Abstract
Alternative splicing multiplies genomic coding capacity and regulates proteomic composition. A well-studied example of this plasticity leads to the synthesis of functionally distinct isoforms of the Fibroblast Growth Factor Receptor-2 (FGFR2). The regulation of this isoform diversity necessitates the silencing of FGFR2 exon IIIb, which is mediated by flanking intronic splicing silencers and the polypyrimidine tract binding protein (PTB). To visualize this splicing decision in vivo, we developed mice harboring a green fluorescent protein construct that reports on the silencing of exon IIIb. The animals also harbor a red fluorescent protein reporter of constitutive splicing as an allelic control. This dual reporter system revealed that in various organs and cell types the silencing of exon IIIb required the intronic silencers. In neurons, which do not express PTB, we observed robust silencer-dependent repression of exon IIIb, suggesting that the neural paralog, brain PTB, can take over this function. In the epidermis, however, the intronic silencers were not required for efficient silencing. This work provides a first glimpse at splicing regulation among different cell types in vivo and promises the drafting of an anatomic map of splicing decisions.
Keywords: alternative splicing, exon silencing, FGFR2, in vivo imaging
INTRODUCTION
Alternative splicing of primary transcripts is a potent and prevalent mechanism driving the diversity of the proteome (Maniatis and Tasic 2002; Johnson et al. 2003). An elegant example of this is the regulated splicing of fibroblast growth factor receptor 2 (FGFR2) transcripts that leads to the production of two functionally distinct receptor isoforms, FGFR2 (IIIb) and FGFR2 (IIIc). Isoform choice requires the coordinate regulation of the alternative inclusion of exons IIIb and IlIc (Del Gatto et al. 1996, 1997; Carstens et al. 1998, 2000; Muh et al. 2002; Wagner and Garcia-Blanco 2002; Baraniak et al. 2003; Hovhannisyan and Carstens 2005; Wagner et al. 2005; Baraniak et al. 2006). Exon IIIb silencing, which is absolutely critical for proper isoform regulation, depends on the combined effect of weak splice sites, an exonic splicing silencer (ESS), and two flanking intronic splicing silencers (ISSs) (Fig. 1A; Del Gatto et al. 1996; Carstens et al. 2000; Wagner et al. 2005). The ISSs, upstream intronic splicing silencer (UISS) and intronic control element (ICE), and the binding sites for the polypyrimidine tract binding protein (PTB) within them, are required for full silencing of the exon (Carstens et al. 2000; Wagner and Garcia-Blanco 2002; Wagner et al. 2005). Recruitment of PTB to these sites leads to exon IIIb skipping, and RNAi-mediated PTB knockdown leads to its inclusion, establishing PTB as a repressor of this exon (Wagner and Garcia-Blanco 2002).
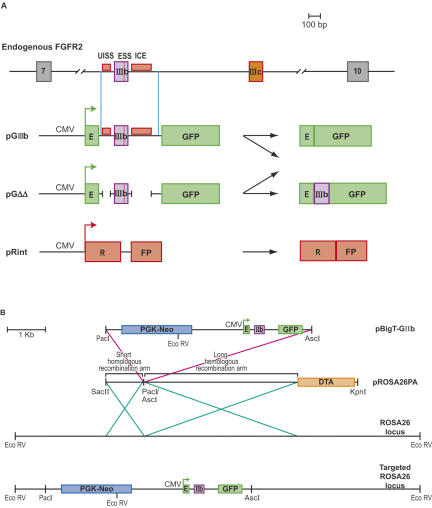
FIGURE 1.
Reporters of FGFR2 exon IIIb silencing in transgenic mice. (A) Schematic representation of the endogenous FGFR2 transcripts and of the reporter constructs used to image exon silencing in transgenic animals and their possible splicing products. (B) Targeting reporter constructs into the ROSA26 genomic locus. Reporter constructs were cloned into vector pBigT, which contains a neomycin resistance cassette, to create plasmid pBigT-GIIIb (same was done for pGΔΔ and pRint). PBigT-GIIIb was then cloned into pROSA26PA vector, which contains homologous sequences to the ROSA26 locus, and a diphtheria toxin gene for negative selection of nonhomologous recombination events. Vectors harboring fluorescent reporters were inserted into the ROSA26 genomic locus by homologous recombination. The targeted locus is shown containing neomycin resistance and the fluorescent reporter.
The great majority of studies of alternative splicing have been carried out in vitro or ex vivo using cell lines. Recently, we and others have created reporters that permit the visualization of splicing outcomes in living cells (Sheives and Lynch 2002; Wagner et al. 2004; Levinson et al. 2006; Newman et al. 2006; for review, see Kemp et al. 2005). These reporters, which usually depend on expression of green fluorescent protein (GFP), have been used predominantly in cell culture. The first use of fluorescence reporters to evaluate splicing outcomes in vivo was carried out by Kole and coworkers to evaluate the efficacy of antisense compounds that targeted a cryptic splice site (Sazani et al. 2002). Subsequently, Ellis et al. showed that transgenic mice harboring reporters for smooth muscle-specific splicing events expressed high levels of GFP only in smooth muscle (Ellis et al. 2004). Although this was an important step in validating cell culture results in vivo, the imaging data per se did not prove sufficiently robust to report on splicing decisions (Ellis et al. 2004). This difficulty was due to large variations in transcript levels between tissues and transgenic lines harboring randomly integrated reporter constructs. Very recently we have used reporters of FGFR2 exon IlIc splicing to evaluate mesenchymal epithelial transitions in prostate tumors (Oltean et al. 2006).
In order to assay FGFR2 exon IIIb silencing and ISS activity in living animals, we used reporters where expression of EGFP (herein GFP) is contingent on ISS-dependent exon IIIb silencing. To minimize variation in transcript level between transgenic lines, control and experimental reporters were integrated by homologous recombination into the ROSA26 locus. Here we show the use of these reporters to visualize exon IIIb silencing and to address the requirement for ISS elements in different cell types and tissues in vivo.
RESULTS
Imaging FGFR2 exon IIIb silencing in whole animals
We developed transgenic mice that harbor the silencing reporter pGIIIb, which contains the GFP ORF bifurcated by a constitutive intron that is itself interrupted by exon IIIb and flanking ISSs (Fig. 1A). The ISSs are deleted in reporter pGΔΔ (Fig. 1A). A control reporter, pRint, encodes the ORF of the DsRed dimer variant RFP (herein RFP) (Campbell et al. 2002) bifurcated by the same constitutive intron in pGIIIb (Fig. 1A). All GFP and RFP transcripts are driven by the CMV-IE promoter. GFP expression from pGIIIb and pGΔΔ transcripts depends on silencing exon IIIb since inclusion of this exon interrupts the GFP ORF (Wagner et al. 2004). We did not include activating elements downstream of ICE in these constructs since these were not designed to visualize the overall regulation of exon IIIb splicing. Thus, we did not expect that the constructs would exactly mimic the regulation of the endogenous FGFR2 transcripts. The constructs were intended to interrogate specifically the silencing of exon IIIb.
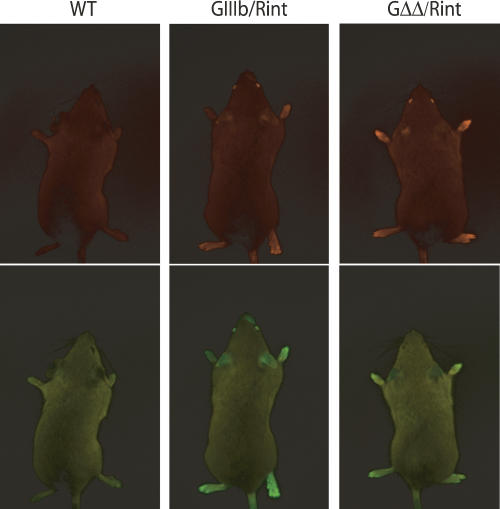
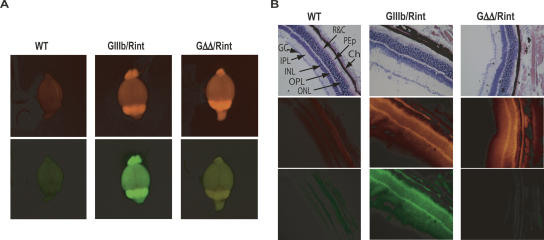
To minimize variation in transcript level between cell types and between transgenic lines, the reporters were integrated into the ROSA26 locus (Zambrowicz et al. 1997; Soriano 1999) by homologous recombination (Fig. 1B and Materials and Methods). The ROSA26 locus has been shown to be transcriptionally active in many tissues, and homozygous ROSA26 disruptions have no apparent phenotype. Two independent lineages from different ES cell targeting events were produced for the GIIIb and GΔΔ reporters, and one was obtained for Rint reporter. This approach permitted the use of an allelic RFP control reporter, essential to interpret the data. We crossed mice harboring GFP reporters to mice harboring the RFP reporter to obtain double reporter mice: 129-Gt(ROSA26)Sor(GIIIb/Rint)BGB (GIIIb/Rint) and 129-Gt(ROSA26)Sor(GΔΔ/Rint)BGB (GΔΔ/Rint). GIIIb/Rint and GΔΔ/Rint animals were imaged in a fluorescence light box using nontransgenic animals (wild type) as a reference for background fluorescence (Fig. 2). RFP expression was detected in the eyes, ears, paws, and tail of GIIIb/Rint and GΔΔ/Rint animals (Fig. 2). GFP was also expressed in the eyes, ears, and whisker region of GIIIb/Rint animals, which contrasted with the loss of detectable GFP in the same tissues of GΔΔ/Rint mice (Fig. 2). This result strongly suggested that these tissues efficiently silenced exon IIIb, and this silencing was sensitive to the deletion of the ISSs. In contrast, the paws and tails of both GIIIb/Rint and GΔΔ/Rint mice expressed GFP (Fig. 2), a surprising result that is discussed in more detail below. Based on whole animal imaging, we concluded that the fluorescence constructs could provide a visual report of alternative splicing in vivo.
FIGURE 2.
Imaging FGFR2 exon IIIb silencing in whole animals. Wild-type (WT), GIIIb/Rint, and GΔΔ/Rint mice were imaged in a fluorescence light box. Images were acquired for 300 msec with a CCD camera using RFP (upper panels) or GFP (lower panels) excitation/emission filters (see Materials and Methods).
It is important to note that all the results shown in this article were identical in GIIIb/Rint and GΔΔ/Rint animals where the ROSA26 GIIIb and GΔΔ alleles derived from two independent ES cells. It is also important to note that the expression of GFP GIIIb/ROSA26 and GΔΔ/ROSA26 animals was the same as that observed in GIIIb/Rint and GΔΔ/Rint indicating that the presence of the RFP allele did not affect the essential findings from the dual reporter animals (data not shown).
Silencing of exon IIIb in the heart and in skeletal muscles
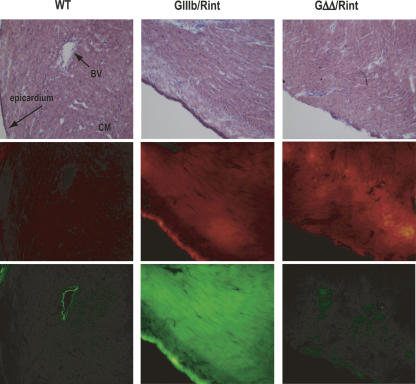
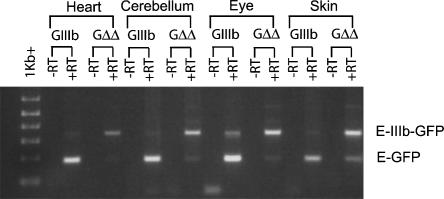
In order to visualize exon IIIb silencing in more detail we examined whole organs and fixed cryosections from GIIIb/Rint and GΔΔ/Rint mice. Expression of GFP and RFP was observed in intact hearts of GIIIb/Rint animals, and was well matched across several different cell types within these hearts (Fig. 3). In contrast, GFP expression in all GΔΔ/Rint heart cell types was barely detectable in the context of robust expression from the allelic RFP control (Fig. 3). RT-PCR analysis using total heart RNA confirmed that GIIIb transcripts skipped exon IIIb while GΔΔ transcripts predominantly included it (Fig. 4). Similar results were obtained with triceps and rectus femoris skeletal muscles (data not shown). These results indicated that both cardiac and skeletal muscle silenced exon IIIb in an ISS-dependent manner.
FIGURE 3.
Silencing of FGFR2 exon IIIb in the heart. Cryosections of hearts from wild-type (WT), GIIIb/Rint, and GΔΔ/Rint animals were stained with Hematoxylin and Eosin Y (H&E) and imaged by light microscopy to detect histological structures (top panels) after they were analyzed by epifluorescence microscopy to detect RFP (middle panels) and GFP expression (bottom panels). Images were acquired at 200× magnification. (BV) blood vessel; (CM) cardiac muscle.
FIGURE 4.
Molecular analysis of pGIIIb and pGΔΔ transcripts. Total RNA from heart, cerebellum, eye, and skin of GIIIb/Rint or GΔΔ/Rint mice was analyzed by RT-PCR for the presence of GFP transcripts and for the inclusion of exon IIIb. −RT lanes were not subjected to reverse transcription. PCR products derived from exon IIIb inclusion and skipping transcripts are indicated by E-IIIb-GFP and E-GFP, respectively.
Silencing of exon IIIb in neural tissues
ISS-dependent silencing of exon IIIb in heart was consistent with expression of PTB in that organ (Lillevali et al. 2001; Gooding et al. 2003; Ladd et al. 2005). Unlike cardiac muscle cells, neurons do not express PTB but instead express brPTB, which has been postulated to be a weaker silencer of exon definition (Markovtsov et al. 2000; Polydorides et al. 2000; Lillevali et al. 2001; McCutcheon et al. 2004). Therefore, we interrogated neural tissues in GIIIb/Rint animals for RFP and GFP expression. Imaging of whole brains in a fluorescence light box revealed widespread and well-matched expression of GFP and RFP in the brains of GIIIb/Rint animals (Fig. 5A). The same was true of eyes and spinal cord (data not shown). Imaging of the same organs from GΔΔ/Rint animals showed robust RFP expression but only background levels of GFP expression (Fig. 5A; data not shown). It must be noted that the yellow signal observed in the GΔΔ/Rint whole brain images obtained with the LT-9MACROIMSYS light box was due to RFP bleed-through, and this was never observed using the more stringent filters in the Olympus IX epifluorescence microscope. High levels of GFP and RFP were observed in neurons in the retina, the ventral horn, and the cerebellar cortex of GIIIb/Rint mice (Fig. 5B; data not shown). Equivalent sections from GΔΔ/Rint mice were RFP positive, but GFP was not detected above background (Fig. 5B; data not shown). We concluded that many neurons regulated exon IIIb repression in an ISS-dependent manner. These data were supported by molecular analysis of RNAs from eyes and cerebellum (Fig. 4).
FIGURE 5.
ISS dependent exon IIIb silencing in neural tissues. (A) Fluorescent images of whole brains from wild-type (WT), GIIIb/Rint, and GΔΔ/Rint animals were obtained using a fluorescence light box and acquired for 300 msec with a CCD camera using RFP (upper panels) or GFP (lower panels) excitation/emission filters. The yellow signal observed in the GFP channel when imaging GΔΔ/Rint was due to bleed-through from RFP; it was not observed in GΔΔ/rosa26+ mice (not shown). (B) Cryosections of the eyes were stained with H&E and imaged by light microscopy to detect histological structures (top panels) after they were analyzed by epifluorescence microscopy to detect RFP (middle panels) and GFP expression (bottom panels). Eye sections illustrate a detailed view of the retina. All images in B were acquired at 200× magnification. (GC) layer of ganglion cells; (IPL) inner plexiform layer; (INL) inner nuclear layer; (OPL) outer plexiform layer; (ONL) outer nuclear layer; (R&C) rods and cone receptors; (PEp) pigment epithelium; (Ch) choroids.
The ISSs flanking exon IIIb are functional in many tissues and cell types
A wide survey of organs and tissues of different animals originating from different ES targeting events was performed. This survey, which included kidney, adrenal gland, testis, pancreas, and stomach, in addition to those organs shown above, revealed that most organs and tissues in GIIIb/Rint mice expressed RFP and that all the tissues that expressed RFP also expressed GFP (data not shown). The same tissues from GΔΔ/Rint mice, which as expected were RFP positive, did not express GFP. These findings led us to conclude that these diverse tissues expressed factors that mediate ISS-dependent silencing of exon IIIb in pGIIIb transcripts.
ISS independent silencing in the epidermis
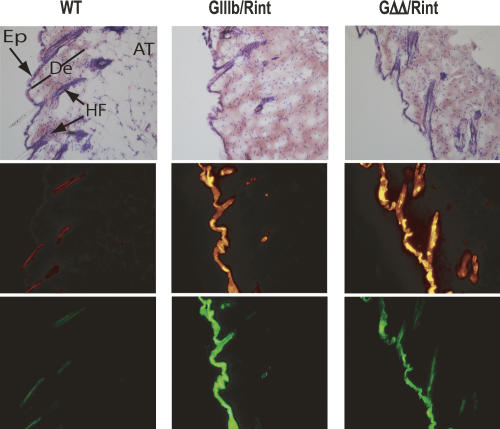
An exception to the widespread requirement for ISSs was suggested by the GFP expression in the skin over the paws and tails of GΔΔ/Rint animals (Fig. 2). Examination of cryosections confirmed that GΔΔ/Rint animals expressed GFP in the epidermis, albeit the levels of expression were significantly lower than those observed in epidermis of GIIIb/Rint animals (Fig. 6). Nonetheless, GFP expression in GΔΔ/Rint epidermis was higher than that observed in all other tissues of these animals. Because keratinized epidermis has been reported to autofluorescence (Rohrschneider et al. 2005), we carefully compared these sections to those from wild-type mice, and this epidermis consistently showed very low levels of autofluorescence (Fig. 6). RT-PCR analysis of skin total RNA showed that pGΔΔ transcripts skipped exon IIIb more frequently than pGΔΔ transcripts in other organs (Fig. 4), which was consistent with the levels of GFP expression. These data suggested that in cells of the epidermis, silencing of exon IIIb did not absolutely require the flanking ISS elements.
FIGURE 6.
ISS independent silencing in the skin. Cryosections of skin from the back of the animals were stained with H&E and imaged by light microscopy to detect histological structures (top panels) after they were analyzed by epifluorescence microscopy to detect RFP (middle panels) and GFP expression (bottom panels). Images were acquired at 200× magnification. (AT) adipose tissue; (De) dermis; (Ep) epidermis; (HF) hair follicle.
DISCUSSION
We have shown here the use of fluorescence reporters to visualize alternative splicing decisions in a wide variety of tissues and in living animals. The results obtained by this system were consistent with molecular analysis obtained by RT-PCR; however, it must be noted that the results obtained using the fluorescence reporters supersedes the molecular analysis, which is an average of many cell types. Using this system we have tested and confirmed the importance of the two flanking ISS elements in tissues and organs. Unexpectedly, we have observed efficient use of these silencing elements in neural tissues.
The inclusion of exon IIIb in pGIIIb transcripts is under the control of at least four cis-acting elements in addition to the splice sites; one enhances exon inclusion and three promote silencing. The two ISSs exert profound repression on the exon in the majority of tissues and must act by binding PTB and other, heretofore uncharacterized, UISS and ICE interacting factors. Whereas several conserved elements within ICE are needed for silencing, the PTB sites are strictly required for effective silencing (Wagner et al. 2005). This requirement is consistent with exon IIIb silencing observed in heart, which has been shown to express both PTB and brPTB (nPTB) (Lillevali et al. 2001; Gooding et al. 2003) mRNAs and PTB protein (Ladd et al. 2005).
Mature neurons, however, do not express PTB (Yamamoto et al. 1999; Markovtsov et al. 2000; Polydorides et al. 2000; Lillevali et al. 2001; Gooding et al. 2003; McCutcheon et al. 2004), and yet we observe strong silencing of exon IIIb that is strictly ISS dependent. Neural tissues express brPTB, but previous work in tissue culture suggests that this PTB paralog is a weak repressor (Markovtsov et al. 2000; Polydorides et al. 2000). Based on our in vivo data we speculate that brPTB can capably silence exon IIIb in neurons in the cerebellum and in the retina. These issues will be conclusively resolved by crossing our reporters into mice where PTB and/or brPTB have been disrupted, when these become available.
The explanation of repression in other tissues is not as readily understood. For instance adult murine skeletal muscles do not express significant levels of PTB, brPTB, or the paralog ROD1 (Yamamoto et al. 1999; Markovtsov et al. 2000; Polydorides et al. 2000; Lillevali et al. 2001). It is not clear whether or not a fourth paralog, smPTB, is expressed in skeletal muscle (Gooding et al. 2003) and can repress exon IIIb via PTB sites. It is also possible that skeletal muscle cells have increased activity of the other factors and can tolerate weaker PTB activity (Singh and Valcarcel 2005). These questions will be tested by creating reporter mice that express constructs with discrete mutations of sub-elements within UISS and ICE.
Epidermal cells, in contrast to those mentioned above, are capable of repressing exon IIIb independently of the two ISS elements, which is not surprising since cells of the dermis and epidermis have been shown to handle the alternative splicing of other transcripts in a unique manner (Barksdale and Baker 1995). The ISS independent silencing could be due to a strong effect exerted by the previously characterized ESS, which is indeed critical for exon IIIb repression in the keratinocyte line SVK14 (Del Gatto-Konczak et al. 1999). The ISS independence can also be due to the presence of additional cis-elements that respond to factors or combinations of factors uniquely found in epidermal cells. These cis-acting elements may be lurking among the many evolutionarily conserved sequences that have no function in a limited panel of cells in vitro (Carstens et al. 1998, 2000; Mistry et al. 2003; Wagner et al. 2005). While others have proposed that different cis-acting sequences within a splicing precursor could have differential importance in different cell types (Modafferi and Black 1999), these data derived from chimerical constructs and cells of uncertain provenance. Our data on the epidermis led us to conclude that different cell types in their normal physiological context make the same alternative splicing decision via widely different mechanisms.
Finally, the data presented in this article foretell the imaging of other normal splicing decisions in normal organs and tissues, the regulation of these choices in normal development, and their alteration in disease states. Without question, the system requires technical improvements (e.g., use of quantitative imaging methods) and biological refinement (e.g., the use of reporters in mice with engineered disruption of trans-acting factors). Nonetheless, we propose that this first report offers an unprecedented view of splicing decision making in cells and tissues in vivo.
MATERIALS AND METHODS
Targeting reporters into the ROSA26 locus
The construction of the reporters has been described previously (Wagner et al. 2004) and in the text (Fig. 1A). Reporters were subcloned into pBigT vector (Srinivas et al. 2001) and subsequently into the ROSA26 targeting vector pROSA26PA (Fig. 1B; Soriano 1999). Both pBigT and pROSA26PA were kindly provided by Dr. Frank Costantini (Columbia University, New York). Alternative splicing regulation of reporters within targeting vectors was verified in murine-cultured cells before proceeding to targeting mouse ES cells (data not shown). Plasmids were linearized using KpnI, gel purified, and transfected into AK 7 ES cells (from 129S4/SvJaeSor mice), which were injected into C57/BL6J blastocysts at the Duke University Transgenic Facility. Chimeras were screened for germline transmission by PCR screen and by fluorescence imaging. Two lines from different ES cell targeting events were produced for each of the GIIIb and GΔΔ reporters, and one line was obtained for Rint reporter. Transgenic GIIIb and GΔΔ reporter mice were crossed to Rint reporter mice to obtain double reporter animals.
Macroscopic imaging
Transgenic mice were euthanized with a lethal injection of euthasol according to guidelines and recommendations of the Duke University Institutional Animal Care and Use Committee. Animals were imaged immediately after euthanasia using an LT-9MACROIMSYS light box (Lightools Research, Inc.). Tissues were then dissected, rinsed in PBS (pH 7.4), and individually imaged in the light box. GFP fluorescence excitation was produced using a 470/40× excitation filter and captured using a 515-nm emission filter, and RFP fluorescence excitation was obtained with a 540/40× filter and detected using a 590-nm emission filter. Images were acquired using a color CCD camera and analyzed using the Image ProPlus v. 4.0 software (Media Cybernetics, Inc.).
Microscopic analysis and histologic staining
Excised organs were washed in PBS (pH 7.4), embedded in TissueTek OCT compound (Sakura Finetek, Inc.), and frozen in dry ice. Samples were stored at −80°C until analyzed. Frozen 14 μm sections were prepared in a cryostat and fixed with 4% (w/v) paraformaldehyde for 30 min at room temperature, rinsed twice in PBS for 10 min, and sealed using gel mount (Biomeda, Inc.). Tissue sections were imaged using an Olympus IX epifluorescence microscope. Images were acquired using an Olympus DP70 digital camera and processed using DP controller software (Olympus Technologies, Ltd.). Coverslips were removed from imaged slides, and tissues were stained using Hematoxylin and Eosin Y using standard procedures to identify cells and histological landmarks.
RT-PCR analysis
Tissues were stored at −80°C in RNA Later (Ambion, Inc.) until processed for molecular analysis. Frozen tissues were ground into powder using a mortar and pestle and liquid nitrogen. RNA was extracted using Trizol (Invitrogen, Inc.), following the recommendations of the manufacturer. RT-PCR was performed using primers designed to amplify either the GFP or the RFP ORF. Primer sequences will be provided upon request.
ACKNOWLEDGMENTS
We thank Dr. Frank Costantini for providing plasmids pBigT and pROSA26PA. We thank present and past members of the Garcia-Blanco laboratory for suggestions and reagents and Robin Davis for technical assistance. We also thank Cheryl Bock, Blanche Capel, Mark Dewhirst, Dan Kiehart, Elwood Lynney, Brian Sorg, and Eric Wagner for helpful discussions and suggestions. We are very grateful to Drs. Blanche Capel, Susan Michalowski, James Pearson, and Robin Wharton for their insightful comments on the manuscript. We acknowledge support from grants from the NCI (1R33 CA97502) and NIH (1RO1 GM63090) to M.A.G.-B., and NIH (1RO1 GM63090 supplement) to V.I.B. These institutes had no role in the design and/or preparation of the study.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.248506.
REFERENCES
- Baraniak, A.P., Lasda, E.L., Wagner, E.J., Garcia-Blanco, M.A. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak, A.P., Chen, J.R., Garcia-Blanco, M.A. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barksdale, S., Baker, C.C. Differentiation-specific alternative splicing of bovine papillomavirus late mRNAs. J. Virol. 1995;69:6553–6556. doi: 10.1128/jvi.69.10.6553-6556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., Tsien, R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens, R.P., McKeehan, W.L., Garcia-Blanco, M.A. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol. Cell. Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens, R.P., Wagner, E.J., Garcia-Blanco, M.A. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7388–7400. doi: 10.1128/mcb.20.19.7388-7400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto, F., Gesnel, M.C., Breathnach, R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto, F., Plet, A., Gesnel, M.C., Fort, C., Breathnach, R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak, F., Olive, M., Gesnel, M.C., Breathnach, R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, P.D., Smith, C.W., Kemp, P. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by α-tropomyosin regulatory elements in transgenic mice. J. Biol. Chem. 2004;279:36660–36669. doi: 10.1074/jbc.M405380200. [DOI] [PubMed] [Google Scholar]
- Gooding, C., Kemp, P., Smith, C.W. A novel polypyrimidine tract-binding protein paralog expressed in smooth muscle cells. J. Biol. Chem. 2003;278:15201–15207. doi: 10.1074/jbc.M210131200. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan, R.H., Carstens, R.P. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol. Cell. Biol. 2005;25:250–263. doi: 10.1128/MCB.25.1.250-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kemp, P.R., Ellis, P.D., Smith, C.W. Visualization of alternative splicing in vivo. Methods. 2005;37:360–367. doi: 10.1016/j.ymeth.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., Stenberg, M.G., Swanson, M.S., Cooper, T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Levinson, N., Hinman, R., Patil, A., Stephenson, C.R., Werner, S., Woo, G.H., Xiao, J., Wipf, P., Lynch, K.W. Use of transcriptional synergy to augment sensitivity of a splicing reporter assay. RNA. 2006;12:925–930. doi: 10.1261/rna.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillevali, K., Kulla, A., Ord, T. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech. Dev. 2001;101:217–220. doi: 10.1016/s0925-4773(00)00566-9. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Markovtsov, V., Nikolic, J.M., Goldman, J.A., Turck, C.W., Chou, M.Y., Black, D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon, I.E., Hentschel, S.J., Fuller, G.N., Jin, W., Cote, G.J. Expression of the splicing regulator polypyrimidine tract-binding protein in normal and neoplastic brain. Neuro-oncol. 2004;6:9–14. doi: 10.1215/S1152851703000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry, N., Harrington, W., Lasda, E., Wagner, E.J., Garcia-Blanco, M.A. Of urchins and men: Evolution of an alternative splicing unit in fibroblast growth factor receptor genes. RNA. 2003;9:209–217. doi: 10.1261/rna.2470903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modafferi, E.F., Black, D.L. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh, S.J., Hovhannisyan, R.H., Carstens, R.P. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2) J. Biol. Chem. 2002;277:50143–50154. doi: 10.1074/jbc.M207409200. [DOI] [PubMed] [Google Scholar]
- Newman, E.A., Muh, S.J., Hovhannisyan, R.H., Warzecha, C.C., Jones, R.B., McKeehan, W.L., Carstens, R.P. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltean, S., Sorg, B.S., Albrecht, T., Bonano, V.I., Brazas, R.M., Dewhirst, M.W., Garcia-Blanco, M.A. Alternative inclusion of fibroblast growth factor receptor 2 exon IIIc in Dunning prostate tumors reveals unexpected epithelial mesenchymal plasticity. Proc. Natl. Acad. Sci. 2006;103:14116–14121. doi: 10.1073/pnas.0603090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polydorides, A.D., Okano, H.J., Yang, Y.Y., Stefani, G., Darnell, R.B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrschneider, L.R., Custodio, J.M., Anderson, T.A., Miller, C.P., Gu, H. The intron 5/6 promoter region of the ship1 gene regulates expression in stem/progenitor cells of the mouse embryo. Dev. Biol. 2005;283:503–521. doi: 10.1016/j.ydbio.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Sazani, P., Gemignani, F., Kang, S.H., Maier, M.A., Manoharan, M., Persmark, M., Bortner, D., Kole, R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- Sheives, P., Lynch, K.W. Identification of cells deficient in signaling-induced alternative splicing by use of somatic cell genetics. RNA. 2002;8:1473–1481. [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas, S., Watanabe, T., Lin, C.S., William, C.M., Tanabe, Y., Jessell, T.M., Costantini, F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1 doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J., Garcia-Blanco, M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J., Baines, A., Albrecht, T., Brazas, R.M., Garcia-Blanco, M.A. Imaging alternative splicing in living cells. Methods Mol. Biol. 2004;257:29–46. doi: 10.1385/1-59259-750-5:029. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J., Baraniak, A.P., Sessions, O.M., Mauger, D., Moskowitz, E., Garcia-Blanco, M.A. Characterization of the intronic splicing silencers flanking FGFR2 exon IIIb. J. Biol. Chem. 2005;280:14017–14027. doi: 10.1074/jbc.M414492200. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., Tsukahara, K., Kanaoka, Y., Jinno, S., Okayama, H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz, B.P., Imamoto, A., Fiering, S., Herzenberg, L.A., Kerr, W.G., Soriano, P. Disruption of overlapping transcripts in the ROSA β geo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]