Abstract
Mammalian H/ACA RNPs are essential for ribosome biogenesis, pre-mRNA splicing, and telomere maintenance. To form mature RNA–protein complexes, one H/ACA RNA associates with four core proteins. In the cell, this process is assisted by at least one nuclear assembly factor, NAF1. Here we report several unanticipated dynamic aspects of H/ACA RNP proteins. First, when overexpressed, NAF1 delocalizes to the cytoplasm. However, its nucleocytoplasmic shuttling properties remain unaffected. These observations demonstrate a subtle equilibrium between NAF1 expression levels and the availability of NAF1 nuclear binding sites. Second, although NAF1 is excluded from mature RNPs in nucleoli and Cajal bodies, NAF1 associates with mature H/ACA RNA in cell lysates. This association occurs post-lysis because it is observed even when NAF1 and the H/ACA RNA are expressed in separate cells. This documents a protein–RNP association in cell lysates that is absent from intact cells. Third, in similar experiments, all H/ACA core proteins, except NAP57, exchange with their exogenous counterparts, portraying an unexpected dynamic picture of H/ACA RNPs. Finally, the irreversible association of only NAP57 with H/ACA RNA and the conundrum that only NAP57 is mutated in X-linked dyskeratosis congenita (even though most core proteins are required for maintaining H/ACA RNAs) may be more than a coincidence.
Keywords: H/ACA RNP, snoRNP, dyskeratosis congenita, nucleocytoplasmic shuttling, RNP dynamics, telomerase, nuclear localization signal
INTRODUCTION
Mammalian H/ACA RNPs function mainly in the pseudouridylation of ribosomal and spliceosomal small nuclear RNAs but also in the processing of preribosomal RNA and in the stabilization of telomerase RNA. They all consist of one of the ∼200 different H/ACA RNAs and the same four evolutionary conserved core proteins, NAP57 (dyskerin or Cbf5p in yeast), NOP10, NHP2 (forming a core trimer), and GAR1 (Smith and Steitz 1997; Kiss 2001; Filipowicz and Pogacic 2002; Decatur and Fournier 2003; Meier 2005). In human cells, formation of these particles and consequent stabilization of the H/ACA RNA depends on at least one nuclear assembly factor, NAF1 (Darzacq et al. 2006; Hoareau-Aveilla et al. 2006).
It is commonly regarded that the four core proteins and the H/ACA RNAs form stable RNPs with no exchange of components. This is based on the following three observations. First, all core proteins, except GAR1, are essential for the stabilization of all H/ACA RNAs (Bousquet-Antonelli et al. 1997; Henras et al. 1998; Lafontaine et al. 1998; Watkins et al. 1998; Dez et al. 2001). Second, all H/ACA RNPs can be precipitated with antibodies directed against any of the core proteins, including GAR1 (Balakin et al. 1996; Ganot et al. 1997). Third, once an H/ACA RNA has assembled into an RNP, it cannot be competed for with an excess of the same RNA (Wang and Meier 2004). Surprisingly, we now demonstrate that the only protein irreversibly associated with H/ACA RNAs is NAP57.
H/ACA RNPs are also of general interest because some of their components are mutated in the often-fatal bone marrow failure syndrome dyskeratosis congenita (Dokal and Vulliamy 2003; Marrone and Mason 2003). The X-linked form of dyskeratosis congenita (X-DC) is caused by mostly missense mutations in NAP57, whereas the milder and more rare autosomal dominant form is caused by mutations in telomerase reverse transcriptase or RNA (Heiss et al. 1998; Mitchell et al. 1999; Vulliamy et al. 2001a; Armanios et al. 2005). Human telomerase RNA is an H/ACA RNA and is stabilized by the proteins of the H/ACA core trimer (Mitchell et al. 1999; Dragon et al. 2000; Dez et al. 2001). A loss of telomerase RNA and shortened telomeres have been observed in cells of X-DC patients (Mitchell et al. 1999; Vulliamy et al. 2001b). Since mutations in H/ACA core proteins could impact the stability of H/ACA RNPs, it has been a mystery as to why X-DC mutations have only been identified in NAP57 but none of the other core proteins (Marrone and Mason 2003). Our findings provide a possible explanation for this enigma.
RESULTS AND DISCUSSION
Differential localization and abundance of endogenous and exogenous NAF1
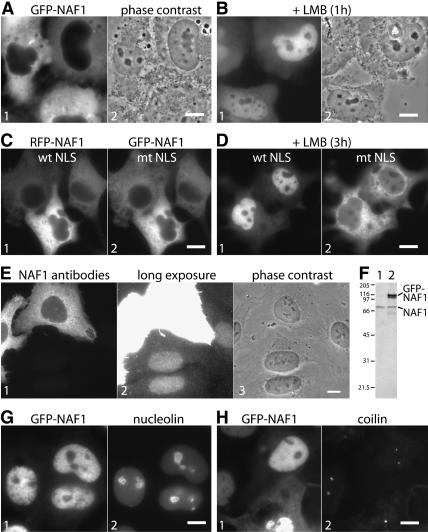
Human NAF1 is concentrated in the nucleus but excluded from sites of mature H/ACA RNPs, the nucleoli and Cajal bodies (Darzacq et al. 2006; Hoareau-Aveilla et al. 2006). Surprisingly, when NAF1 was fused to the green fluorescent protein (GFP) and transiently expressed, it accumulated mostly in the cytoplasm (Fig. 1A). To test if this discrepancy reflected a mere difference in steady-state distribution and if GFP-NAF1 shuttled between the nucleus and cytoplasm like endogenous NAF1 (Darzacq et al. 2006), we blocked CRM1-mediated nuclear export by incubation with leptomycin B (LMB). As a consequence, GFP-NAF1 accumulated in the nucleoplasm, suggesting that it was rapidly imported but prevented from returning to the cytoplasm (Fig. 1B).
FIGURE 1.
Localization and shuttling of transiently expressed NAF1. (A) Direct fluorescence of COS cells transiently transfected with GFP-NAF1 (panel 1) and corresponding phase contrast image (panel 2). (B) Same as A but 1 h after leptomycin B (LMB) addition. (C) Direct double fluorescence of cotransfected wild-type RFP-NAF1 and GFP-NAF1 containing a mutated NLS. (D) Same as in C but 3 h after LMB addition. Note the nuclear accumulation of the tagged NAF1 containing a wild-type (panel 1) compared with a mutated NLS (panel 2). (E) Indirect immunofluorescence of U2OS cells transfected with untagged NAF1 and probed with affinity purified NAF1 peptide antibodies (panel 1), a longer exposure of the same image (panel 2, note the nuclear localization of the endogenous NAF1), and the corresponding phase contrast image (panel 3). (F) Western blot of extracts from U2OS cells untransfected (lane 1) or transfected (lane 2) with GFP-NAF1, probed with NAF1 antibodies, and developed by enhanced chemiluminescence. The migrating positions of endogenous NAF1, the transfected GFP-NAF1, and the molecular weight markers in kilodaltons are indicated. (G) Double fluorescence of GFP-NAF1 (panel 1) transfected COS cells probed with nucleolin antibodies (panel 2) 3 h after LMB addition. Note the exclusion of GFP-NAF1 from nucleoli. (H) Same as G but probed with coilin antibodies. Note the lack of enrichment of GFP-NAF1 in Cajal bodies identified by coilin. Bars, 10 μm.
CRM1 exports nuclear export signal containing proteins (Fornerod et al. 1997). Indeed, scanning the amino acid sequence of human NAF1 revealed several potential nuclear export signal consensus sequences. However, there was only one classical bipartite nuclear localization signal (NLS) present consisting of two monopartite NLSs, 324 KQRKKSQIQGRKKLK 338. To test if this NLS was responsible for nuclear import of GFP-NAF1, four lysines were mutated to alanines 324KQRAASQIQGRAALK338. When this GFP-NAF1 with the mutated NLS was cotransfected with wild-type NAF1 fused to RFP (Fig. 1C) and the cells were incubated with LMB to monitor nuclear import, all of the wild-type RFP-NAF1 was imported, but GFP-NAF1 with the mutated NLS remained mostly excluded from the nuclei (Fig. 1D, cf. panels 1 and 2). This finding indicates that the above-identified NLS in NAF1 is at least partially responsible for the nuclear import of NAF1. Other factors that may contribute to its nuclear import could be cryptic or nonclassical NLSs or, more interestingly, its binding partner NAP57, which contains several classical and functional NLSs of its own (Meier and Blobel 1994; Heiss et al. 1999). Specifically, NAF1 and NAP57 may enter the nucleus in a complex aided by the NLSs of both proteins. Activity of NLSs from both proteins may be crucial for the import of the larger complex of the core trimer that includes the NLS-less NOP10 and NHP2 (Wang and Meier 2004; Darzacq et al. 2006).
The differential localization of the exogenous GFP-NAF1 and the endogenous NAF1 was surprising because GFP tagging of a protein in most cases does not interfere with its proper localization. To test if indeed the tag was responsible for the mislocalization of the exogenously expressed NAF1, we replaced the GFP tag with the lacI repressor or the much shorter hemagglutinin epitope. Since none of these tags changed the cytoplasmic mislocalization of exogenous NAF1 (data not shown; Darzacq et al. 2006), untagged NAF1 was transfected and its localization monitored using NAF1 antibodies (Fig. 1E). Untagged NAF1 accumulated predominantly in the cytoplasm with a small amount detectable in the nucleus (Fig. 1E, panel 1). The nuclear fraction of exogenous NAF1, although barely visible at this short exposure, represented a much higher concentration than that of the endogenous NAF1, which was visible in the nuclei of untransfected cells only after a longer exposure (Fig. 1E, panel 2). On Western blots of whole-cell extracts, the transfected GFP-NAF1 was much more abundant than endogenous NAF1 even though the tagged protein was only expressed in ∼10%–20% of all cells (Fig. 1F, lane 2). Therefore, transfected NAF1 is overexpressed and appears to saturate limiting nuclear binding sites for NAF1 causing a “cytoplasmic overflow.” For example, such nuclear binding sites could be represented by NAP57 that has not yet incorporated into RNPs and that may depend on NAF1 for stabilization (Darzacq et al. 2006). As endogenous NAF1 was solely nuclear and as transfected GFP-NAF1 was cytoplasmic as early as 6 h post-transfection even in low expressing cells, NAF1 appears to be a low-abundance protein. Indeed, a small amount of NAF1 should be sufficient to chaperone the small number of newly synthesized NAP57 molecules before incorporation into RNPs at the site of H/ACA RNA transcription (Darzacq et al. 2006).
Exchange of H/ACA RNP proteins in cell lysates
We previously showed that NAF1, when tethered to a chromosomal locus in the nucleus, recruits NAP57 but not H/ACA RNAs (Darzacq et al. 2006). This lack of cellular association of NAF1 with mature H/ACA RNPs is further supported by the failure of NAF1 to concentrate in nucleoli and Cajal bodies, the sites of mature H/ACA RNPs (Darzacq et al. 2006; Hoareau-Aveilla et al. 2006). To test if overexpression of NAF1 in the nucleus could overcome this lack of association, GFP-NAF1 was transiently expressed in COS cells and concentrated in the nuclei by addition of LMB to prevent its export. Even under these excessively concentrated conditions, GFP-NAF1 remained excluded from nucleoli, marked by nucleolin, and failed to accumulate in Cajal bodies, identified by coilin (Fig. 1G,H). As previously noted in yeast and mammalian cells, therefore, NAF1 associates only to a very minor degree, if at all, with H/ACA RNA containing RNPs (Dez et al. 2002; Fatica et al. 2002; Yang et al. 2002; Darzacq et al. 2006). Unexpectedly, a GFP-NAF1 interaction with H/ACA RNA was observed when analyzed biochemically.
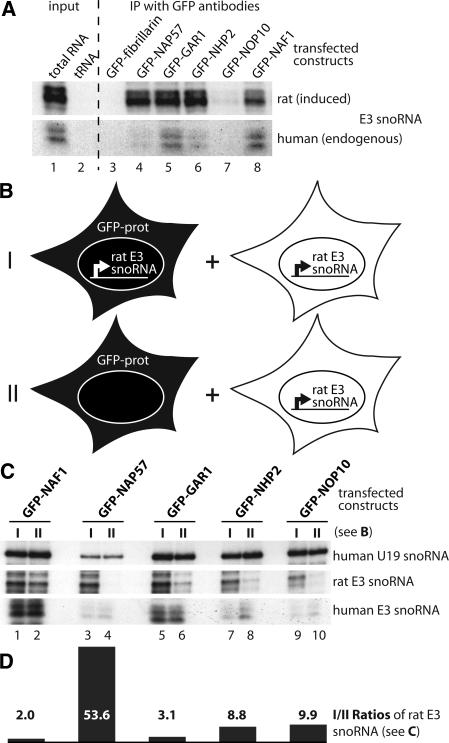
We established a stable human U2OS cell line that expresses the rat H/ACA RNA E3 from an inducible promoter (Darzacq et al. 2006). To test if H/ACA core proteins associated with this RNA, GFP-tagged proteins were transiently transfected into this cell line and H/ACA RNA transcription was induced, followed by cell lysis and immunoprecipitation of the fusion proteins with antibodies to GFP. The presence of the induced rat E3 (Fig. 2A, upper panel) and the endogenous human E3 snoRNA (lower panel) in the precipitates was analyzed by an RNase protection assay (Darzacq et al. 2006). Most GFP-H/ACA core proteins, but not the C/D RNP control protein GFP-fibrillarin, efficiently precipitated the full-length induced H/ACA RNA E3 (Fig. 2A, upper panel, lanes 3–7). The stabilization of the full-length rat E3 snoRNA suggested that these core proteins were correctly incorporated into an RNP. This conclusion was supported by our previous observation that knockdown of NAP57 causes a destabilization and consequent loss of the E3 snoRNA in these cells (Darzacq et al. 2006). The core protein GFP-NOP10 exhibited a reduced association likely caused by the size of the GFP tag, which was about four times that of NOP10 (Fig. 2A, lane 7). Specifically, the GFP tag may slow down NOP10 association with newly synthesized snoRNAs, as in the case of the induced E3 snoRNA (Fig. 2A, lane 7), but eventually incorporate into the snoRNP, as in the case of the snoRNP U19 (Fig. 2C, lanes 9,10). Nevertheless, GFP-NAF1, despite its lack of association with mature H/ACA RNPs in intact cells, precipitated the full-length E3 snoRNA as efficiently as did the H/ACA core proteins (Fig. 2A, lane 8).
FIGURE 2.
Association of GFP-tagged proteins with H/ACA RNAs in cell lysates. (A) Autoradiograph of induced rat (upper panel) and endogeous human E3 snoRNA (lower panel) detected by RNase A/T1 protection assay in total U2OS cell RNA 24 h after induction of the rat E3 transgene (lane 1) and, as a negative control, in yeast tRNA (lane 2). Lanes 3–8 show the same as lane 1, but the RNA was isolated from immunoprecipitates with GFP antibodies from E3 cell lysates transfected with the indicated GFP constructs and induced for rat E3 expression. One-tenth cell equivalent was used in lane 1 compared to lanes 3–8. (B) Schematic of the two GFP-construct transfection and rat E3 snoRNA induction approaches (I,II) used in C for immunoprecipitation with GFP antibodies followed by snoRNA analysis. Briefly, I was a repetition of the experiment in A, but in II the GFP-tagged constructs were transfected into parent U2OS cells lacking the rat E3 snoRNA transgene. In both cases the transfected cells were mixed with an equal number of untransfected U2OS cells (but induced for transgene expression) and lysed for immunoprecipitation with anti-GFP antibodies. (C) RNase protection analysis of the H/ACA snoRNAs in the immunoprecipitates of the GFP-tagged constructs (indicated on top) following the experimental approach (I, odd lanes) and (II, even lanes) in B. Note that in the cell lysates, only GFP-NAP57 failed to associate with the rat E3 snoRNA expressed in separate cells (lane 4). (D) Graph of the ratios between odd and even lanes from the center panel (rat E3 snoRNA) of C corrected for human E3 snoRNA. The values represent the average from two independent RNase protection assays.
Given the efficient association of GFP-NAF1 with mature E3 particles in cell lysates but not in intact cells, we tested if it bound to the induced H/ACA RNA only after cell lysis. For this purpose, we compared the outcome of two experimental approaches (Fig. 2B, I,II). In the first approach, we basically repeated the previous experiment (Fig. 2A) by transfecting the GFP-tagged constructs into U2OS cells expressing the rat E3 snoRNA followed by immunoprecipitation of the GFP-fusion proteins (Fig. 2B, I). In the second approach, the GFP-tagged constructs were transfected into parent U2OS cells (lacking the rat E3 gene), whereas the rat E3 snoRNA was expressed in separate cells (Fig. 2B, II). In both cases, 2 d post-transfection, an equal number of transfected and untransfected (but rat E3 snoRNA induced) cells were mixed, lysed, and processed for immunoprecipitation with anti-GFP antibodies. Endogenous and induced H/ACA snoRNAs were detected using RNase protection analysis (Fig. 2C). In both cases, all GFP-fusion proteins associated with the endogenous human H/ACA snoRNAs U19 and E3 (Fig. 2C, top and bottom panels). In addition, GFP-NAF1 precipitated the induced rat E3 snoRNA regardless of whether it was transfected into cells expressing E3 or not (Fig. 2C, center panel, cf. lanes 1 and 2). The latter indicated that GFP-NAF1 associated with the rat E3 snoRNA after cell lysis, presumably by binding to NAP57 and by replacing GAR1 in the H/ACA RNP as predicted from our competitive in vitro and in vivo binding studies (Darzacq et al. 2006). In agreement with this interpretation, GFP-GAR1 also exchanged between mature rat E3 RNPs (Fig. 2C, center panel, cf. lanes 5 and 6). In contrast, GFP-NAP57 only associated with rat E3 snoRNA if it was transfected into the cells expressing rat E3 (Fig. 2C, center panel, cf. lanes 3 and 4). This demonstrated that the major core protein and pseudouridylase NAP57 bound only to newly synthesized H/ACA RNA, consistent with its presence at the H/ACA RNA transcription site (Darzacq et al. 2006), but not to that in preexisting RNPs. Consequently, NAP57 was unable to exchange post-RNP assembly. After transfection into non-E3 expressing U2OS cells, GFP-NHP2 and GFP-NOP10 also associated with the rat E3 snoRNA expressed in separate cells (Fig. 2C, center panel, cf. lanes 7 and 8, lanes 9 and 10, respectively). Specifically, when the amount of precipitated E3 snoRNA was adjusted relative to that of the endogenous E3 and U19 snoRNAs, exchange of GFP-NHP2 and GFP-NOP10 between mature snoRNPs was over twofold less than that of GFP-GAR1 and GFP-NAF1 (Fig. 2C,D). This finding was consistent with NOP10 and NHP2 forming part of a core trimer with NAP57. Nevertheless, NAP57 alone was primarily responsible for the stability of H/ACA RNPs and for our previously documented lack of RNA exchange between H/ACA RNPs (Wang and Meier 2004).
Protein dynamics of H/ACA RNPs
In summary, our observations show that H/ACA RNPs are dynamic RNA–protein complexes that allow exchange of all core proteins except for NAP57. NAP57 is the largest and catalytic subunit of the RNP protein moiety. It associates tightly with each newly synthesized H/ACA RNA, likely requiring de novo synthesis of both for every new particle. The tight NAP57-H/ACA RNA association marries substrate recognition by the RNA with catalysis by the pseudouridylase mirroring the unification of these two activities in pseudouridylases that only consist of a single protein. Therefore, this tight RNA–protein association in H/ACA RNPs may be rooted in the evolution of pseudouridylases. Conversely, the exchange of the other core proteins could reflect a possible rearrangement or breathing of the complex during catalysis. For example, recent crystal structures of partial archaeal H/ACA protein complexes demonstrate how NOP10 brackets the catalytic domain of NAP57 (Hamma et al. 2005; Manival et al. 2006; Rashid et al. 2006). During catalysis this NOP10–NAP57 interaction may need to be loosened to allow access to and/or release of substrate RNAs.
Unexpectedly, our studies reveal a differential localization of the exogenously and endogenously expressed assembly factor NAF1 and an H/ACA RNP association in cell lysates not observed in vivo. The differential localization was independent of protein tags and apparently caused by overexpression of the low-abundance NAF1. Meanwhile, the NAF1 association with H/ACA RNPs in cell lysates may well be due to the loss of cellular compartmentalization in the lysates. Alternatively or as a result of decompartmentalization, a post-translational modification of NAF1 or NAP57 that is usually sequestered (e.g., a phosphorylation or dephosphorylation event) could be responsible for the in vitro phenomenon. Regardless, our findings signify a caveat when working with RNPs in cell extracts, particularly in the light of similar observations with the RNA binding protein HuR and its interaction with c-fos mRNA (Mili and Steitz 2004).
Finally, our findings may shed light on a long-standing conundrum concerning the bone marrow failure syndrome X-linked dyskeratosis congenita. Two hallmarks of cells from patients with X-DC are shortened telomeres and loss of telomerase RNA (Mitchell et al. 1999). Similar to other H/ACA RNAs, human telomerase RNA is stabilized by the H/ACA core proteins (Mitchell et al. 1999; Dragon et al. 2000; Dez et al. 2001). Genetic depletion studies in yeast demonstrated that all H/ACA core proteins, except GAR1, are essential for the stability of H/ACA RNAs (Bousquet-Antonelli et al. 1997; Henras et al. 1998; Lafontaine et al. 1998; Watkins et al. 1998). Nevertheless, X-DC mutations have only been identified in NAP57 but none of the other core proteins (Marrone and Mason 2003). Therefore, the tight association of NAP57 with H/ACA RNAs may make this bimolecular complex particularly sensitive to perturbations and may provide an explanation for why mutations in NAP57 result in X-DC.
MATERIALS AND METHODS
Tissue culture, transfection, and cell lines
Hela and COS-1 cells were cultured in high-glucose DMEM (GIBCO) containing 10% fetal bovine serum (FBS, GIBCO). The rat E3 snoRNA inducible U2OS cells were generated as described previously (Darzacq et al. 2006) and cultured in low-glucose DMEM (GIBCO) containing 10% FBS. Cells were transiently transfected with expression plasmids using FuGene 6 (Roche) for 24–30 h according to the manufacturer's protocol. Transcription of the transgene was induced using 6 μg/mL doxycycline overnight. For shuttling studies, 10 ng/mL LMB (Biomol) was added.
DNA/RNA constructs
The following plasmids were used for transient transfection and have been described previously: GFP-NAF1 (pNK16, NAF1 fused to monomeric GFP), RFP-NAF1 (pNK31, NAF1 fused to monomeric RFP), and NAF1 (pNK40) (Darzacq et al. 2006); GFP-NAP57, NOP10-GFP, NHP2-GFP, and GAR1-GFP (Pogacic et al. 2000); and GFP-fibrillarin (Platani et al. 2000). GFP-NAF1 with the mutated NLS (pTM184) was generated based on pNK16 by pairwise replacement of two lysines by alanines using the QuickChange II kit (Stratagene) according to the manufacturer's instructions. The probes for RNase protection assays of rat and human E3 snoRNA (Darzacq et al. 2006) and for U19 (Kiss et al. 1996) were as published.
Fluorescence methods and immunochemicals
For indirect immunofluorescence, cells were permeabilized and processed as we described previously (Isaac et al. 1998), and the following primary antibodies were used (dilutions and source given in parentheses): polyclonal anti-NAF1 IgGs (CRX6 at 1:100) (Darzacq et al. 2006), anti-coilin ascites fluid (5P10 at 1:1000) (Almeida et al. 1998), and nucleolin ascites fluid (7G2 at 1:1000) (Pinol-Roma, 1999). The following fluorescently labeled secondary antibodies were used: goat anti-rabbit IgGs at 1:200 (rhodamine, Chemicon) and goat anti-mouse IgGs at 1:1000 (rhodamine, Jackson ImmunoResearch).
Imaging was performed at the Analytical Imaging Facility of the Albert Einstein College of Medicine. A 60×, 1.4 numerical aperture planapo objective was used on an Olympus IX 81 inverted microscope containing automatic excitation and emission filter wheels connected to a Sensicam QE air-cooled charge-coupled device camera (Roper Scientific) run by IPLab Spectrum software (Scanalytics). Images were processed for contrast and brightness using Adobe Photoshop CS2 (Adobe Systems Inc.).
RNase protection assay and RNP precipitation
RNase A/T1 protection analysis was performed essentially as described (Goodall et al. 1990). Anti-sense probes were synthesized in the presence of [α-32P]-CTP using the Maxiscript kit (Ambion) and gel purified; 1–5 μg of total RNA (prepared with TRIzol, Invitrogen) was mixed with probe (50,000 cpm) for hybridization and digestion with 40 μg/mL RNase A (Roche) and 2 μg/mL RNase T1 (Calbiochem). Typically, half of the samples were analyzed in 6% polyacrylamide–8 M urea sequencing gels followed by autoradiography. Bands were quantitated using a PhosphorImager (Molecular Dynamics).
RNPs were immunoprecipitated essentially as described (Darzacq et al. 2002). Briefly, after transfection of GFP-tagged constructs with FuGene 6 (Roche) into 1.2 × 106 cells and simultaneous induction of the transgene with 6 μg/mL doxycycline for 24 h, the cells were lysed and sonicated in 500 μL of 40 mM Tris-HCl (pH 7.4), 300 mM NaCl, 0.01% Triton X-100, and the RNPs precipitated with anti-GFP agarose beads (Medical & Biological Laboratories). After proteinase K treatment of the RNPs, the snoRNAs were extracted and analyzed by RNase protection.
ACKNOWLEDGMENTS
We thank Maria Carmo-Fonseca, Angus Lamond, and Serafin Pinol-Roma for contributing the referenced reagents. We acknowledge the services of the Albert Einstein Analytical Imaging Facility. This work was supported by grants from the National Institutes of Health HL079566 to U.T.M. and EB2060 to R.H.S.
Footnotes
Artilce published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.249306.
REFERENCES
- Almeida, F., Saffrich, R., Ansorge, W., Carmo-Fonseca, M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 1998;142:899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios, M., Chen, J.L., Chang, Y.P., Brodsky, R.A., Hawkins, A., Griffin, C.A., Eshleman, J.R., Cohen, A.R., Chakravarti, A., Hamosh, A., et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl. Acad. Sci. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin, A.G., Smith, L., Fournier, M.J. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Henry, Y., Gélugne, J.-P., Caizergues-Ferrer, M., Kiss, T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 1997;16:4770–4776. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq, X., Jady, B.E., Verheggen, C., Kiss, A.M., Bertrand, E., Kiss, T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq, X., Kittur, N., Roy, S., Shav-Tal, Y., Singer, R.H., Meier, U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W.A., Fournier, M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- Dez, C., Henras, A., Faucon, B., Lafontaine, D., Caizergues-Ferrer, M., Henry, Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p, and Nop10p. Nucleic Acids Res. 2001;29:598–603. doi: 10.1093/nar/29.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez, C., Noaillac-Depeyre, J., Caizergues-Ferrer, M., Henry, Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002;22:7053–7065. doi: 10.1128/MCB.22.20.7053-7065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal, I., Vulliamy, T. Dyskeratosis congenita: Its link to telomerase and aplastic anaemia. Blood Rev. 2003;17:217–225. doi: 10.1016/s0268-960x(03)00020-1. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Pogacic, V., Filipowicz, W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000;20:3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., Dlakic, M., Tollervey, D. Naf1p is a box H/ACA snoRNP assembly factor. RNA. 2002;8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W., Pogacic, V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Caizergues-Ferrer, M., Kiss, T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Goodall, G.J., Wiebauer, K., Filipowicz, W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- Hamma, T., Reichow, S.L., Varani, G., Ferre-D'Amare, A.R. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat. Struct. Mol. Biol. 2005;12:1101–1107. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- Heiss, N.S., Knight, S.W., Vulliamy, T.J., Klauck, S.M., Wiemann, S., Mason, P.J., Poustka, A., Dokal, I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- Heiss, N.S., Girod, A., Salowsky, R., Wiemann, S., Pepperkok, R., Poustka, A. Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum. Mol. Genet. 1999;8:2515–2524. doi: 10.1093/hmg/8.13.2515. [DOI] [PubMed] [Google Scholar]
- Henras, A., Henry, Y., Bousquet-Antonelli, C., Noaillac-Depeyre, J., Gelugne, J.P., Caizergues-Ferrer, M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau-Aveilla, C., Bonoli, M., Caizergues-Ferrer, M., Henry, Y. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA. 2006;12:832–840. doi: 10.1261/rna.2344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac, C., Yang, Y., Meier, U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T., Bortolin, M.L., Filipowicz, W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol. Cell. Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D.L.J., Bousquet-Antonelli, C., Henry, Y., Caizergues-Ferrer, M., Tollervey, D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manival, X., Charron, C., Fourmann, J.B., Godard, F., Charpentier, B., Branlant, C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 2006;34:826–839. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone, A., Mason, P.J. Dyskeratosis congenita. Cell. Mol. Life Sci. 2003;60:507–517. doi: 10.1007/s000180300042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, U.T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, U.T., Blobel, G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili, S., Steitz, J.A. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., Wood, E., Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma, S. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani, M., Goldberg, I., Swedlow, J.R., Lamond, A.I. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic, V., Dragon, F., Filipowicz, W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid, R., Liang, B., Baker, D.L., Youssef, O.A., He, Y., Phipps, K., Terns, R.M., Terns, M.P., Li, H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21:249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Smith, C.M., Steitz, J.A. Sno storm in the nucleolus: New roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P.J., Dokal, I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001a;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- Vulliamy, T.J., Knight, S.W., Mason, P.J., Dokal, I. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol. Dis. 2001b;27:353–357. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- Wang, C., Meier, U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N.J., Gottschalk, A., Neubauer, G., Kastner, B., Fabrizio, P., Mann, M., Lührmann, R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA- binding protein, are present together with Gar1p in all box H/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P.K., Rotondo, G., Porras, T., Legrain, P., Chanfreau, G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002;277:45235–45242. doi: 10.1074/jbc.M207669200. [DOI] [PubMed] [Google Scholar]