relevant k values in table 1.
We consider a reasonably localized offset power-law kernel, so that heterogeneous mixing is appreciable compared to mass-action, and parameters well within the regime in which moment and correlation equations are most useful. We plot the spatial model under power-3 closure, since the collapse of power-1 closure for very small infective densities (Bolker 1999; Filipe et al. 2004) also extends to the SIR case.
We find that the pair equations match the spatial simulation extremely well in the early stages of the epidemic up to around 15–20 days and in particular when k is close to the value predicted by (3.6), improving significantly on predictions of the non-spatial model and moment equations. While the moment and pair equations overestimate exact equilibrium densities, both capture well the qualitative rate at which the epidemic declines, although fail to accurately reproduce the epidemic tail and underestimate the number of uninfected individuals remaining at the end of the epidemic (). One can heuristically argue that the stochastic nature of the full model means that pockets of susceptibles may form at long distances from epidemic foci, so that the epidemic may extinguish globally due to a lack of susceptibles to infect locally at the epidemic ‘wavefront’: will be lower in pair equations since they fail to capture such intermediate scale phenomena.
6. Conclusions
The use of moment methods to capture spatial and network epidemic models allows complex stochastic systems to be more thoroughly and reliably investigated by deriving deterministic equations capturing ensemble behaviour. Motivated by heuristic considerations of spatial and network models by connecting individuals according to a spatial kernel, we develop a useful means of capturing the former by the latter, demonstrating that pair approximations allow complex spatial models to be mapped onto more analytically tractable frameworks.
We show that reducing spatial moment equations to a set of ODEs allows spatial models to be mapped onto network approximations by mapping moments of their respective distributions by
| (6.1) |
where A is the effective uniform neighbourhood size of a given kernel and m refers to the order of the moment of interest (so that m=1 corresponds to the expected number of individuals of type X ([X]) and the first spatial moment , m=2 the number of X–Y pairs [XY] and second moment , and so on). This mapping of moment closure approximations is also consistent with the more general mapping derived in §2, demonstrating that n, τ and ϕ may be determined directly from the kernel and hence that field data on U(r) is sufficient to parameterize an approximating network model.
For systems where mixing is not too divergent from mass-action, we demonstrate that pair equations provide an excellent approximation to spatial models. That this approximation holds independent of the moment closure we adopt implies that a more general mapping between full spatial and network models exists and this is currently being pursued.
Our methodology differs significantly from the approach of Bauch & Galvani (2003) by capturing the local saturation of contact processes (as quantified by the parameters n and ϕ), in considering the analytically more tractable case of equal-weight links in the network model used and by avoiding the introduction of a cut-off radius R.
While we consider a simple SIR model to illustrate the methodology, the framework we develop is analytically robust in allowing extensions to more complex models. This will permit more realistic spatial models in future to be reduced to more analytically tractable forms, yet still retaining a fundamental link with explicit space, and therefore allowing an improved understanding of key determinants driving epidemic spread. One such application is to the modelling of foot and mouth disease (FMD) and we are currently extending the framework developed here and in past work (Ferguson et al. 2001a,b) to include spatially targeted control measures (namely vaccination and culling) in a more rigorous manner, together with a better account of long-distance jumps in seeding new infection foci. We are also examining how pair models might be extended to capture more accurately the dynamics of spatial processes with short-range kernels.
Note that we do not explicitly consider the relative appropriateness of spatial and network models with varying neighbourhood size n here, but consider for clarity and illustrative purposes only a fairly typical localized regime in which one expects moment closure models to be of most use: sufficiently deviant from homogeneous mixing to motivate a spatial approach, yet avoiding parameter regimes where the kernel is highly localized and moment closure approximations break down due to the increasingly important effect of higher-order correlations. Future work may therefore consider the effect of varying n on the translation between the models.
Note that while a variety of approaches may be developed for modelling the between-farm spread of FMD, the multiple potential pathways of infection, along with aerosol transmission over a wide range of spatial scales, lends itself particularly well to models incorporating continuous spatial kernels and indeed this approach was used in modelling the 2001 epidemic in the UK (Ferguson et al. 2001a,b; Keeling 2001). Moreover, while moment closure models represent an intuitive and analytically simpler approach than more complex and computationally intensive approaches (Morris 2001), enabling better real-time use of mathematical models to aid policy decisions, further research is required to compare the validity of different spatial approaches to modelling FMD and this is also currently being examined.
Acknowledgements
This work was funded by BBSRC (P.E.P.) and the Royal Society (N.M.F.). The authors would like to thank two anonymous referees for their helpful comments and suggestions for improving the manuscript.
Supplementary Material
Appendix A considers in more detail how the connectivity (or transitivity) of the approximating network model architecture may be directly calculated from the spatial transmission kernel U(r) used to construct the network. Appendix B highlights how the basic reproduction number R0 may be calculated for the spatial and network SIR models with two competing hazards, whilst appendices C and D demonstrate how analytical expressions for the spatial clustering parameter k may be estimated for a given U(r) and we consider the cases where the assumption of separability may or may not be adopted. Appendix E considers how the functional form of the pair correlation model on the network may be demonstrated to give an excellent approximation to the spatial moment closure model in the regime where deviation from homogeneous mixing is limited.
References
- Albert R, Barabási A.-L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. doi:10.1103/RevModPhys.74.47 [Google Scholar]
- Anderson R.M, Gupta S. The significance of sexual partner contact networks for the transmission dynamics of HIV. J. Acquir. Immune Defic. Syndr. 1990;3:417–429. [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1992. Infectious diseases of humans: dynamics and control. [Google Scholar]
- Barrat A, Barthelemy M, Pastor-Satorras R, Vespignani A. The architecture of complex weighted networks. Proc. Natl Acad. Sci. USA. 2004;101:3747–3752. doi: 10.1073/pnas.0400087101. doi:10.1073/pnas.0400087101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauch C.T, Galvani A.P. Using network models to approximate spatial point-process models. Math. Biosci. 2003;184:101–114. doi: 10.1016/s0025-5564(03)00042-7. doi:10.1016/S0025-5564(03)00042-7 [DOI] [PubMed] [Google Scholar]
- Bauch C, Rand A. A moment closure model for sexually transmitted disease transmission through a concurrent partnership network. Proc. R. Soc. B. 2000;267:2019–2027. doi: 10.1098/rspb.2000.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker B.M. Analytic models for the patchy spread of plant disease. Bull. Math. Biol. 1999;61:849–874. doi: 10.1006/bulm.1999.0115. doi:10.1006/bulm.1999.0115 [DOI] [PubMed] [Google Scholar]
- Bolker B.M. Combining endogenous and exogenous spatial variability in analytical population models. Theor. Popul. Biol. 2003;64:255–270. doi: 10.1016/s0040-5809(03)00090-x. doi:10.1016/S0040-5809(03)00090-X [DOI] [PubMed] [Google Scholar]
- Bolker B, Grenfell B. Space, persistence and dynamics of measles epidemics. Phil. Trans. R. Soc. B. 1995;348:309–320. doi: 10.1098/rstb.1995.0070. [DOI] [PubMed] [Google Scholar]
- Bolker B, Pacala S.W. Using moment equations to understand stochastically driven spatial pattern formation in ecological systems. Theor. Popul. Biol. 1997;52:179–197. doi: 10.1006/tpbi.1997.1331. doi:10.1006/tpbi.1997.1331 [DOI] [PubMed] [Google Scholar]
- Bolker B.M, Pacala S.W. Spatial moment equations for plant competition: Understanding spatial strategies and the advantages of short dispersal. Am. Nat. 1999;153:575–602. doi: 10.1086/303199. doi:10.1086/303199 [DOI] [PubMed] [Google Scholar]
- Bolker B.M, Pacala S.W, Levin S.A. Moment methods for ecological processes in continuous space. In: Dieckmann U, Law R, Metz J.A.J, editors. The geometry of ecological interactions. Cambridge University Press; Cambridge, UK: 2000. pp. 388–411. ch. 20. [Google Scholar]
- Diekmann O, Heesterbeek J.A.P. Wiley; New York: 2000. Mathematical epidemiology of infectious diseases: model building, analysis and interpretation. [Google Scholar]
- Dieckmann U, Law R. Relaxation projections and the method of moments. In: Dieckmann U, Law R, Metz J.A.J, editors. The geometry of ecological interactions—simplifying spatial complexity. Cambridge University Press; Cambridge, UK: 2000. pp. 412–455. ch. 21. [Google Scholar]
- Dieckmann U, Law R, Metz J.A.J, editors. The geometry of ecological interactions: simplifying spatial complexity. Cambridge University Press; Cambridge, UK: 2000. [Google Scholar]
- Dorogovtsev S.N, Mendes J.F.F. Evolution of networks. Adv. Phys. 2002;51:1079–1187. doi:10.1080/00018730110112519 [Google Scholar]
- Durrett R, Levin S.A. The importance of being discrete (and spatial) Theor. Popul. Biol. 1994;46:363–394. doi:10.1006/tpbi.1994.1032 [Google Scholar]
- Erdös P, Rényi A. On random graphs. Publ. Math. Debrecen. 1959;6:290–297. [Google Scholar]
- Ferguson N.M, Garnett G.P. More realistic models of sexually transmitted disease transmission dynamics: sexual partnership networks, pair models, and moment closure. Sex. Transm. Dis. 2000;27(10):600–609. doi: 10.1097/00007435-200011000-00008. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Donnelly C.A, Anderson R.M. The foot-and-mouth epidemic in great Britain: pattern of spread and impact of interventions. Science. 2001;292:1155–1160. doi: 10.1126/science.1061020. doi:10.1126/science.1061020 [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Donnelly C.A, Anderson R.M. Transmission intensity and impact of control policies on the foot and mouth epidemic in great Britain. Nature. 2001;413:542–548. doi: 10.1038/35097116. doi:10.1038/35097116 [DOI] [PubMed] [Google Scholar]
- Filipe J.A.N, Maule M.M, Gilligan C.A. On ‘analytical models for the patchy spread of plant disease’. Bull. Math. Biol. 2004;66:1027–1037. doi: 10.1016/j.bulm.2003.11.001. doi:10.1016/j.bulm.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Gloster J, Blackall R.M, Sellers R.F, Donaldson A. Forecasting the airborne spread of foot-and-mouth disease. Vet. Record. 1981;108:370–374. doi: 10.1136/vr.108.17.370. [DOI] [PubMed] [Google Scholar]
- Green, D. M., Kiss, I. Z. & Kao R. R. In press. Parameterization individual-based models: comparisons with deterministic mean-field models. J. Theor. Biol [DOI] [PubMed]
- Hagenaars T.J, Donnelly C.A, Ferguson N.M. Spatial heterogeneity and the persistence of infectious diseases. J. Theor. Biol. 2004;229:349–359. doi: 10.1016/j.jtbi.2004.04.002. doi:10.1016/j.jtbi.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Hastings A. Spatial heterogeneity and ecological models. Ecology. 1990;71:426–428. [Google Scholar]
- Hugh-Jones M.E, Wright P.B. Studies on the 1967–8 foot-and-mouth disease epidemic. The relation of weather to the spread of disease. J. Hyg. 1970;68:253–271. doi: 10.1017/s0022172400028722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareiva P. Space: the final frontier for ecological theory. Ecology. 1994;75:1. [Google Scholar]
- Keeling M. Spatial models of interacting populations. In: McGlade J, editor. Advanced ecological theory: principles and applications. Blackwell Science; Oxford: 1999. pp. 64–99. ch. 3. [Google Scholar]
- Keeling M.J. The effects of local spatial structure on epidemiological invasions. Proc. R. Soc. B. 1999;266:859–867. doi: 10.1098/rspb.1999.0716. doi:10.1098/rspb.1999.0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, Eames K.T.D. Networks and epidemic models. J. R. Soc. Interface. 2005;2:295–307. doi: 10.1098/rsif.2005.0051. doi:10.1098.rsif.2005.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, Rand D.A, Morris A.J. Correlation models for childhood epidemics. Proc. R. Soc. B. 1997;264:1149–1156. doi: 10.1098/rspb.1997.0159. doi:10.1098/rspb.1997.0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, et al. Dynamics of the 2001 UK foot and mouth epidemic: Stochastic dispersal in a heterogeneous landscape. Science. 2001;294:813–817. doi: 10.1126/science.1065973. doi:10.1126/science.1065973 [DOI] [PubMed] [Google Scholar]
- Kirkwood J.G. Statistical mechanics of fluid mixtures. J. Chem. Phys. 1935;3:300–313. doi:10.1063/1.1749657 [Google Scholar]
- Kretzschmar M, Morris M. Measures of concurrency in networks and the spread of infectious disease. Math. Biosci. 1996;133:165–195. doi: 10.1016/0025-5564(95)00093-3. doi:10.1016/0025-5564(95)00093-3 [DOI] [PubMed] [Google Scholar]
- Law R, Murrell D.J, Dieckmann U. Population growth in space and time: the spatial logistic equations. Ecology. 2003;84:252–262. [Google Scholar]
- Lloyd A.L, May R.M. Spatial heterogeneity in epidemic models. J. Theor. Biol. 1996;179:1–11. doi: 10.1006/jtbi.1996.0042. doi:10.1006/jtbi.1996.0042 [DOI] [PubMed] [Google Scholar]
- May R.M, Lloyd A.L. Infection dynamics on scale-free networks. Phys. Rev. E. 2001;64:066112. doi: 10.1103/PhysRevE.64.066112. doi:10.1103/PhysRevE.64.066112 [DOI] [PubMed] [Google Scholar]
- Mollison D. Proc. Sixth Berkeley Symp. on Mathematical Statistics and Probability. vol. 3. 1972. The rate of spatial propagation of simple epidemics. pp. 579–614. [Google Scholar]
- Mollison D. Spatial contact models for ecological and epidemic spread. J. R. Stat. Soc. B. 1977;39:283–326. [Google Scholar]
- Morris R.S, et al. Predictive spatial modelling of alternative control strategies fot the foot-and-mouth disease epidemic in Great Britain, 2001. Vet. Rec. 2001;149:137–144. doi: 10.1136/vr.149.5.137. [DOI] [PubMed] [Google Scholar]
- Murrell D.J, Law R. Beetles in fragmented woodlands: a formal framework for dynamics of movement in ecological landscapes. J. Anim. Ecol. 2000;69:471–483. doi:10.1046/j.1365-2656.2000.00409.x [Google Scholar]
- Newman M.E.J. Spread of epidemic disease on networks. Phys. Rev. E. 2002;66:16–128. doi: 10.1103/PhysRevE.66.016128. [DOI] [PubMed] [Google Scholar]
- Newman M.E.J. The structure and function of complex networks. SIAM Rev. 2003;45(2):167–256. doi:10.1137/S003614450342480 [Google Scholar]
- Newman M.E.J, Strogatz S.H, Watts D.J. Random graphs with arbitrary degree distributions and their applications. Phys. Rev. E. 2001;64:26–118. doi: 10.1103/PhysRevE.64.026118. [DOI] [PubMed] [Google Scholar]
- Rand D.A. Correlation equations and pair approximations for spatial ecologies. In: McGlade J, editor. Advanced ecological theory—principles and applications. Blackwell Science; Oxford: 1999. pp. 100–142. ch. 4. [Google Scholar]
- Read J.M, Keeling M.J. Disease evolution on networks: the role of contact structure. Proc. R. Soc. B. 2003;270:699–708. doi: 10.1098/rspb.2002.2305. doi:10.1098/rspb.2002.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Kareiva P, editors. Spatial ecology: the role of space in population dynamics and interspecific interactions. Princeton University Press; Princeton, NJ: 1997. [Google Scholar]
- Wallinga J, Edmunds W.J, Kretzschmar M. Human contact patterns and the spread of airborne infectious diseases. Trends Microbiol. 1999;7:372–377. doi: 10.1016/s0966-842x(99)01546-2. doi:10.1016/S0966-842X(99)01546-2 [DOI] [PubMed] [Google Scholar]
- Watts D.J, Strogatz S.H. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. doi:10.1038/30918 [DOI] [PubMed] [Google Scholar]
- Ziman J.M. Cambridge University Press; Cambridge, UK: 1979. Models of disorder: the theoretical physics of homogeneously disordered systems. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A considers in more detail how the connectivity (or transitivity) of the approximating network model architecture may be directly calculated from the spatial transmission kernel U(r) used to construct the network. Appendix B highlights how the basic reproduction number R0 may be calculated for the spatial and network SIR models with two competing hazards, whilst appendices C and D demonstrate how analytical expressions for the spatial clustering parameter k may be estimated for a given U(r) and we consider the cases where the assumption of separability may or may not be adopted. Appendix E considers how the functional form of the pair correlation model on the network may be demonstrated to give an excellent approximation to the spatial moment closure model in the regime where deviation from homogeneous mixing is limited.