Abstract
Squalene synthase (SS) represents a putative branch point in the isoprenoid biosynthetic pathway capable of diverting carbon flow specifically to the biosynthesis of sterols and, hence, is considered a potential regulatory point for sterol metabolism. For example, when plant cells grown in suspension culture are challenged with fungal elicitors, suppression of sterol biosynthesis has been correlated with a reduction in SS enzyme activity. The current study sought to correlate changes in SS enzyme activity with changes in the level of the corresponding protein and mRNA. Using an SS-specific antibody, the initial suppression of SS enzyme activity in elicitor-challenged cells was not reflected by changes in the absolute level of the corresponding polypeptide, implicating a post-translational control mechanism for this enzyme activity. In comparison, the absolute level of the SS mRNA did decrease approximately 5-fold in the elicitor-treated cells, which is suggestive of decreased transcription of the SS gene. Study of SS in intact plants was also initiated by measuring the level of SS enzyme activity, the level of the corresponding protein, and the expression of SS gene promoter-reporter gene constructs in transgenic plants. SS enzyme activity, polypeptide level, and gene expression were all localized predominately to the shoot apical meristem, with much lower levels observed in leaves and roots. These later results suggest that sterol biosynthesis is localized to the apical meristems and that apical meristems may be a source of sterols for other plant tissues.
Sterols are essential molecules for all eukaryotic organisms, and many genetic mutations that eliminate enzymatic steps in sterol biosynthesis are lethal. For example, severe mutations in the squalene synthase (SS) gene in yeast (Saccharomyces cerevisiae) are lethal, and these mutants require exogenous ergosterol to survive (Karst and Lacroute, 1977). Mice that are homozygous for an SS knockout mutation also do not survive past mid-gestation (Tozawa et al., 1999). The essentiality for sterols is based on the many different functions sterols contribute to cellular physiology. Sterols are found predominantly in cell membranes and are thought to contribute to the proper functioning of membranes by controlling the fluidity characteristics of the membranes (Hartmann and Benveniste, 1987; Hartmann, 1998). Sterols also function as biosynthetic precursors for steroid hormones such as testosterone and estrogen in mammals, ecdysteroids in insects, and brassinosteroids in plants (Fujioka et al., 1997; Noguchi et al., 1999). Sterols are suspected of serving novel functions as well (Nes, 1987). For instance, sterols have been implicated as ligands for nuclear receptors directly affecting transcription and signal transduction pathways (Edwards and Ericsson, 1999). At least one regulatory protein controlling a developmental program is now known to be activated upon esterification with sterols (Porter et al., 1996). Jang et al. (2000) and Schrick et al. (2000) have more recently suggested that sterols other than brassinolides may serve as novel signals controlling cell fate during plant embryogenesis.
Sterols are synthesized via the isoprenoid biosynthetic pathway with SS catalyzing the first enzymatic step committing carbon away from the central isoprenoid pathway toward sterol biosynthesis (Goldstein and Brown, 1990). SS catalyzes the condensation of two molecules of farnesyl diphosphate to form the linear 30 carbon compound squalene (Popjak et al., 1961a, 1961b), and this activity has been localized to the smooth endoplasmic reticulum (ER; Popjak and Agnew, 1979; Stamellos et al., 1993). Additional studies have suggested that the carboxy-terminal portion of the SS protein anchors the protein to the ER membrane, whereas the catalytic site of the enzyme is associated with the amino-terminal portion of the protein found on the cytoplasmic face of the ER (Robinson et al., 1993).
Given the putative positioning of SS at a key branch point in the isoprenoid biosynthetic pathway, a regulatory role for SS in controlling sterol biosynthesis has been investigated in animals (Faust et al., 1979; Guan et al., 1998), yeast (Kennedy et al., 1999; Kennedy and Bard, 2001), and plants (Threlfall and Whitehead, 1988; Vögeli and Chappell, 1988; Chappell et al., 1989; Devarenne et al., 1998). For example, when tobacco (Nicotiana tabacum) cell cultures are challenged with fungal elicitor, sterol biosynthesis and accumulation are suppressed, and this reduction in sterol biosynthesis has been correlated with a dramatic decrease in the SS enzyme activity (Threlfall and Whitehead, 1988; Vögeli and Chappell, 1988). Other plants also terminate sterol biosynthesis in response to pathogen or elicitor challenge (Brindle et al., 1988; Vanderheijden et al., 1988, 1989; Zook and Kuc, 1991; Fulton et al., 1993; Haudenschild and Hartmann, 1995). Potato (Solanum tuberosum) tuber slices accumulate prodigious amounts of steroidal glycoalkaloids as part of their wound response. Interestingly, addition of the elicitor arachidonic acid to potato tuber discs arrests steroid glycoalkaloid accumulation and suppresses SS enzyme activity similar to that of tobacco (Shih et al., 1973; Brindle et al., 1988; Zook and Kuc, 1991). It is still unclear how the reduction in SS enzyme activity is brought about in tobacco, potato, and other plants responding to pathogens and elicitors. Initial measurement of the SS steady-state mRNA levels in elicitor-treated tobacco (Devarenne et al., 1998) and potato (Yoshioka et al., 1999) indicate that the mRNA levels remain fairly constant or decline slightly. Such results have suggested that regulation of SS might occur at many levels from transcriptional to post-translational, at least in plants responding to pathogen challenge.
In contrast to those studies of sterol metabolism in pathogen/elicitor challenged plants, regulation of sterol biosynthesis in the context of normal cellular development has been more limited. Most studies have dealt with the identification and characterization of the sterol composition of developing and germinating seeds and measuring sterol biosynthetic capacity over these developmental times by measuring the incorporation of labeled precursors into sterols as well as a few select enzyme activities (Kemp et al., 1967; Kemp and Mercer, 1968; Garg and Nes, 1987; Heupel et al., 1987; Fenner et al., 1989; Guo et al., 1995). Several investigations have also sought to determine the contribution of specific biosynthetic steps either by selection of genetic mutants or by the use of genetic engineering approaches (Maillotvernier et al., 1991; Chappell et al., 1995; Diener et al., 2000). These studies have, for the most part, demonstrated the importance of enzymes in sterol biosynthesis but have not directly addressed how the sterol biosynthetic machinery is regulated. Other investigators have examined the mRNA steady-state levels or gene-promoter activities for several central isoprenoid and sterol-specific biosynthetic genes, but more with an emphasis on the developmental regulation of these genes rather than a correlation with sterol or other isoprenoid metabolism (Narita and Gruissem, 1989; Enjuto et al., 1994, 1995; Rodríguez-Concepción and Gruissem, 1999; Diener et al., 2000). In more direct studies, Chappell et al. (1989) observed a 4-fold transient induction of SS enzyme activity at the same time that sterol biosynthesis and accumulation were maximal during a growth cycle of a tobacco cell suspension culture.
As part of an ongoing effort to investigate the regulation of sterol biosynthesis, the current work was aimed at better understanding how SS enzyme activity may be regulated in plants. In particular, a tobacco SS (TSS)-specific antibody was developed to assist in determining whether SS was regulated post-translationally in elicitor-treated cell cultures, and transgenic lines harboring SS promoter-β-glucuronidase (GUS) reporter gene constructs were developed as a means for assessing transcriptional control of SS gene expression and for identifying in which tissues and cells sterol biosynthesis was occurring.
RESULTS
Development and Validation of a TSS Antibody
Our previous attempts at developing antibodies specific for TSS have met with limited success. Hanley and Chappell (1992) used a partially purified TSS preparation to elicit immunological responses in several mice, and then screened the sera for their ability to inhibit TSS enzyme activity in vitro. One of these sera did show a dose-dependent inhibition of the enzyme activity. However, this serum detected a number of tobacco microsomal proteins, limiting its utility for identifying and monitoring a specific TSS peptide. More importantly, the previously obtained sera could not detect any SS protein produced upon expression of the corresponding tobacco gene in Escherichia coli (T.P. Devarenne and J. Chappell, unpublished data). We, therefore, developed an alternative method for the preparation of a suitable antibody.
Earlier studies demonstrated that expression of a full-length TSS cDNA (TSS-1.2) in E. coli did not yield high levels of TSS protein or enzyme activity, presumably because of the hydrophobic carboxy terminus that normally anchors the protein to the ER membrane. Removal of the 24 carboxy-terminal amino acids that comprise the membrane-anchoring domain through manipulation of the cDNA (TSS-1.1), however, allowed for low level expression of a soluble, enzymatically active TSS protein (Devarenne et al., 1998). To obtain even greater quantities of a more soluble form of the TSS protein, the first one-half of the TSS cDNA, corresponding to a 23-kD hydrophilic domain of the amino terminus was engineered into a suitable E. coli expression vector, pET-28b+, and the recombinant vector was transformed into BL21(DE3) host cells. Expression of the partial TSS cDNA in E. coli resulted in the accumulation of a new 23-kD peptide, which was subsequently purified to near homogeneity based on a His affinity tag located at the carboxy terminus of the TSS peptide. Eight rabbits were immunized with the TSS 23-kD peptide, and the sera from all eight rabbits were screened for their ability to detect, by western blot, a protein of predicted size for TSS in tobacco microsomes and for their ability to inhibit TSS enzyme activity in vitro. Although none of the sera was capable of inhibiting TSS enzyme activity, serum from one animal did detect a protein of predicted size for TSS at 47 kD (see below), and was chosen for further characterization. All further comparisons were performed with IgG fractions from both the preimmune and immune sera purified by protein-A affinity chromatography.
Although the antibodies from the immune serum did not inhibit TSS activity in an in vitro assay, the preliminary detection of predictably sized proteins by western blotting suggested that immunoprecipitation types of experiments could further validate the antibodies. The immunoprecipitation experiments were, however, problematic to perform properly with an integral membrane protein like SS. For instance, it was not possible to use the tobacco microsomes as a source of SS for the immunoprecipitation experiments. Even without adding any antibodies to the tobacco microsomes, SS enzyme activity readily pelleted from the solution upon centrifugation, as called for in a typical immunoprecipitation protocol. Use of SS solubilized from these membrane preparations also was not feasible. High concentrations of detergents were needed to solubilize TSS, and these detergents adversely affected the immunoprecipitation assays. As an alternative, soluble TSS enzyme was obtained by expression of TSS-1.1 in E. coli (Devarenne et al., 1998; Fig. 1A). Substitution of a Tris buffer for the potassium phosphate buffer used in the earlier work also resulted in approximately 10 times more SS activity recovered in the E. coli extracts.
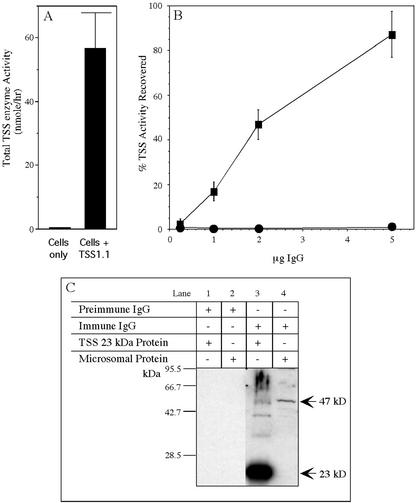
Figure 1.
A, Validation of an antibody with immunospecificity for TSS. A carboxy-truncated TSS cDNA (TSS-1.1) was expressed in E. coli and the soluble SS activity used for immunoprecipitation assays. B, Aliquots of the soluble SS were incubated with IgGs isolated from preimmune serum (●) or serum of a rabbit immunized with a 23-kD amino-terminal fragment of the TSS enzyme (▪) and the immunoprecipitable SS activity determined. C, Duplicate samples of 200 ng of the 23-kD TSS peptide and 10 μg of total microsomal protein were separated by 8% to 16% (w/v) SDS-PAGE and transferred to polyvinylidene difluoride membranes, and the membranes probed with either IgGs purified from preimmune serum (lanes 1 and 2) or immune serum (lanes 3 and 4).
Addition of the IgGs purified from a rabbit immunized with the 23-kD TSS protein precipitated SS enzyme activity in a dose-dependent manner (Fig. 1B). Five micrograms of anti-TSS IgG was capable of immunoprecipitating 80% of the soluble TSS enzyme used in this assay, whereas identical amounts of IgGs purified from preimmune sera were incapable of precipitating any SS activity. Using western blots, the IgGs purified from the preimmune sera also did not detect any proteins in extracts from E. coli expressing the TSS 23-kD peptide or any proteins in tobacco microsomes (Fig. 1C). In contrast, the IgGs from the immunized animal did react strongly with the 23-kD TSS peptide expressed in the E. coli and readily detected a 47-kD protein in microsomes isolated from tobacco cell cultures (Fig. 1C). Conceptual translation of the full-length TSS cDNA predicts a protein of 47 kD. Immunodetection of either the 23-kD TSS peptide or the 47-kD microsomal protein was eliminated if the anti-TSS IgGs were pre-incubated with purified 23-kD peptide (data not shown).
TSS Enzyme Activity, mRNA, and Protein Levels in Response to Fungal Elicitor
Based on limited measurements of the TSS mRNA in elicitor-treated tobacco cells, Devarenne et al. (1998) previously suggested that the decrease in TSS enzyme activity occurred via a post-transcriptional mechanism. Yoshioka et al. (1999) performed similar experiments with potato and concluded that the suppression of SS was at least partially subject to transcriptional control. To gain a better understanding of how SS might be regulated in elicitor-treated tobacco cells, the levels of TSS protein (Fig. 2B) and mRNA (Fig. 2C) were, therefore, determined relative to the corresponding enzyme activity (Fig. 2A). The data in Figure 2 are plotted as a percentage relative to that measured in control cells at the start of this experiment to facilitate this comparison.
Figure 2.
Suppression of TSS enzyme activity in elicitor-treated tobacco cell suspension cultures is accompanied by changes in the level of the TSS protein and mRNA. Cells were collected at the indicated time points after addition of 1 μg of P. parasitica elicitin per milliliter of cell suspension culture and kept frozen until analyzed for TSS enzyme activity (A), TSS protein (B), and TSS mRNA (C). Microsomes isolated from the cells were used to measure SS enzyme activity and to quantify TSS protein by chemiluminescent immunodetection, whereas TSS mRNA was detected by RNA-blot hybridization using a full-length cDNA to probe total RNA isolated from aliquots of the collected cells.
Cell cultures in their rapid growth phase were used for this experiment with an approximate tripling of cell mass over the 72-h time course. This increase in the control culture mass corresponded to a doubling of SS enzyme activity by 48 h, which was matched by a parallel increase in the absolute level of the SS protein (compare white symbols in Fig. 2, A and B). The increase in enzyme activity and protein was preceded by a transient, 3-fold induction of the SS mRNA with the maximum occurring about 12 h after initiation of the experiment and returning to near starting levels by 48 h (Fig. 2C, white symbols).
As reported earlier (Threlfall and Whitehead, 1988; Vögeli and Chappell, 1988), SS enzyme activity was rapidly suppressed in response to elicitor treatment of the tobacco cell cultures (Fig. 2A, black symbols). The activity declined 65% to 70% between 8 to 14 h after elicitor treatment and remained more or less at this reduced level throughout the remainder of the experiment. In contrast, the SS protein level was constant for the first 36 h and then declined to barely detectable levels by 48 h (Fig. 2B, black symbols). SS mRNA also changed significantly under these conditions with a decline in the mRNA level mirroring that for the enzyme activity. The mRNA level declined by approximately 80% within the first 14 h of elicitor-treatment and remained at this level for the duration of the experiment (Fig. 2C, black symbols).
Expression of SS in Planta
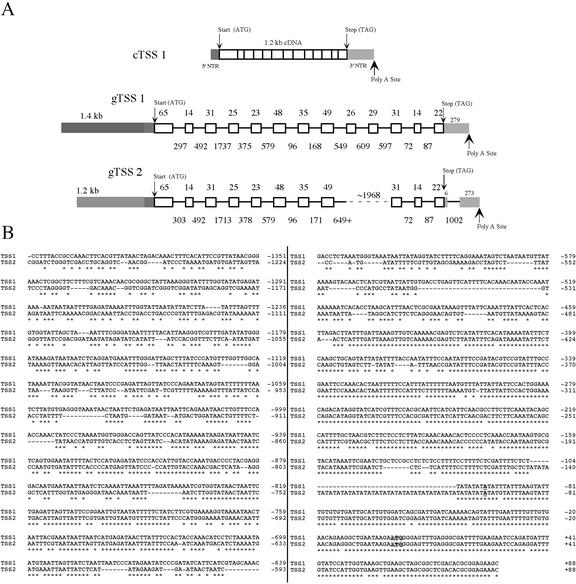
Because the above results indicated that TSS enzyme activity could be partially correlated with the level of the TSS mRNA, more direct experiments to examine the transcriptional control exerted by the TSS promoter were undertaken. Although a genomic clone for one TSS gene (gTSS-1) had previously been isolated and sequenced (Devarenne et al., 1998), Southern-blot analysis indicated the presence of another copy of the SS gene in the tobacco genome. The tobacco genomic library was, therefore, re-screened using a TSS cDNA. Multiple clones were isolated and initially differentiated into only two classes based on the presence or absence of a 1.0-kb intron in the 3′-non-translated region (NTR) region (Fig. 3A). Further sequence analysis confirmed only the two different genomic clones, gTSS-1 and gTSS-2, which could be distinguished by several silent nucleotide substitutions within the coding portions of the genes. Although the full gTSS-2 DNA has not been sequenced, the sequence information obtained so far indicates that, whereas the exon-intron organization is absolutely conserved between gTSS-1 and gTSS-2 (except for the additional 3′ intron in gTSS-2), four of the 10 introns examined vary in size from 3 to 24 bp (Fig. 3A).
Figure 3.
A, The intron-exon organization of the two TSS genomic clones is nearly identical. Numbers above exons indicate the number of amino acids encoded by the exonic DNA. Numbers below introns indicate the number of base pairs in an intron. Start and stop codons and polyadenylation sites are noted as such. Also shown for comparison is a full-length TSS cDNA. The DNA sequences for the genes reported here have been deposited in GenBank: cTSS-1, U60057; gTSS-1, U59683; and gTSS-2, AY096801. B, The 5′ regions of gTSS-1 and gTSS-2 corresponding to their promoter regions are aligned and annotated for comparison. The ATG translation start codon is outlined, and the adenosine of the ATG start codon is designated as +1 nucleotide. The transcription start site, determined previously by Devarenne et al. (1998), is in bold and underlined. Identical sequence alignments are noted by an asterisk below the two sequences.
Regions from both TSS genes corresponding to the 5′-promoter domains were isolated by PCR, sequenced, and analyzed for sequence similarities (Fig. 3B). With reference to the translation start site as +1, the transcription start site maps 96 bp upstream to an adenosine based on the primer extension data of Devarenne et al. (1998). The identical transcription start site is predicted for both TSS genes because this 5′-NTR region of both genes differs by only six nucleotide substitutions, and because only one primer extension product was observed previously. Seven nucleotides further upstream of the transcription start site, the promoters diverge by a 41 nucleotide AT repeat inserted into the TSS-2 promoter (−104 to −145). Immediately upstream of this insert sequence, the promoters again share a great deal of identity for approximately 360 nucleotides, after which the identity is isolated to small groups of nucleotides. Alignments of the TSS promoters with the SS promoters from Arabidopsis (AtSS; T.P. Devarenne and J. Chappell, unpublished data), human (HSS; Guan et al., 1995), and yeast (YSS; Kennedy et al., 1999) were attempted as well. An AtSS genomic clone, including the AtSS promoter, was isolated from an Arabidopsis genomic library using the AtSS cDNA (Nakashima et al., 1995) as a probe (T.P. Devarenne and J. Chappell, unpublished data). Over approximately 1,400 bp of promoter region, the TSS promoters were only 23% identical to the AtSS promoter and less so for either the HSS or the YSS promoters (data not shown). The key cis regions required for the sterol-mediated feedback regulation in the HSS promoter, the SRE-1 regions (Guan et al., 1995, 1997, 1998), and the repressor (URS) or activator (UAS) elements found within the YSS promoter (Kennedy and Bard, 2001) were not found in either the TSS or the AtSS promoters.
To functionally characterize the transcriptional activities controlled by the two TSS promoters and to define general regions harboring elements important for these transcriptional activities, full-length promoters for the TSS-1 and TSS-2 and several promoter deletion constructs were fused to the GUS reporter gene. Full-length promoter size was arbitrarily set at approximately 1,400 bp and 5′-deletion mutants corresponding to three-fourths, one-half, one-fourth, and one-eighth of the full-length promoter were prepared for each as well. The 3′ end of each promoter construct ended at the ATG translation start codon and was fused with the corresponding start site of the GUS reporter gene. At least 10 independent lines were generated for each promoter construct. R1 seeds from each transgenic line were germinated on kanamycin selection plates and first scored for the segregation of kanamycin resistance after 10 to 15 d. The kanamycin resistant plants were then transferred to non-selection media and grown for an additional 15 to 20 d. The aerial portions of 10 to 12 seedlings per line were then harvested, combined, and used to determine total GUS activity (Fig. 4A). In general, the overall level of GUS activity observed for any of the promoter-reporter lines was relatively low and approximately 1/100 to 1/250 of the levels typically seen with the 35S cauliflower mosaic virus constitutive promoter (data not shown). For the TSS-1 promoter, the activity of the full-length promoter was approximately 65% to 75% as active as the 1.0- and 0.722-kb promoter elements, respectively. Deletion of another approximately 350 bp from the 5′ end of the promoter resulted in a 2-fold reduction in measurable GUS activity, which was reduced another 2.5-fold upon deletion of the next 170 bp. Expression levels of the GUS reporter gene driven by the TSS-2 promoter were similar to the TSS-1 promoter. Truncation of 5′ end of the TSS-2 full-length promoter from 1.2 to 1.0 or 0.722 kb, however, did not appear to stimulate overall GUS activity as observed for the TSS-1 promoter, but further truncation to the 350-bp promoter construct decreased activity similarly about 2-fold.
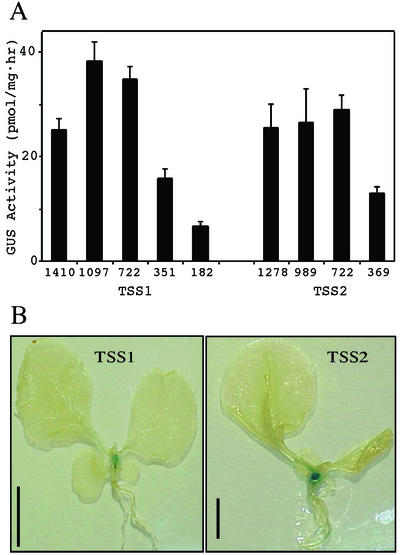
Figure 4.
The TSS-1 and TSS-2 promoters direct expression of the GUS reporter gene in transgenic tobacco plants. Full-length promoter and 5′-deletion constructs were prepared by fusing the indicated promoter fragments to the GUS reporter gene in the pBI101 vector and engineering these gene fusions into transgenic tobacco via standard Agrobacterium tumefaciens-mediated transformation. Promoter length designations refer to the 5′-end point of a construct relative to the translation start site at +1. R1 seeds were germinated in the presence of kanamycin, and only 30-d-old plantlets able to grow on kanamycin for the first 15 d were evaluated quantitatively (A) or qualitatively (B) for GUS activity. The values presented in A represent the averaged data for a minimum of 10 independent transgenic lines for each construct. Representative transgenic plants harboring the 1.4-kb TSS1 or 1.2-kb TSS2 promoter-GUS reporter constructs were stained histochemically for GUS expression for 48 h. Error bars in A represent the sds, whereas the inset bar in B represents 1 cm.
The TSS promoter-GUS reporter transgenic lines were also assessed for tissue-specific expression by histochemical staining of intact plants (Fig. 4B). In all cases, no histochemical staining was observed with promoter constructs less than 722 bp, and no apparent expression of GUS activity was detected in roots regardless of promoter length. One-month-old transgenic lines harboring full-length promoters for either TSS-1 or TSS-2 (Fig. 4B) exhibited identical staining patterns with very intense GUS expression in the apical stem region, some expression in the base of the leaf petioles and very faint expression in leaf margins. Similar, if not identical, staining patterns were observed in a minimum of five independent transgenic lines for the full-length, three-fourths, and one-half-length promoter constructs.
The results of the histochemical staining for the TSS promoter:GUS transgenic plants indicated that the TSS genes are expressed predominately in the meristematic regions of stems. To confirm this data, 30-d-old non-transgenic plants were dissected into specific tissues, and the SS enzyme activity and protein levels in each were determined (Fig. 5). Consistent with the histochemical staining for TSS promoter-reporter activity in transgenic plants, SS enzyme activity was highest in stem, apical meristem, and leaf petioles (Fig. 5D), with much lower levels of activity present in roots and leaf tissues. The level of enzyme activity was also positively correlated with the relative levels of SS protein as determined by western-blot analysis (Fig. 5E). The level of SS protein unfortunately could not be determined in leaf samples because the antibodies also contained a strong reactivity with Rubisco, which overlaps in size with SS and obscured our ability to accurately detect the SS peptide. The relatively high level of SS enzyme activity and protein in stem tissue was not unexpected given that only the extreme distal tips of the stem were cut-off during tissue preparation to isolate shoot meristems, and a significant proportion of the shoot meristem remained with the stem tissue.
Figure 5.
Analysis of 1-month-old wild-type tobacco plant tissues for TSS enzyme activity and protein levels. A 1-month-old wild-type tobacco plant (A) was dissected into its component parts as indicated (B and C), and the level of TSS enzyme activity (D) and enzyme protein (E) was determined. Dashed lines indicate how organs were excised into specific tissues; LB, leaf blade; LM, leaf margin; PT, petiole; AM, apical meristem; ST, stem; and RT, roots. The inset bars in A through C equal 1 cm. Microsomes prepared from each of the tissues were used simultaneously to measure TSS enzyme activity (D) and the level of the TSS protein by chemiluminescent immunodetection (E).
DISCUSSION
Regulation of TSS in Elicitor-Treated Cells
Despite impressive progress in elucidating the biochemical steps and complexities of sterol metabolism (Guo et al., 1995; Diener et al., 2000; Jang et al., 2000; Schrick et al., 2000), very little is actually known about how plant cells monitor their sterol content and regulate sterol biosynthesis. This is in contrast to the in-depth appreciation of the mechanistic details for how sterol metabolism in mammalian (Brown and Goldstein, 1997) and fungal systems (Karst and Lacroute, 1977; Kennedy and Bard, 2001) is regulated. In the current work, we have attempted to take advantage of the general observation that sterol biosynthesis is suppressed in pathogen or elicitor-treated plant cells or tissue (Shih et al., 1973; Brindle et al., 1988; Threlfall and Whitehead, 1988; Vögeli and Chappell, 1988; Vanderheijden et al., 1989; Zook and Kuc, 1991; Fulton et al., 1993; Haudenschild and Hartmann, 1995) to investigate the molecular regulation of SS, a putative branch point diverting carbon from the central isoprenoid pathway to sterols, as just one possible means by which sterol biosynthesis may be controlled. The data presented here suggests that the suppression of SS in elicitor-treated tobacco cells is regulated at several different levels.
Within the first 15 h of elicitation, the TSS enzyme activity is suppressed, and there is a corresponding decrease in the TSS mRNA. The decline in the mRNA level is consistent with a decrease in the transcription rate of the TSS gene, an increase in the turnover rate of the TSS mRNA, or some combination of these two. We were not, however, able to use the transgenic lines harboring the TSS promoter-GUS reporter gene to distinguish between these possibilities. Although the GUS reporter enzyme is suitable for measuring inducible expression directed by a gene promoter, the relatively long half-life of the GUS reporter enzyme obscures its usefulness in measuring transcriptional inactivation. Hence, we can only conclude from the current data that the TSS mRNA level is subject to transcriptional or post-transcriptional control in elicitor-treated cells. Equally important, precedence for both types of mechanisms has been reported in plants. In 1993, Newman et al. described sequences found within the 3′-NTR of mRNAs that are correlated with the destabilization of mRNAs. These sequences, also known as DST elements, are associated with rapidly turned-over mRNAs in plants but are not obvious within the 3′-NTR of the TSS mRNAs. In contrast, the data from Yoshioka et al. (1999) suggested that SS is under transcriptional control in elicitor-treated potato tissue. In examining the SS mRNA levels in potato tuber slices, these investigators noted that the induction of SS mRNA by wounding was suppressed if the tuber slices were immediately challenged with an elicitor. More direct experimentation, like nuclear run-on transcription assays (Chappell and Chrispeels, 1986), will be necessary to clarify whether SS gene expression is subject to transcriptional suppression or mRNA turnover in elicitor-treated cells.
The rapid decline in TSS enzyme activity within the first 30 h of the elicitor-treatment does not occur because of a loss or degradation of the TSS protein (Fig. 2, compare A and B). This suggests that TSS enzyme activity is regulated by some post-translational modification of the SS protein. Perhaps one of the best examples for post-translational regulation of an enzyme is that for 3-hydroxy-3-methyl glutaryl-CoA reductase (HMGR), an early, key step in the isoprenoid biosynthetic pathway (Goldstein and Brown, 1990). In animals, selective phosphorylation of HMGR has been demonstrated to attenuate enzyme activity (Omkumar et al., 1994; Omkumar and Rodwell, 1994), and likewise in plants, a single phosphorylation of the Arabidopsis HMGR at Ser-577 completely inactivates enzyme activity (Dale et al., 1995; Toroser and Huber, 1998). TSS activity may also be regulated by phosphorylation. TSS contains a casein kinase II consensus phosphorylation site centered at Thr-78 and adjacent to domain II of the protein, a region that contributes to the formation of the active site. Preliminary experiments have demonstrated that the 23-kD peptide fragment is readily phosphorylated in vitro by casein kinase II (T.P. Devarenne and J. Chappell, unpublished data). Whether this phosphorylation occurs in vivo and whether it or some other phosphorylation site might be regulating enzyme activity within the first 30 h of elicitation is currently not known.
Thirty hours after elicitor-treatment, immunodetectable TSS protein is rapidly lost or degraded within a 15- to 20-h period (Fig. 2B). This could represent a selective degradation of the TSS enzyme or a more general turnover of cellular proteins in response to the elicitation treatment. If this does reflect an orchestrated event, then it is tempting to speculate on the involvement of a PEST sequence found at amino acids 75 to 89 within TSS. PEST sequences were originally identified by Rogers et al. (1986) as being involved in targeting proteins for rapid turnover. PEST sequences have subsequently been shown to be involved in regulated degradation of many proteins, but not necessarily enzymes of isoprenoid biosynthesis (Chun and Simoni, 1992; Ravid et al., 2000). Assessing the relative importance of the TSS PEST sequence for regulating targeted protein degradation can be readily tested by fusing it to the amino terminus of a reporter protein and then determining the turnover rate of the reporter activity before and after elicitor treatment. As an alternative, epitope-tagged TSS could be used in a similar fashion as described by Korth et al. (2000). These investigators were able to measure changes in the half-life of HMGR upon shifting plants from light to dark conditions.
Regulation of TSS in Planta
The current results demonstrate that both the TSS1 or TSS2 promoters are sufficient to direct high-level expression of the GUS reporter gene in the apical regions of shoots with much lower levels of expression in any other tissue (Fig. 4). The results also suggest that 722 bp of the promoters are sufficient to direct this expression and that there are probably at least two cis-elements that contribute quantitatively and qualitatively to this expression. The two elements appear to reside between positions −722 to −351 bp and −350 to −182 bp relative to the translation start site in the native gene. The sequences within these regions have been carefully examined for several types of sterol response elements, consensus elements documented to be important for the regulation of the sterol LDL receptor gene (Osborne, 1995), HMGR (Osborne, 1995) and SS in mammals (Guan et al., 1997), and two cis elements recently mapped within the YSS promoter (Kennedy and Bard, 2001). Although no identical matches were observed, the rudiments of several SRE elements could be discerned with matches of five to seven nucleotides of eight to 10 nucleotides per element. Obviously, more detailed mapping of the important cis-elements within the TSS promoter will be necessary before functionality can be ascribed to any cis-sequence.
The localized expression of the GUS reporter gene expression to the shoot meristems as directed by the TSS promoter was unexpected. We initially presumed that all meristematic zones as well as those undergoing cell expansion would require sterol biosynthesis to support the accompanying membrane biogenesis. Importantly, the almost exclusive expression of the TSS promoter in shoot meristems was independently corroborated by an identical distribution of endogenous TSS enzyme activity and protein (Fig. 5). Other investigators examining the expression pattern of other isoprenoid biosynthetic genes by RNA-blot analysis or promoter-reporter gene constructs have made similar observations. However, these investigators have not, in our opinion, fully recognized the significance of their findings. For example, the groups of Boronat (Caelles et al., 1989; Enjuto et al., 1994) and Fink (Learned and Fink, 1989) were among the first to clone the HMGR genes from Arabidopsis and to document the presence of two copies of this gene in this species. In experiments measuring the level of the corresponding mRNA in various plant tissues, Enjuto et al. (1994) suggested that the HMGR 1 gene was constitutively expressed, whereas HMGR 2 gene expression was described as more tissue specific. Closer inspection of their data does indicate that the HMGR 1 mRNA is present in many tissue types, but the level in tissues representing apical meristems is very much higher than in any other tissues. In a follow-up publication, Enjuto et al. (1995) reported that the expression of the Arabidopsis HMGR2 gene was restricted to shoot meristems and floral (stigma) tissues based on histological staining of transgenic tobacco plants harboring HMGR2 promoter-GUS reporter constructs. Likewise, expression of Fackel, a C-14 sterol reductase gene in Arabidopsis, appears to be specific to shoot and root meristems (Jang et al., 2000; Schrick et al., 2000). Even though these various studies have clearly localized expression of genes essential for sterol biosynthesis to the shoot and root meristems, the implications of these results were not discussed.
Studies into sterol metabolism have been subtly influenced by what we suggest is a general assumption that all cells have the capacity to monitor and regulate their own sterol biosynthesis. This is obviously true to some extent; otherwise, tissue culture systems would not be viable for example. However, the current results suggest that the shoot meristem represents a biosynthetic center for sterols, which can then be distributed to daughter cells, neighboring cells, and distal tissues throughout the plant. Although there is some support for this suggestion in the literature, high sterol biosynthetic rates in apical shoots (Bhatt and Bhatt, 1984), and sterol/triterpenoid transport (Adler and Grebenok, 1995; Trojanowska et al., 2000), much remains to be tested to validate this speculative interpretation.
MATERIALS AND METHODS
Development of SS Antibodies
The amino terminus region of the TSS protein is considerably more hydrophilic than the carboxy terminus; therefore, the 5′ region of the cDNA was chosen for bacterial expression and immunization purposes. Using standard PCR methodology, a 5′ region of the full-length TSS cDNA was amplified using a forward primer corresponding to the start ATG codon (5′-GCGGGAATTCCATGGGGAGTTTGAG GGCGATT-3′, NcoI restriction site underlined) and a reverse primer terminating at nucleotide 594 (5′-CACCTCGAGTGAATTCGAGAGAGAATCTGAAGCCAG-3′, XhoI restriction site underlined and nucleotide 594 in bold). The amplified DNA fragment was digested with NcoI and XhoI, ligated into the corresponding restriction sites of the pET 28b+ plasmid vector (Novagen, Madison, WI), and transformed into the Escherichia coli strain TB-1 for propagation of plasmid DNA. Proper construction of this expression vector was verified by standard DNA sequencing using an ABI 310 sequencer (PE Applied Biosystems, Foster City, CA) and the plasmid subsequently named TSS-594 pET 28b+.
Expression of the TSS-594 pET 28b+ vector in E. coli and purification of the encoded His-tagged TSS 23-kD peptide was performed using nickel affinity chromatography as follows. A 2-mL overnight culture of BL21 (DE3) cells harboring the expression vector was diluted into 100 mL of fresh culture media, the culture was grown to an optical density of 1.5 at 600 nm and then shifted to 28°C, and isopropylthio-β-galactoside was added to a final concentration of 0.3 mm. After an additional incubation for 5 h, the cells were collected by centrifugation, resuspended in 4 mL of binding buffer (5 mm imidazole, 0.5 m NaCl, and 20 mm Tris-HCl, pH 7.9), sonicated three times for 30 s, and centrifuged at 39,000g for 20 min. The resulting supernatant was filtered through a 0.45-μm filter and applied to a 0.5-mL His-Bind column prepared according to the manufacturer's instructions (Novagen). The column was washed with 5 mL of binding buffer and 5 mL of wash buffer (60 mm imidazole, 0.5 m NaCl, and 20 mm Tris-HCl, pH 7.9) and then eluted with 3 mL of 100 mm imidazole buffer (100 mm imidazole, 0.5 m NaCl, and 20 mm Tris-HCl, pH 7.9) followed by 3 mL of 1 m imidazole buffer (1 m imidazole, 0.5 m NaCl, and 20 mm Tris-HCl, pH 7.9). The eluent from each step was checked for the presence of the TSS 23-kD peptide by 15% (w/v) SDS-PAGE. The majority of the TSS 23-kD peptide was consistently found in the eluent from the 100 mm imidazole buffer. This fraction was subsequently concentrated to 100 μL, and the buffer was exchanged with 50 mm Tris-HCl, pH 8.0, using an ultrafree-4 centrifugal filter with a 5-kD cut-off membrane (Millipore, Bedford, MA). Several of these partially purified samples were pooled and further purified by preparative gel electrophoresis. The samples were separated by a 15% (w/v) SDS-PAGE and transferred to nitrocellulose (MSI, Inc, Westborough, MA), and after visualizing the proteins using ponceau S (Sigma, St. Louis), that region of the membrane corresponding to the TSS 23-kD peptide was cut out of the membrane, washed with water to eliminate the ponceau S stain, and then dissolved in 500 μL of dimethyl sulfoxide.
Before inoculation of rabbits with the purified TSS peptide, preimmune serum was obtained from each rabbit and screened for cross-reactivity to the TSS 23-kD peptide and plant microsomal proteins via standard western blotting techniques. The purified TSS 23-kD peptide dissolved in dimethyl sulfoxide was first diluted with an equal volume of phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 2 mm KH2PO4, pH 7.4) and then mixed with an equal volume of Complete Freund's adjuvant (Sigma). Each rabbit was initially injected subcutaneously with 1 mg of the purified TSS peptide followed by booster injections of 500 mg in incomplete Freund's adjuvant at 2-week intervals. Serum samples were tested 1 week after booster injections until the desired immune reaction was detected by western blotting. The final serum samples were obtained by allowing the collected blood to clot at 37°C for 1 h and freezing the serum supernatant fraction after centrifugation at 10,000g for 10 min. Total IgG was purified from the preimmune and immune serum by protein A affinity chromatography (Nab Spin columns, Pierce, Rockford, IL).
Validation of SS Antibodies
A previously described carboxy-terminal truncated TSS cDNA (TSS-1.1 pET11d) expressed in E. coli (Devarenne et al., 1998) served as a source of SS activity for immunoprecipitation experiments. After isopropylthio-β-galactoside induction of 2-mL cultures of E. coli harboring the TSS1.1 construct for 5 h, the cells were collected by centrifugation, resuspended in 100 μL of 50 mm Tris-HCl, pH 7.5, 40 mm MgCl2, 26.4 mm β-mercaptoethanol, 150 mm NaCl, 1 mm dithiothreitol, 2 mm phenylmethylsulfonyl fluoride, and 0.01% (v/v) Tween 20, sonicated three times for 5 s each time, and cooled on ice between sonications. The sonicate was centrifuged for 10 min at 4°C, and the supernatant was used for the immunoprecipitation assays. Aliquots of the supernatant were mixed with increasing amounts of purified IgG from either the preimmune or immune serums, incubated for 3 h at 4°C on a rotating platform, followed by an additional 1-h incubation after adding approximately 10 mg of protein A Sepharose. The samples were then centrifuged at 1,000g in a microcentrifuge at room temperature, and the pellets were washed three times with 50 mm Tris-HCl, pH 7.5, 40 mm MgCl2, 26.4 mm β-mercaptoethanol, 150 mm NaCl, and 0.01% (v/v) Tween 20. The final pellets were resuspended directly in TSS reaction buffer and assayed for enzyme activity.
SS Assay
The SS assay is based on the conversion of radiolabeled farnesyl diphosphate to squalene, which can be separated from one another by thin-layer chromatography and quantified by scintillation counting of the corresponding thin-layer chromatography zones. The assays were performed as described previously (Vögeli and Chappell, 1988; Chappell et al., 1989; Devarenne et al., 1998) except using 50 mm Tris-HCl, pH 7.5, 40 mm MgCl2, and 5 mm β-mercaptoethanol as the assay buffer. Protein levels were measured by the Bradford method (Bio-Rad, Hercules, CA).
SDS-PAGE and Immunodetection
For routine analyses, protein samples were size separated by SDS-PAGE (15% [w/v] acrylamide) and stained with Coomassie Blue. Samples for immunodetection were separated on 8% to 16% (w/v) precast gels (Bio-Rad) in SDS-PAGE running buffer and transferred to polyvinylidene difluoride membranes (Amersham, Arlington Heights, IL). Membranes were blocked with standard Tween 20/Tris-buffered saline (TTBS) blocking solution (Towbin and Gordon, 1984) before adding the anti-TSS antibodies. Membranes were incubated for 12 h at room temperature with shaking, washed three times with TTBS, and then challenged with goat anti-rabbit IgG conjugated with horseradish peroxidase (Sigma) in TTBS blocking solution for another hour at room temperature. Chemiluminescent detection of the secondary antibody was performed with the ECL-plus reagent according to the manufacturer's instructions (Amersham). The exposed x-ray film was quantified using laser densitometry.
Cell Culture and Elicitor Treatment
Cell suspension cultures of tobacco (Nicotiana tabacum cv Kentucky 14) were maintained in Murashige-Skoog medium on a weekly subculturing cycle according to Chappell and Nable (1987). Cultures in the rapid phase of growth (approximately 4 d after subculturing) were elicited by the addition of 1 μg of recombinant Phytophthora parasitica elicitin per milliliter of cell culture.
Isolation and Sequencing of Genomic TSS
A genomic clone for one TSS gene was previously isolated and described earlier by Devarenne et al. (1998). A second genomic clone was isolated for the current work using identical methodology. Both genomic clones were isolated from a tobacco genomic library packaged in lambda EMBL 3 (CLONTECH, Palo Alto, CA) by probing the library with a TSS cDNA. Overlapping subfragments of the genomic clones were obtained by PCR, blunt-end ligated into the EcoRV site of pBluescript II KS + (Stratagene, La Jolla, CA), and subjected to automated DNA sequencing.
Promoter Constructs, Plant Transformation, and Reporter Gene Expression
Full-length and 5′-deletion mutants of the TSS1 and TSS2 promoters were generated by PCR using primers having HindIII and BamHI restriction sites and inserting the amplified DNAs into the corresponding restriction sites flanking the GUS reporter gene in pBI101.1 (CLONTECH). Constructs verified by automated DNA sequencing were electroporated into Agrobacterium tumefaciens strain GV3850 and the resulting bacteria used for leaf dip transformation and regeneration of tobacco KY160 according to Yin et al. (1997). Seeds from Ro plants were germinated on medium containing 100 mg L−1 kanamycin, and the 3- to 4-week-old kanamycin-resistant plants were used to measure the expression of the GUS reporter gene.
Expression of the GUS reporter gene was measured quantitatively and qualitatively. For quantitative analyses, kanamycin resistant seedling were ground in 50 mm NaPO4, pH 7, 0.05% (v/v) Triton X-100, and 0.1% (v/v) β-mercaptoethanol and the level of GUS enzyme activity (methylumbelliferone glucuronidase activity) in the 10,000g supernatant determined as described by Yin et al. (1997). Plants were also stained histochemically for GUS expression by submerging the seedling in 50 mm NaPO4, pH 7, 0.5 mm potassium ferricyanide, 10 mm EDTA, and 0.05% (v/v) Triton X-100 with approximately 35 mg of 5-bromo-4-chloro-3-indolyl β-glucuronide per 25 mL of buffer for 24 to 72 h at 37°C. Plants were subsequently destained in 75% to 100% (v/v) ethanol.
ACKNOWLEDGMENTS
We thank Dr. Subbarro Bondada for his excellent advice and support with the immunological work and Mr. Wes Davis for his assistance in evaluating the transgenic lines.
Footnotes
This work was supported by a grant from the National Science Foundation and by the Kentucky Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001438.
LITERATURE CITED
- Adler JH, Grebenok RJ. Biosynthesis and distribution of insect-molting hormones in plants: a review. Lipids. 1995;30:257–262. doi: 10.1007/BF02537830. [DOI] [PubMed] [Google Scholar]
- Bhatt PN, Bhatt DP. Regulation of sterol biosynthesis in Solanum species. J Exp Bot. 1984;35:890–896. [Google Scholar]
- Brindle PA, Kuhn PJ, Threlfall DR. Biosynthesis and metabolism of sesquiterpenoid phytoalexins and triterpenoids in potato cell-suspension cultures. Phytochemistry. 1988;27:133–150. [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Caelles C, Ferrer A, Balcells L, Hegardt FG, Boronat A. Isolation and structural characterization of a cDNA-encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme-A reductase. Plant Mol Biol. 1989;13:627–638. doi: 10.1007/BF00016018. [DOI] [PubMed] [Google Scholar]
- Chappell J, Chrispeels MJ. Transcriptional and posttranscriptional control of phaseolin and phytohemagglutinin gene-expression in developing cotyledons of Phaseolus vulgaris. Plant Physiol. 1986;81:50–54. doi: 10.1104/pp.81.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Nable R. Induction of sesquiterpenoid biosynthesis in tobacco cell suspension cultures by fungal elicitor. Plant Physiol. 1987;85:469–473. doi: 10.1104/pp.85.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Vonlanken C, Vogeli U, Bhatt P. Sterol and sesquiterpenoid biosynthesis during a growth-cycle of tobacco cell suspension cultures. Plant Cell Rep. 1989;8:48–52. doi: 10.1007/BF00735777. [DOI] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C. Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl Coenzyme-A reductase a rate-limiting step for isoprenoid biosynthesis in plants. Plant Physiol. 1995;109:1337–1343. doi: 10.1104/pp.109.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun KT, Simoni RD. The role of the membrane domain in the regulated degradation of 3-hydroxy-3-methylglutaryl Coenzyme A reductase. J Biol Chem. 1992;267:4236–4246. [PubMed] [Google Scholar]
- Dale S, Arro M, Becerra B, Morrice NG, Boronat A, Hardie DG, Ferrer A. Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem. 1995;233:506–513. doi: 10.1111/j.1432-1033.1995.506_2.x. [DOI] [PubMed] [Google Scholar]
- Devarenne TP, Shin DH, Back K, Yin SH, Chappell J. Molecular characterization of tobacco squalene synthase and regulation in response to fungal elicitor. Arch Biochem Biophys. 1998;349:205–215. doi: 10.1006/abbi.1997.0463. [DOI] [PubMed] [Google Scholar]
- Diener AC, Li HX, Zhou WX, Whoriskey WJ, Nes WD, Fink GR. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell. 2000;12:853–870. doi: 10.1105/tpc.12.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem. 1999;68:157–185. doi: 10.1146/annurev.biochem.68.1.157. [DOI] [PubMed] [Google Scholar]
- Enjuto M, Balcells L, Campos N, Caelles C, Arro M, Boronat A. Arabidopsis thaliana contains 2 differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc Natl Acad Sci USA. 1994;91:927–931. doi: 10.1073/pnas.91.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M, Lumbreras V, Marin C, Boronat A. Expression of the Arabidopsis HMG2 gene, encoding 3-hydroxy-3-methylglutaryl Coenzyme-A reductase, is restricted to meristematic and floral tissues. Plant Cell. 1995;7:517–527. doi: 10.1105/tpc.7.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust JR, Brown MS, Goldstein JL. Squalene synthetase activity in human fibroblasts: regulation via the low-density lipoprotein receptor. Proc Natl Acad Sci USA. 1979;76:5018–5022. doi: 10.1073/pnas.76.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner GP, Patterson GW, Lusby WR. Developmental regulation of sterol biosynthesis in Cucurbita maxima l. Lipids. 1989;24:271–277. [Google Scholar]
- Fujioka S, Li JM, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Kroon PA, Matern U, Threlfall DR, Whitehead IM. Inhibition of phytosterol biosynthesis in elicitor-treated cultures of Ammi majus. Phytochemistry. 1993;34:139–145. [Google Scholar]
- Garg VK, Nes WD. Changes in sterol biosynthesis from [2-14C]mevalonic acid during development of Cucurbita maxima seedlings. In: Stumpf PK, Mudd JB, Nes WD, editors. The Metabolism, Structure, and Function of Plant Lipids. New York: Plenum Press; 1987. pp. 87–89. [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Guan GM, Dai PH, Osborne TF, Kim JB, Shechter I. Multiple sequence elements are involved in the transcriptional regulation of the human squalene synthase gene. J Biol Chem. 1997;272:10295–10302. doi: 10.1074/jbc.272.15.10295. [DOI] [PubMed] [Google Scholar]
- Guan GM, Dai PH, Shechter I. Differential transcriptional regulation of the human squalene synthase gene by sterol regulatory element-binding proteins (SREBP) 1a and 2 and involvement of 5′ DNA sequence elements in the regulation. J Biol Chem. 1998;273:12526–12535. doi: 10.1074/jbc.273.20.12526. [DOI] [PubMed] [Google Scholar]
- Guan GM, Jiang GJ, Koch RL, Shechter I. Molecular cloning and functional analysis of the promoter of the human squalene synthase gene. J Biol Chem. 1995;270:21958–21965. doi: 10.1074/jbc.270.37.21958. [DOI] [PubMed] [Google Scholar]
- Guo DA, Venkatramesh M, Nes WD. Developmental regulation of sterol biosynthesis in Zea mays. Lipids. 1995;30:203–219. doi: 10.1007/BF02537823. [DOI] [PubMed] [Google Scholar]
- Hanley K, Chappell J. Solubilization, partial-purification, and immunodetection of squalene synthetase from tobacco cell suspension cultures. Plant Physiol. 1992;98:215–220. doi: 10.1104/pp.98.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann MA. Plant sterols and the membrane environment. Trends Plant Sci. 1998;3:170–175. [Google Scholar]
- Hartmann MA, Benveniste P. Plant membrane sterols: isolation, identification, and biosynthesis. Methods Enzymol. 1987;148:632–650. [Google Scholar]
- Haudenschild C, Hartmann MA. Inhibition of sterol biosynthesis during elicitor-induced accumulation of furanocoumarins in parsley cell suspension cultures. Phytochemistry. 1995;40:1117–1124. [Google Scholar]
- Heupel RC, Nes WD, Verbeke JA. Developmental regulation of sterol and pentacyclic triterpene biosynthesis and composition: a correlation with sorghum floral initiation. In: Stumpf PK, Mudd JB, Nes WD, editors. The Metabolism Structure, and Function of Plant Lipids. New York: Plenum Press; 1987. pp. 53–56. [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takatsuto S, Ishii A, Aida M, Yoshida S, Sheen J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000;14:1485–1497. [PMC free article] [PubMed] [Google Scholar]
- Karst F, Lacroute F. Ergosterol biosynthesis in Saccharomyces cerevisiae: mutants deficient in the early steps of the pathway. Mol Gen Genet. 1977;154:269–277. doi: 10.1007/BF00571282. [DOI] [PubMed] [Google Scholar]
- Kemp R, Goad L, Mercer E. Changes in the levels and composition of the esterified and unesterified sterols of maize seedlings during germination. Phytochemistry. 1967;6:1609–1615. [Google Scholar]
- Kemp RJ, Mercer EI. The sterol esters of maize seedlings. Biochem J. 1968;110:111–118. doi: 10.1042/bj1100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MA, Barbuch R, Bard M. Transcriptional regulation of the squalene synthase gene (erg9) in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1445:110–122. doi: 10.1016/s0167-4781(99)00035-4. [DOI] [PubMed] [Google Scholar]
- Kennedy MA, Bard M. Positive and negative regulation of squalene synthase (erg9), an ergosterol biosynthetic gene, in Saccharomyces cerevisiae. Biochim Biophys Acta. 2001;1517:177–189. doi: 10.1016/s0167-4781(00)00246-3. [DOI] [PubMed] [Google Scholar]
- Korth KL, Jaggard DAW, Dixon RA. Developmental and light-regulated post-translational control of 3-hydroxy-3-methylglutaryl-CoA reductase levels in potato. Plant J. 2000;23:507–516. doi: 10.1046/j.1365-313x.2000.00821.x. [DOI] [PubMed] [Google Scholar]
- Learned RM, Fink GR. 3-Hydroxy-3-methylglutaryl-Coenzyme-A reductase from Arabidopsis thaliana is structurally distinct from the yeast and animal enzymes. Proc Natl Acad Sci USA. 1989;86:2779–2783. doi: 10.1073/pnas.86.8.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillotvernier P, Gondet L, Schaller H, Benveniste P, Belliard G. Genetic-study and further biochemical-characterization of a tobacco mutant that overproduces sterols. Mol Gen Genet. 1991;231:33–40. doi: 10.1007/BF00293818. [DOI] [PubMed] [Google Scholar]
- Nakashima T, Inoue T, Oka A, Nishino T, Osumi T, Hata S. Cloning, expression, and characterization of cDNAs encoding Arabidopsis thaliana squalene synthase. Proc Natl Acad Sci USA. 1995;92:2328–2332. doi: 10.1073/pnas.92.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita JO, Gruissem W. Tomato hydroxymethylglutaryl-coa reductase is required early in fruit-development but not during ripening. Plant Cell. 1989;1:181–190. doi: 10.1105/tpc.1.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD. Multiple roles for plant sterols. In: Stumpf PK, Mudd JB, Nes WD, editors. The Metabolism, Structure, and Function of Plant Lipids. New York: Plenum Press; 1987. pp. 3–9. [Google Scholar]
- Newman TC, Ohmetakagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Takatsuto S, Sakurai A, Yoshida S, Li JM, Chory J. Arabidopsis det2 is defective in the conversion of (24r)-24-methylcholest-4-en-3-one to (24r)-24-methyl-5 alpha-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 1999;120:833–839. doi: 10.1104/pp.120.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omkumar RV, Darnay BG, Rodwell VW. Modulation of syrian-hamster 3-hydroxy-3-methylglutaryl-CoA reductase-activity by phosphorylation: role of serine-871. J Biol Chem. 1994;269:6810–6814. [PubMed] [Google Scholar]
- Omkumar RV, Rodwell VW. Phosphorylation of ser(871) impairs the function of his(865) of syrian-hamster 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1994;269:16862–16866. [PubMed] [Google Scholar]
- Osborne TF. Transcriptional control mechanisms in the regulation of cholesterol balance. Crit Rev Eukaryot Gene Expr. 1995;5:317–335. doi: 10.1615/critreveukargeneexpr.v5.i3-4.50. [DOI] [PubMed] [Google Scholar]
- Popjak G, Agnew WS. Squalene synthetase. Mol Cell Biochem. 1979;27:97–116. doi: 10.1007/BF00218354. [DOI] [PubMed] [Google Scholar]
- Popjak G, Goodman DS, Cornforth JW, Cornforth RH, Ryhage R. Mechanism of squalene biosynthesis from mevalonate and farnesyl pyrophosphate. Biochem Biophys Res Commun. 1961a;4:138–142. doi: 10.1016/0006-291x(61)90363-1. [DOI] [PubMed] [Google Scholar]
- Popjak G, Goodman DS, Cornforth JW, Cornforth RH, Ryhage R. Studies on the biosynthesis of cholesterol. J Biol Chem. 1961b;236:1934–1947. [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-Coenzyme A reductase. J Biol Chem. 2000;275:35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Tsay YH, Kienzle BK, Smithmonroy CA, Bishop RW. Conservation between human and fungal squalene synthetases: similarities in structure, function, and regulation. Mol Cell Biol. 1993;13:2706–2717. doi: 10.1128/mcb.13.5.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M, Gruissem W. Arachidonic acid alters tomato hmg expression and fruit growth and induces 3-hydroxy-3-methylglutaryl coenzyme a reductase-independent lycopene accumulation. Plant Physiol. 1999;119:41–48. doi: 10.1104/pp.119.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the pest hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G. Fackel is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Shih M, Kuc J, Williams EB (1973) Suppression of steroid glycoalkaloid accumulation as related to rishitin accumulation in potato tubers. 63: 821–826.
- Stamellos KD, Shackelford JE, Shechter I, Jiang GJ, Conrad D, Keller GA, Krisans SK. Subcellular-localization of squalene synthase in rat hepatic cells: biochemical and immunochemical evidence. J Biol Chem. 1993;268:12825–12836. [PubMed] [Google Scholar]
- Threlfall DR, Whitehead IM. Coordinated inhibition of squalene synthetase and induction of enzymes of sesquiterpenoid phytoalexin biosynthesis in cultures of Nicotiana tabacum. Phytochemistry. 1988;27:2567–2580. [Google Scholar]
- Toroser D, Huber SC. 3-Hydroxy-3-methylglutaryl-Coenzyme A reductase kinase and sucrose-phosphate synthase kinase activities in cauliflower florets: Ca2+ dependence and substrate specificities. Arch Biochem Biophys. 1998;355:291–300. doi: 10.1006/abbi.1998.0740. [DOI] [PubMed] [Google Scholar]
- Towbin H, Gordon J. Immunoblotting and dot immunobinding: current status and outlook. J Immunol Methods. 1984;72:313–340. doi: 10.1016/0022-1759(84)90001-2. [DOI] [PubMed] [Google Scholar]
- Tozawa R, Ishibashi S, Osuga J, Yagyu H, Oka T, Chen Z, Ohashi K, Perrey S, Shionoiri F, Yahagi N et al. Embryonic lethality and defective neural tube closure in mice lacking squalene synthase. J Biol Chem. 1999;274:30843–30848. doi: 10.1074/jbc.274.43.30843. [DOI] [PubMed] [Google Scholar]
- Trojanowska MR, Osbourn AE, Daniels MJ, Threlfall DR. Biosynthesis of avenacins and phytosterols in roots of Avena sativa cv. Image Phytochemistry. 2000;54:153–164. doi: 10.1016/s0031-9422(00)00062-5. [DOI] [PubMed] [Google Scholar]
- Vanderheijden R, Threlfall DR, Verpoorte R, Whitehead IM. Regulation and enzymology of pentacyclic triterpenoid phytoalexin biosynthesis in cell-suspension cultures of Tabernaemontana divaricata. Phytochemistry. 1989;28:2981–2988. [Google Scholar]
- Vanderheijden R, Verheij ER, Schripsema J, Svendsen AB, Verpoorte R, Harkes PAA. Induction of triterpene biosynthesis by elicitors in suspension cultures of Tabernaemontana species. Plant Cell Rep. 1988;7:51–54. doi: 10.1007/BF00272977. [DOI] [PubMed] [Google Scholar]
- Vögeli U, Chappell J. Induction of sesquiterpene cyclase and suppression of squalene synthetase activities in plant cell cultures treated with fungal elicitor. Plant Physiol. 1988;88:1291–1296. doi: 10.1104/pp.88.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin SH, Mei L, Newman J, Back K, Chappell J. Regulation of sesquiterpene cyclase gene expression: characterization of an elicitor- and pathogen-inducible promoter. Plant Physiol. 1997;115:437–451. doi: 10.1104/pp.115.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Yamada N, Doke N. cDNA cloning of sesquiterpene cyclase and squalene synthase, and expression of the genes in potato tuber infected with Phytophthora infestans. Plant Cell Physiol. 1999;40:993–998. doi: 10.1093/oxfordjournals.pcp.a029633. [DOI] [PubMed] [Google Scholar]
- Zook MN, Kuc JA. Induction of sesquiterpene cyclase and suppression of squalene synthetase-activity in elicitor-treated or fungal-infected potato-tuber tissue. Physiol Mol Plant Pathol. 1991;39:377–390. [Google Scholar]