Abstract
Alternative pre-mRNA splicing may be the most efficient and widespread mechanism to generate multiple protein isoforms from single genes. Here, we describe the genomic analysis of one of the most frequent types of alternative pre-mRNA splicing, alternative 5′- and 3′-splice-site selection. Using an EST-based alternative splicing database recording >47,000 alternative splicing events, we determined the frequency and location of alternative 5′- and 3′-splice sites within the human genome. The most common alternative splice sites used in the human genome are located within 6 nucleotides (nt) of the dominant splice site. We show that the EST database overrepresents alternative splicing events that maintain the reading frame, thus supporting the concept that RNA quality-control steps ensure that mRNAs that encode for potentially harmful protein products are destroyed and do not serve as templates for translation. The most frequent location for alternative 5′-splice sites is 4 nt upstream or downstream from the dominant splice site. Sequence analysis suggests that this preference is a consequence of the U1 snRNP binding sequence at the 5′-splice site, which frequently contains a GU dinucleotide 4 nt downstream from the dominant splice site. Surprisingly, ∼50% of duplicated 3′-YAG splice junctions are subject to alternative splicing. This high probability of alternative 3′-splice-site activation in close proximity of the dominant 3′-splice site suggests that the second step of the splicing may be prone to violate splicing fidelity.
Keywords: alternative splice-site activation, bioinformatics, human genome, pre-mRNA
INTRODUCTION
The splicing of nuclear pre-mRNAs is a fundamental process required for the expression of most metazoan genes. It is carried out by the spliceosome, which recognizes splicing signals and catalyzes the removal of noncoding intronic sequences to assemble protein-coding sequences into mature mRNA (Black 2003). Splicing signals are sequence elements that are located at the 5′- and 3′-splice sites, the polypyrimidine tract, and the branchpoint sequence upstream of the 3′-splice site. The 5′-splice site is characterized by a poorly conserved consensus sequence, YAG/guragu, at the exon/intron boundary (where Y stands for pyrimidines, R for purines, capital letters for exon, lowercase letters for intron, and the slash indicates the exon/intron boundary). The 3′-splice site is recognized by the polypyrimidine tract and by its YAG consensus sequence at the intron/exon junction (Black 2003). It is still unclear why particular splice sites are recognized and chosen while others are not utilized. Recognition and utilization of splice sites requires snRNPs to bind to the splicing signals. The first step of the splicing reaction is the assembly of the E complex (early or commitment complex) and is characterized by the association of U1 snRNP with the 5′-splice-site consensus sequence U2AF to the intron/exon junction and the polypyrimidine tract, and a loose association of U2 snRNP with the branchpoint (Reed and Palandjian 1997). After stabilization of U2 snRNP with the pre-mRNA and splice-site pairing during A complex formation (Lim and Hertel 2004), U1 snRNA is displaced by U6 snRNA at the 5′-splice site after rearrangements in the B complex (Krämer 1996). Two sequential transesterification reactions in the C complex result in intron excision. In the first step, the 2′-hydroxyl group of the branchpoint adenosine attacks the 5′-splice site to generate lariat and upstream exon intermediates. U5 snRNP holds the 5′- and 3′-exons in proximity to promote their ligation in a second transesterification reaction in which the 3′-hydroxyl of the 5′-exon attacks the phosphodiester linkage at the 3′-splice site to generate ligated exons (Konarska and Query 2005).
Alternatively spliced transcripts are created when different splice sites are chosen during pre-mRNA splicing. Of the ∼25,000 genes encoded by the human genome (International Human Genome Sequencing Consortium 2004), >60% are thought to produce transcripts that are alternatively spliced (Johnson et al. 2003). Thus, alternative splicing leads to the production of multiple mRNA isoforms from a single pre-mRNA, exponentially enriching the proteomic diversity of higher eukaryotic organisms. One of the most common forms of alternative splicing is alternative splice-site activation. Alternative splice-site activation can occur at the 5′- and/or 3′-splice sites.
Here, we performed a computational analysis using the Alternative Splicing Database (ASD) (Thanaraj et al. 2004) to evaluate alternative 5′- and 3′-splice-site selection and usage within the human genome. ASD is derived from EST entries and reports the use of >15,000 alternative splice sites within the human transcriptome. A high frequency of alternative 5′-splice-site activation is observed 4 nucleotides (nt) upstream or downstream from the dominant splice site. Sequence analysis suggests that this bias originates from sequence requirements for U1 snRNP binding to the 5′-splice site. The most common alternative 3′-splice-site usage occurs within 6 nt of the dominant splice site, supporting a mechanism that promotes plasticity in AG choice at the 3′-splice site. Flanking each splice site, we observe an increase in the probability of alternative splicing every 3 nt from the dominant splice site, revealing a reading frame bias. This bias can be explained by a combination of two factors: a dinucleotide bias within coding exons, and mRNA quality-control mechanisms.
RESULTS AND DISCUSSION
Alternative 5′-splice-site selection
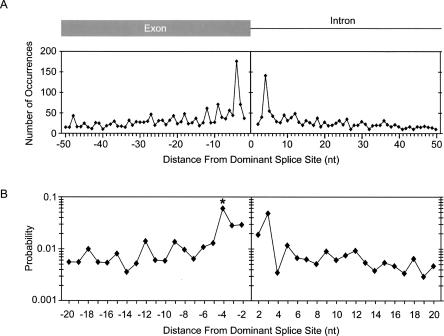
Alternative splice-site usage accounts for ∼40% (≈20,000 events) of all alternative splicing (Thanaraj et al. 2004). To determine the frequency of alternative splice-site activation relative to the dominant splice site in humans, we extracted and aligned all alternative splicing events reported in the ASD for internal exons (see Materials and Methods). The incidence of alternative 5′-splice-site usage is greatest close to the dominant splice site, and decreases farther away from the dominant splice site (Fig. 1A). The most common positions of alternative 5′-splice-site utilization within this distribution are located 4 nt upstream and downstream from the dominant splice site. This observation is surprising when considering that the deletion or insertion of 4 nt would cause a shift in the protein reading frame. However, the 5′-splice-site consensus sequence, YAG/guragu, provides the sequence context to support this unusually high level of alternative 5′-splice-site activation. The major spliceosome requires the presence of a GU dinucleotide at the 5′ end of the intron. Thus, position +4 of the 5′-splice site consensus could serve as an alternative exon/intron junction if the downstream nucleotides also conform to the 5′-splice-site consensus (Fig. 2A). Similarly, position −4 could serve as an alternative exon/intron junction if the surrounding nucleotides also conform to the 5′-splice-site consensus. To determine the probability of alternative 5′-splice-site activation 4 nt upstream or downstream from the dominant splice site, we computed the fraction of EST-verified alternative splicing over the total number of GU dinucleotides that are found at these positions within the human genome. While the frequency of alternative splice-site usage +4 or −4 nt from the dominant splice site is roughly equivalent (Fig. 1A), the probabilities of alternative GU selection are drastically different between the two positions (Fig. 1B). A GU dinucleotide 4 nt upstream from the dominant splice site has a 6.1% chance that it will be used as an alternative 5′-splice site, while most other positions only have ∼1% chance (Fig. 1B). In contrast, a GU dinucleotide located 4 nt downstream from the dominant splice site displays one of the lowest probabilities calculated (0.34%). These observations suggest that strong alternative 5′-splice sites located 4 nt downstream of the dominant splice site are selected against, most likely because intron sequences are not under protein-coding pressure. However, within the exon, as is the case for alternative splicing events 4 nt upstream from the dominant splice site, it is likely that the selection pressure to prevent the formation of a competing alternative 5′-splice site competes with the pressures to maintain the encoded amino acid.
FIGURE 1.
Distribution of alternative 5′-splice sites relative to the dominant splice site. (A) The frequency of alternative 5′-splice-site activation is shown in nucleotide resolution. The Y-axis indicates the number of alternative splice sites. The X-axis indicates the nucleotide distance between alternative and dominant splice sites. Alternative 5′-splice sites upstream from the exon/intron boundary are denoted as negative, those downstream are positive. (B) The probability that a GU is activated as an alternative 5′-splice site is plotted as a function of nucleotide distance from the dominant splice site. Each point was calculated by dividing the number of alternative splice-site usage events at a particular distance from the dominant splice site by the total number of times GU occurs at that same distance from splice sites within the human genome (∼125,000 exons). The Y-axis is shown in log scale. The asterisk at the −4 position denotes that the probability calculation at this position does not include the probability of 0.016 that a GC dinucleotide is used as a splice donor, which accounts for 46% of all events at this position.
FIGURE 2.
Alignment of alternative 5′-splice sites. (A) Sequence context around dominant and alternative 5′-splice sites. (B) Comparison of the nucleotide bias of constitutive 5′-splice sites (∼125,000 exons) with alternative splice-site activation at positions −4 and +4. The green bars indicate exon sequences of the dominant splice site. The red dashed bars designate exon sequences of the alternative splice site. The numbers to the right indicated the sample size used to generate the sequence pictograms.
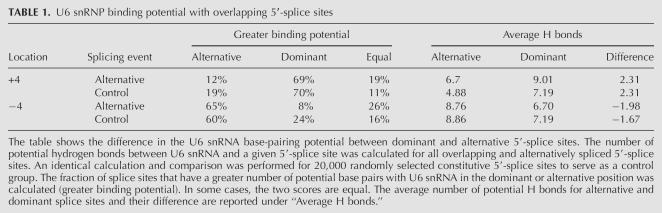
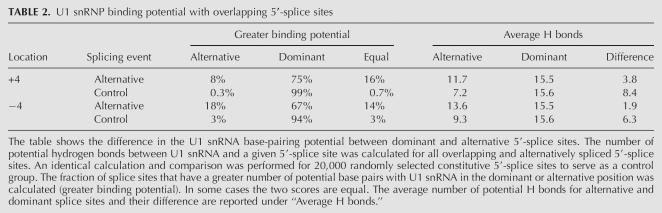
It is possible that alternative 5′-splice-site selection +4 or −4 nt from the dominant splice site results from overlapping U1 snRNA binding sites, or from differential interactions of U6 snRNA with the 5′-splice site, as recently described (Brackenridge et al. 2003). To evaluate these possibilities, we computed the U1 snRNA and U6 snRNA base-pairing potential for each alternative splice site. The differences in base-pairing potential were then compared with corresponding values generated from 20,000 randomly selected constitutive 5′-splice sites. On average, no significant differences in the U6 snRNA binding potential were observed between alternatively and constitutively spliced 5′-splice sites (Table 1). In contrast, when compared to the control group, our base-pair interaction analysis shows that the strength of U1 snRNP interaction in the +4 or −4 position is significantly stronger for alternatively spliced examples (p < 0.000001). On average, a difference of four to five hydrogen bonds is observed between alternative and constitutive 5′-splice sites (Table 2). Although we cannot exclude the possibility that differential U6 snRNA alignment promotes the selection of alternative 5′-splice sites for certain exons, the results of this comparative analysis suggest that the most common mechanism of alternative 5′-splice-site activation is based on differential U1 snRNA binding.
TABLE 1.
U6 snRNP binding potential with overlapping 5′-splice sites
TABLE 2.
U1 snRNP binding potential with overlapping 5′-splice sites
Surprisingly, 46% of the alternative 5′-splice sites located 4 nt upstream from the dominant splice site contain a G in their +1 position and a C in their +2 position instead of the canonical GU (Fig. 2B). This is a highly significant enrichment considering that only 0.37% of constitutive splice sites utilize a GC in their +1/+2 position (p < 0.0000001). Previous studies demonstrated that a GU at the exon/intron junction can be replaced by a GC and still allow accurate cleavage, however, at considerably slower rates (Aebi et al. 1987). Thus, it is plausible that a GC at the alternative splice site may create a kinetic advantage for splicing to the dominant splice site. Sequence analysis of all internal exons indicates that GC dinucleotides at positions −4/−3 relative to the dominant 5′-splice site are five times more likely to occur than GU dinucleotides. The probability of GU dinucleotides to occur at other upstream positions within the exon is 4%–5%. However, at the −4/−3 position, GU dinucleotides are observed with only a 1.4% probability. In contrast, the probability of GC in the −4/−3 position is increased from 6% to 8.4%. Therefore, it seems that GU dinucleotides are selected against at the −4/−3 position.
We hypothesized that coding pressure may account for the striking enrichment of GC dinucleotides at the −4/−3 position. To test this, we computed how frequently a GC-to-GU transition would result in an amino acid change at this position. This probability was then compared with the probability of an amino acid change when a GU is replaced with a GC at the same position, or equivalent transitions at GCs or GUs throughout the coding regions of randomly selected proteins. To do so, we used Ensembl to extract the complete nucleotide and amino acid sequence information for mRNA isoforms associated with −4 alternative splicing events. These sequences were then analyzed for their mutability at the alternative splice site. C-to-U transitions in the first and second codon positions always result in an amino acid change. In contrast, a C-to-U transition at the wobble position is always silent. Interestingly, within the group of alternative splicing events that utilize GC dinucleotides, mutating the GC to a GU produced a different amino acid 68% of the time, a frequency essentially identical to the mutability across the coding region (68% at the −4 position versus 66% within the coding region). This result is consistent with the interpretation that the preservation of the encoded amino acid dictates the nucleotide context at the alternative splice site rather than the pressure to avoid a GU at this position. In contrast, within the group of events using GU dinucleotides at the alternative site, the mutability was observed to be lower than the mutability across the coding region (54% at the −4 position versus 66% within the coding region; p = 0.01). Although the majority of GU-to-GC transitions at the −4/−3 position would result in amino acid changes, this comparison demonstrates that there is a significant enrichment of U in the third codon position. These results suggest that coding and splice-site selection pressures influence the nucleotide context for alternative splicing events at the −4 position that use GU dinucleotides. We conclude that the unusually high representation of GC dinucleotides at alternative splice sites 4 nt upstream from the dominant splice site is mainly influenced by coding pressure.
It is unclear from this analysis whether alternative 5′-splice-site selection 4 nt from the dominant splice site is a regulated and biologically relevant splicing event, or whether it occurs by chance. Representation alone within the EST database suggests that the transcripts have undergone positive selection to produce a message without a pre-termination stop codon, even though it is expected that a 4-nt addition or deletion would result in a reading frameshift. Indeed, >60% of +4 or −4 alternative transcripts produce protein products without triggering nonsense-mediated decay (NMD) (data not shown). Thus, it is likely that additional alternative splicing events may be involved to prevent the formation of a pre-termination stop codon. It is also possible that the alternative splicing event is located toward the 3′ end of the gene, such that NMD is not activated.
A comparison of alternative splicing probabilities at the +4 and −4 positions may also provide insights into the possibility of regulation. At the +4 position we determined one of the lowest probabilities measured within the series tested (0.34%) (Fig. 1B). Combined with the statistics that 75% of all alternative splice sites have a lower U1 snRNP binding potential (Table 2), these observations are more consistent with the proposal that specific activation of these events occur, and therefore, suggest regulation. A similar analysis for the −4 position is even less clear cut, because the probability of alternative 5′-splice-site activation at this position is significantly higher compared to the frequency at the +4 position (6.1% at −4 versus 0.34% at +4; p < 0.0025) (Fig. 1B), and only 67% of all alternative splice sites have a lower U1 snRNP binding potential (Table 2). However, the mutability analysis described above demonstrated that for the group of alternative splice sites using GU dinucleotides, splice-site selection pressures also contribute to the sequence context at the alternative splice site. In summary, these considerations suggest that some alternative 5′-splice-site selection events 4 nt from the dominant splice site may likely be under regulatory control.
Alternative 3′-splice-site selection
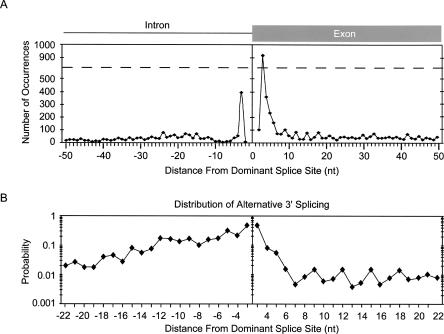
Splicing to the 3′-splice site by the spliceosome depends on three sequence elements; the 3′-splice-site consensus, which is designated by the trinucleotide YAG, the polypyrimidine tract immediately upstream of the intron/exon junction, and the branch-site sequence upstream from the polypyrimidine tract (Reed 2000). Depending on the location with respect to the dominant splice site, our analysis of alternative 3′-splice sites suggests different populations of alternative splicing events (Fig. 3A). The most common alternative splice sites activated are located within 6 nt of the dominant 3′-splice site. However, alternative AG activation close to the dominant splice site occurs more frequently within the exon than the intron, presumably because the requirement for a polypyrimidine tract immediately upstream from the dominant intron/exon junction lowers the representation of AG dinucleotides within this region.
FIGURE 3.
Distribution of alternative 3′-splice sites relative to the dominant splice site. (A) The frequency of alternative 3′-splice-site activation is shown in nucleotide resolution. The X-axis indicates the nucleotide distance between alternative and dominant splice sites. The Y-axis indicates the number of alternative splice sites. The dashed line indicates that the Y-scale is split. (B) The plot represents the probability that a YAG is activated as an alternative splice site. Each point was calculated by dividing the number of alternative splice-site usage events at a particular distance from the dominant splice site by the total number of times YAG occurs at that same distance from splice sites within the human genome (∼125,000 exons). YAG occurs in >95% of alternative and constitutive splice sites. The Y-axis is shown in log scale.
To determine the probability of alternative 3′-splice-site activation upstream or downstream of the dominant splice site, we computed the fraction of EST-verified alternative splicing events utilizing YAG over the total number of YAG trinucleotides that are found at every position within the human genome (Fig. 3B). Extending previous findings (Hiller et al. 2004), the most frequent alternative AG utilization was observed 3 nt on either side of the dominant splice site. However, if an alternative YAG is located within the polypyrimidine tract, there is a significantly greater probability that it will be used than if it is the same distance from the dominant splice site in the downstream direction.
Interactions of U2AF with the polypyrimidine tract and U2 snRNP with the branch site are essential for initial splice-site recognition (Black 2003). In many cases, the small subunit of U2AF interacts with the AG dinucleotide at the intron/exon junction (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). However, the first step of catalysis can occur without direct involvement of the AG dinucleotide at the 3′-intron/exon junction (Anderson and Moore 1997; Hertel and Maniatis 1999). Prior to the second step of catalysis, hSlu7 and SPF45 select the AG dinucleotide at the 3′-exon/intron junction (Chua and Reed 2001; Lallena et al. 2002), while Prp8-mediated rearrangements align the upstream exon with the AG dinucleotide (Konarska et al. 2006). During this step of the splicing reaction, the AG dinucleotide is essential for proper formation of the active site.
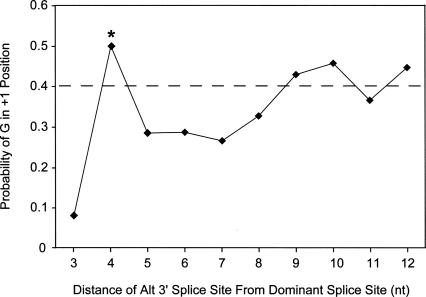
Alternative 3′-splice-site activation is usually mediated through differential U2AF binding. However, it is also possible that alternative 3′-splice-site activation within 6 nt of the dominant splice site can result from alternative AG selection and alignment during the second step of catalysis. That is, the first step of catalysis is carried out through branch-site activation of the dominant splice site, and an alternative intron/exon junction is selected during the alignment of the AG prior to the second step of catalysis (Chua and Reed 2001). Consistent with this hypothesis is the observation that the alternative 3′-splice sites that are located within 8 nt downstream from the dominant 3′-splice site do not contain a well-defined polypyrimidine tract. However, the emergence of a second polypyrimidine tract can be observed when the distance between the dominant and the alternative splice site increases past 8 nt (Figs. 4, 5) This trend is mirrored when evaluating the nucleotide bias at the +1 position of the dominant 3′-splice site. In general, there is a 40% prevalence of G at the +1 position (Figs. 4, 6). This bias is observed for both the alternative and dominant splice sites when the splice sites are sufficiently separated. However, the bias for G at the +1 position of the dominant splice site is lost when alternative splicing occurs near the dominant splice site, presumably because this position is under more selective pressure to favor the emergence of a polypyrimidine tract for the alternative splice site. Guanosine re-emerges as the preferred nucleotide in the +1 position of the dominant splice site after a polypyrimidine tract of sufficient length is formed for the alternative downstream splice site (Fig. 6). Although these analyses do not exclude the possibility that the selection of closely spaced splice acceptors occurs during the initial steps of exon definition, alternative selection of the AG dinucleotide prior to the second step of catalysis constitutes an attractive model to explain the distinct populations of alternative AG selection as observed in Figure 3A.
FIGURE 4.
Alignment of alternative downstream 3′-splice sites. Sequence comparison of constitutive 3′-splice sites (top) with alternative splice-site activation events at positions ranging from +3 to +12 nt from the dominant splice site. The green bars indicate exon sequences of the dominant splice site. The red dashed bars designate exon sequences of the alternative splice site. The numbers to the right indicated the sample size used to generate the sequence pictograms.
FIGURE 5.
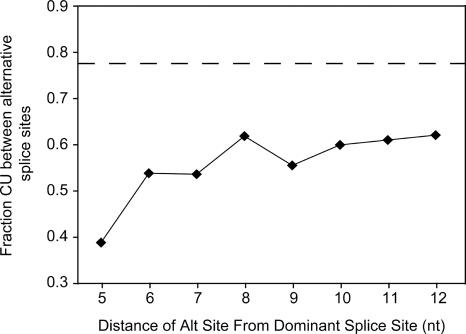
Pyrimidine content between dominant and alternative downstream 3′-splice sites. The average pyrimidine content (Y-axis) was calculated for nucleotides between the dominant and alternative 3′-splice sites and correlated to the nucleotide difference between the alternative splice sites (X-axis). The dashed line represents the average pyrimidine content of constitutively spliced 3′-splice sites. The sequence information required for this analysis was taken from the same data set as described for Figure 4.
FIGURE 6.
Conservation of guanosine in the first exon position of the dominant splice site. The probability of a G in the +1 position at the dominant 3′-splice site depends on the distance between the alternative and dominant splice sites. The X-axis displays the fraction guanosine at the +1 position of the dominant splice site, and the Y-axis represents the nucleotide distance between the splice sites. The dashed line represents the fraction of G at the +1 position of constitutive splice sites. The asterisk at the 4 position denotes an apparent irregularity within the +1 G bias plot. This is because the fourth nucleotide upstream of any 3′-splice junction is under no sequence constraints (see Fig. 4, constitutive). Thus, for alternative splicing 4 nt downstream from the dominant splice site, the constitutive +1 G bias (∼40%) reappears.
The high degree of alternative AG selection close to the dominant splice site implies that the spliceosome is flexible at accommodating differential AG selection during catalysis. Although the actual number of alternative upstream or downstream 3′-splice-site selections differs significantly (Fig. 3A), the probabilities of upstream and downstream events are comparable (Fig. 3B). Our analysis shows that AG dinucleotides within 6 nt upstream or downstream from the dominant splice site have up to a 48% chance to be activated as alternative splice sites. For alternative splicing events in close proximity to the dominant splice site, this frequent occurrence indicates that differential AG selection is more likely to occur than differential GU selection prior to the first step of catalysis. Assuming that final AG selection is carried out between the first and second steps of catalysis, these observations suggest that the spliceosome is more flexible at accommodating and aligning alternative splice acceptors during the second step of the splicing reaction. Alternative AG selections at a greater distance from the dominant splice site display a background level of ∼1% (Fig. 3B), similar in magnitude to what is observed for alternative 5′-splice-site selection at the same distance (Fig. 1B). These comparisons demonstrate that alternative 5′- and 3′-splice-site activations at greater distances from the dominant splice site have comparable probabilities.
As argued above, it is difficult to discern whether EST-verified differential splicing events are the product of regulation, or whether they are mainly the product of erroneous or stochastic splicing. This is because various processes, such as regulated splicing, inaccuracies of the splicing reaction, differential mRNA isoform stabilities, or differential cloning efficiencies, may influence the representation of an mRNA isoform in the EST database. Furthermore, it is reasonable to expect that the EST databases upon which ASD is based are incomplete in their representation of alternative splicing events. To deduce whether the frequent alternative splicing observed at duplicated splice acceptor sites in the +3 or −3 position is likely to be caused by regulation, we considered three independent parameters: their probability of occurrence, their conservation across species, and their tissue specificity. In a recent analysis of duplicated 3′-splice sites, Hiller et al. (2004) demonstrated that five out of 15 analyzed alternative splicing events displayed tissue-specific differential mRNA isoform expression. Significantly, these EST-based observations were supported by experimental verification, consistent with the hypothesis that some alternative splicing events at duplicated YAG trinucleotides are regulated.
We performed a genomic comparison between human and mouse alternative splicing to determine the conservation of YAGYAG intron/exon junctions and to evaluate their involvement in alternative splicing. Only 18% of YAGYAG intron/exon junctions are conserved between the species. Interestingly, the probability of alternative splicing within conserved and nonconserved groups of YAGYAG intron/exon junctions was calculated to be different (49% for conserved and 39% for nonconserved; p = 0.0026), demonstrating that conserved YAGYAG intron/exon junctions are slightly more likely to be targets of alternative splicing. As conservation of gene structure and alternative splicing has been associated with regulation, these observations provide additional support for the notion that at least a subset of conserved tandem 3′-splice sites may be regulated (Akerman and Mandel-Gutfreund 2006).
The probability that a YAGYAG intron/exon junction will occur suggests a different and equally feasible interpretation. Within the human genome, ∼50% of duplicated YAG trinucleotides at the 3′-splice site undergo alternative splice-site selection. Considering that the EST databases are still incomplete, it is expected that there are more alternative splicing events at this position than reported. Thus, the ∼50% chance of alternative alignment suggests that most, if not all, duplicated 3′-intron/exon junctions undergo alternative splice-site selection. Significantly, a similarly high probability of alternative 3′-splice-site activation was observed within the mouse genome (45%), regardless of conservation with the human transcriptome. These data are consistent with the interpretation that alternative splicing of many duplicated 3′-intron/exon junctions may be the consequence of flexible AG selection during the splicing reaction. Because of the high probability of splice-site usage, we propose that a significant fraction of alternative splicing at duplicated splice acceptor sites is likely to be stochastic rather than regulated.
Codon bias in alternative splicing
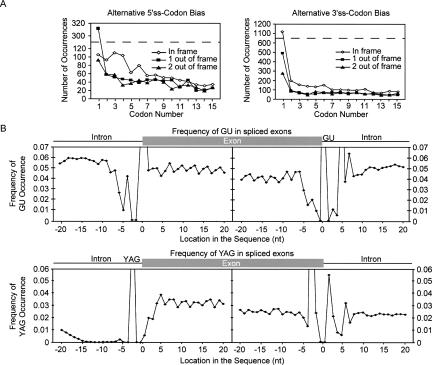
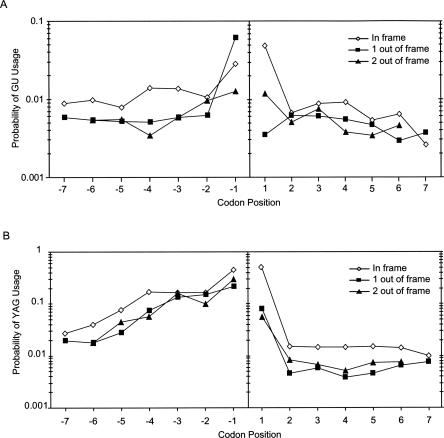
Close inspection of the alternative splicing profiles reveals a cyclic 3-nt character in the abundance of alternative splice-site usage, indicating a bias for translation frame preservation (Figs. 1A, 3A). Indeed, a comparison of in-frame with out-of-frame splicing events shows that the EST database overrepresents mRNAs that maintain the reading frame (Fig. 7A). These observations are consistent with an analysis demonstrating protein reading frame preservation for a subset of conserved alternative exon inclusion/exclusion events (Philipps et al. 2004; Resch et al. 2004; Magen and Ast 2005; Yeo et al. 2005). They are also consistent with recent microarray analyses showing that alternative exon inclusion/exclusion mRNA isoforms that are expected to trigger nonsense-mediated decay through the recognition of premature stop codons are detected only at low cellular concentrations (Pan et al. 2006). To control for sequence bias within the coding region that may cause the apparent frame preservation, we aligned all splice sites and determined the probability of YAG or GU within 20 nt upstream and downstream from each splice site (Fig. 7B). In agreement with previous work (Eskesen et al. 2004), we observe a periodicity of GU or YAG occurrence every 3 nt from the 5′- or 3′-splice site. However, after normalization to this coding region periodicity, a prominent in-frame bias for alternative 5′- and 3′-splice-site usage is still observed (Fig. 8). At positions closer than 6 nt downstream from the 3′-splice site, we still observe periodicity; however, it is overshadowed by the bias toward splice-site activation close to the dominant splice site. We conclude that quality-control steps, such as nonsense-mediated decay, extensively limit the expression of out-of-frame transcripts and prevent the translation of potentially harmful protein isoforms.
FIGURE 7.
Nucleotide bias within internal exons. (A) The number of in-frame and out-of-frame mRNAs generated through alternative 5′- or alternative 3′-splice-site activation is displayed as a function of codon distance from the dominant splice site. The codon number (X-axis) is defined as the number of 3-nt codons from the dominant splice site. (Open diamond) Represents alternative splicing events that do not change the reading frame. (Closed squares and triangles) Represent alternative splicing events that shift the reading frame by 1 and 2 nt, respectively. The dashed line indicates that the Y-scale is split. (B) Within exons the dinucleotide GU and the trinucleotide sequence YAG display a cyclic 3-nt distribution. Intron sequences are devoid of this cyclic character. The X-axis displays the nucleotide distance from the 3′- and 5′-splice site; the Y-axis represents the frequency (fraction) of occurrence.
FIGURE 8.
Reading frame bias of alternative splice-site usage. The probability of alternative (A) 5′- or (B) 3′-splice-site activation is shown as a function of codon number from the dominant splice site. Because each point was calculated by dividing the number of alternative splice-site usage events at a particular distance from the dominant splice site by the total number of times (A) GU or (B) YAG occurs at that same distance from splice sites, this probability calculation normalizes for the exonic sequence bias shown in Figure 7. (Open diamonds) Represent alternative splicing events that do not change the reading frame. (Closed squares and triangles) Represent alternative splicing events that shift the reading frame by 1 and 2 nt, respectively. The Y-axis is shown in log scale. On average, alternative splicing events that do not change the reading frame are approximately twofold more likely to be represented in ASD.
Here we show that the most common form of alternative splice-site activation in the human genome occurs close to the dominant splice site. Our analysis suggests that the U1 snRNP binding consensus sequence at the 5′-splice site is responsible for the most frequent form of alternative 5′-splice-site usage. The most frequent alternative 3′-splice-site activation could be explained by alternative alignment of duplicated YAG trinucleotides prior to the second step of catalysis. Although quality-control steps limit the expression of mRNA isoforms that generate out-of-frame proteins, extensive alternative pre-mRNA splicing promotes the evolution of novel functional isoforms.
MATERIALS AND METHODS
Frequency of alternative splicing events and codon bias
The Alternative Splicing Database was used as the source for genome and alternative splicing information (Thanaraj et al. 2004). For the analysis of the human genome, the reference transcript and the alternative splicing events files were downloaded from ASD. Alternative 5′- and 3′-splicing events were extracted from the exon and intron isoform files, and split into two files containing alternative 5′- or 3′-splicing events. The resulting alternative 5′- and 3′-splicing events files contained information for 9000 and 11,700 events, respectively. For each of these events, the distance of the alternative splice site from the dominant splice site was recorded. To derive frequency information, the number of times a splicing event occurred in relation to the distance from the dominant splice site was counted (Altn).
To evaluate whether ASD overrepresented alternative splicing events that maintained the reading frame, the alternative 5′- and 3′-splicing event files were split up into three separate files: in frame (multiple of three from the dominant splice site), 1 out of frame, and 2 out of frame.
Probability of YAG and GU utilization
The alternative splicing event file was compared to an ASD list of all genes in the human genome to determine which exons are not associated with any type of alternative splicing events to generate an annotated list of constitutively spliced exons. Taking advantage of the gene and exon annotations, the nucleotide sequence of the corresponding exon/intron junctions was extracted from Ensembl and recorded for each exon. This sequence information was used to compute the actual number of YAG trinucleotides (for 3′-splice sites) or GU dinucleotides (for 5′-splice sites) occurring at each position within a 25-nt window of the exon/intron junction of all constitutively spliced exons (Constn). From the resulting sequence information, the probability of alternative 5′- or 3′-splice-site utilization at various distances from the dominant splice site was calculated according to: (Altn)/[(Altn) + (Constn)]. This calculation was performed for every position within the 25-nt window of the dominant splice site.
Prediction of U1 snRNP and U6 snRNP binding potential to alternative 5′-splice sites
To evaluate the U1 snRNP and U6 snRNP binding potential to overlapping 5′-splice sites, the number of potential hydrogen-bond interactions between each snRNA and each 5′-splice site was determined (Fig. 2). As recently described, this approach has been successfully applied to compare and predict the strength of U1 snRNP binding to the 5′-splice site (Freund et al. 2003). The U1 snRNA sequence 5′-ACUUACCUG-3′ was tested for complementarity to each queried 5′-splice site sequence spanning nucleotides −3 to +6. Similarly, the U6 snRNA sequence 5′-AUACAGAGAA-3′ was compared to each queried 5′-splice site sequence spanning nucleotides −2 to +8. In each case, the binding potential was estimated by calculating the sum of hydrogen bonds from all possible base-pair interactions. Three hydrogen bonds were allocated for a GC interaction, and two hydrogen bonds were allocated for an AU interaction (Freund et al. 2003). This calculation was performed for +4 and −4 alternative 5′-splice sites and their respective dominant splice sites. As a control group, 20,000 randomly selected constitutive 5′-splice sites were also analyzed. For this group, hydrogen bonds were calculated for the constitutive 5′-splice site and for the equivalent U1 and U6 snRNA alignments 4 nt upstream or downstream from the constitutive splice site.
Sequence analysis
Sequence data for each alternative 5′- and 3′-splicing event were retrieved from the Ensembl genome database. To validate the correct alternative splicing event, the length of the intron and exon affected by the alternative spicing event was matched to our alternative splicing event file. Nucleotide frequencies were calculated for verified alternative splicing events with sequence information. Alternative splicing events were grouped based on their distance from the dominant splice site. From these files, sequence consensus pictograms were created using a Web site tool made available by C. Burge's laboratory, http://genes.mit.edu/pictogram.html.
To evaluate the relative strength of the polypyrimidine tracts upstream from alternative 3′-splices, an average pyrimidine concentration was calculated. Sequence information for all downstream alternative 3′-splicing events was derived from Ensembl. The alternative splicing events were grouped into files depending on the distance of the alternative site from the dominant site. The fraction of (C + U)/(number of positions) was then calculated for every set of alternative splicing events. Calculations for the average percent CU were initiated at the +1 position relative to the dominant splice site and the −5 position relative to the alternative splice site.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM 62287 (to K.J.H.). We thank members of the Hertel Laboratory for critical reading of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.151106.
REFERENCES
- Aebi, M., Hornig, H., Weissmann, C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- Akerman, M., Mandel-Gutfreund, Y. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res. 2006;34:23–31. doi: 10.1093/nar/gkj408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K., Moore, M.J. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Brackenridge, S., Wilkie, A.O., Screaton, G.R. Efficient use of a ‘dead-end’ GA 5′ splice site in the human fibroblast growth factor receptor genes. EMBO J. 2003;22:1620–1631. doi: 10.1093/emboj/cdg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, K., Reed, R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskesen, S.T., Eskesen, F.N., Kinghorn, B., Ruvinsky, A. Periodicity of DNA in exons. BMC Mol. Biol. 2004;5 doi: 10.1186/1471-2199-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund, M., Asang, C., Kammler, S., Konermann, C., Krummheuer, J., Hipp, M., Meyer, I., Gierling, W., Theiss, S., Preuss, T., et al. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 2003;31:6963–6975. doi: 10.1093/nar/gkg901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel, K.J., Maniatis, T. Serine-arginine (SR)-rich splicing factors have an exon-independent function in pre-mRNA splicing. Proc. Natl. Acad. Sci. 1999;96:2651–2655. doi: 10.1073/pnas.96.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M., Huse, K., Szafranski, K., Jahn, N., Hampe, J., Schreiber, S., Backofen, R., Platzer, M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Query, C.C. Insights into the mechanisms of splicing: More lessons from the ribosome. Genes & Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Vilardell, J., Query, C.C. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Krämer, A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lallena, M.J., Chalmers, K.J., Llamazares, S., Lamond, A.I., Valcarcel, J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Lim, S.R., Hertel, K.J. Commitment to splice site pairing coincides with A complex formation. Mol. Cell. 2004;15:477–483. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Magen, A., Ast, G. The importance of being divisible by three in alternative splicing. Nucleic Acids Res. 2005;33:5574–5582. doi: 10.1093/nar/gki858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino, L., Guth, S., Bilbao, D., Martinez, C., Valcarcel, J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Saltzman, A.L., Kim, Y.K., Misquitta, C., Shai, O., Maquat, L.E., Frey, B.J., Blencowe, B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipps, D.L., Park, J.W., Graveley, B.R. A computational and experimental approach toward a priori identification of alternatively spliced exons. RNA. 2004;10:1838–1844. doi: 10.1261/rna.7136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- Reed, R., Palandjian, L. Spliceosome assembly. In: Krainer A.R., editor. Eukaryotic mRNA processing. IRL Press; Oxford: 1997. pp. 103–129. [Google Scholar]
- Resch, A., Xing, Y., Alekseyenko, A., Modrek, B., Lee, C. Evidence for a subpopulation of conserved alternative splicing events under selection pressure for protein reading frame preservation. Nucleic Acids Res. 2004;32:1261–1269. doi: 10.1093/nar/gkh284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj, T.A., Stamm, S., Clark, F., Riethoven, J.J., Le Texier, V., Muilu, J. ASD: The Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., Green, M.R. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Yeo, G.W., Van Nostrand, E., Holste, D., Poggio, T., Burge, C.B. Identification and analysis of alternative splicing events conserved in human and mouse. Proc. Natl. Acad. Sci. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio, D.A., Blumenthal, T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans . Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]