Abstract
Phosphate (Pi) and its analog phosphite (Phi) are acquired by plants via Pi transporters. Although the uptake and mobility of Phi and Pi are similar, there is no evidence suggesting that plants can utilize Phi as a sole source of phosphorus. Phi is also known to interfere with many of the Pi starvation responses in plants and yeast (Saccharomyces cerevisiae). In this study, effects of Phi on plant growth and coordinated expression of genes induced by Pi starvation were analyzed. Phi suppressed many of the Pi starvation responses that are commonly observed in plants. Enhanced root growth and root to shoot ratio, a hallmark of Pi stress response, was strongly inhibited by Phi. The negative effects of Phi were not obvious in plants supplemented with Pi. The expression of Pi starvation-induced genes such as LePT1, LePT2, AtPT1, and AtPT2 (high-affinity Pi transporters); LePS2 (a novel acid phosphatase); LePS3 and TPSI1 (novel genes); and PAP1 (purple acid phosphatase) was suppressed by Phi in plants and cell cultures. Expression of luciferase reporter gene driven by the Pi starvation-induced AtPT2 promoter was also suppressed by Phi. These analyses showed that suppression of Pi starvation-induced genes is an early response to addition of Phi. These data also provide evidence that Phi interferes with gene expression at the level of transcription. Synchronized suppression of multiple Pi starvation-induced genes by Phi points to its action on the early molecular events, probably signal transduction, in Pi starvation response.
Phosphate (Pi) is one of the major plant nutrients that influences virtually all the biochemical processes and developmental phases of plants. In the absence of Pi, plants exhibit characteristic deficiency symptoms including anthocyanin accumulation, enhanced root growth, and increased root to shoot ratio. The ability of plants to acquire Pi also increases during this period. Molecular dissection of responses to Pi starvation has provided evidence for coordinated expression of genes, including Pi transporters (Raghothama, 1999, 2000). Pi transporters are involved in acquiring Pi against the concentration gradient by an energy-mediated proton cotransport mechanism (Ullrich-Eberius et al., 1981). They are also known to transport ions such as arsenate, vanadate, and phosphite (Phi; Guest and Grant, 1991; Marschner, 1995).
Phi (HPO32−), also referred to as phosphorous acid or phosphonate, is an isostere of the Pi anion in which one of the oxygens bound to the P atom is replaced by hydrogen. The term Phi is used here to describe the alkali metal salts of phosphorous acid as suggested by Carswell et al. (1996). Phi is used extensively as a fungicide and also sold as a superior source of Pi (Guest and Grant, 1991; Rickard, 2000; McDonald et al., 2001a). Phi is rapidly absorbed and translocated within the plant (Guest and Grant, 1991). The uptake is pH dependent and subject to competition by Pi (Ouimette and Coffey, 1990). Furthermore, mobility of Phi in both xylem and phloem is similar to that of Pi (Ouimette and Coffey, 1989). Despite having similar structure and mobility, the published data indicate that Phi is a non-metabolizable form of Pi and plants cannot use this as the sole source of P (Sukarno et al., 1993; Carswell et al., 1996; Forster et al., 1998). One of the distinct differences between Pi and Phi is that Pi can be assimilated into organic P compounds within minutes of uptake, whereas plants lack the ability to assimilate Phi (MacIntire et al., 1950; Guest and Grant, 1991). Furthermore, enzymes that catalyze the transfer of Pi groups can discriminate between Pi and Phi (Guest and Grant, 1991). The observed nutritional effects of Phi are likely due to its oxidation to Pi by microbes (Ohtake et al., 1996). This biological conversion certainly makes Phi an important component of the global P cycle but not a direct source of nutrient for plants.
Phi exhibits various biological activities on fungi and plants (Coffey and Joseph, 1985; Plaxton and Carswell, 1999). Fungi belonging to oomycetes, particularly Phytophthora citricola and Phytophthora cinnamomi, are sensitive to Phi (Guest and Grant, 1991; El-Hamalawi et al., 1995; Wilkinson et al., 2001). It is quite stable and persists in plants for months serving as a deterrent to fungal attack (Ouimette and Coffey, 1989). There are several mechanisms by which Phi inhibits fungal growth. The toxicity of Phi is believed to be due to an increase in inorganic pyrophosphate, which inhibits key phosphorylation reactions in fungi (Niere et al., 1994). Adenylate synthesis appears to be one of the primary sites of action of Phi in fungi (Griffith et al., 1990). It is also shown to compete for the Pi-binding catalytic sites of phosphorylating enzymes (Barchietto et al., 1992). Pi-repressible acid phosphatase, a marker of Pi starvation response in yeast (Saccharomyces cerevisiae), was repressed in the presence of Phi under Pi stress (McDonald et al., 2001b). Furthermore, the nucleotide pools and pentose Pi metabolism in P. citrophthora were altered by phosphonate (Barchietto et al., 1992). Thus, the toxicity of Phi against fungal species may be due to differing sensitivities of phosphorylating enzymes, and/or metabolic pathways regulated by Pi. In addition, activation of plant defense responses by Phi has also been proposed (Guest and Grant, 1991).
Several studies have provided evidence for the negative effects of Phi on growth and development of plants (Sukarno et al., 1993; Carswell et al., 1996; Forster et al., 1998; Ticconi et al., 2001). Both commercial and technical grade Phi were effective in controlling the root and crown rot caused by Phytophthora capsici, but they did not serve as a source of plant nutrient (Forster et al., 1998). Root growth of onion (Allium cepa) and Brassica nigra was inhibited by application of Phi (Sukarno et al., 1993; Carswell et al., 1996). It also suppressed Pi starvation-induced root growth, root hair initiation, and elongation in Arabidopsis (Ticconi et al., 2001). In addition, Phi has been shown to interfere with Pi starvation responses of Brassica napus L. cv Jet Neul by down-regulating the induction of enzymes such as acid phosphatase, phosphoenolpyruvate phosphatase, inorganic pyrophosphate-dependent phosphofructokinase, and the high-affinity Pi translocator (Carswell et al., 1997). In Arabidopsis, the Pi starvation-induced nucleolytic enzyme activities and expression of Pi starvation-induced genes were suppressed by Phi (Ticconi et al., 2001). One common observation in most of these studies is that the negative effects of Phi could be overcome by Pi supplementation. Because multiple responses induced by Pi deficiency are suppressed by Phi, it was postulated that the signal transduction pathway leading to Pi starvation responses might be compromised (Carswell et al., 1997). This important observation has led to the present study in our laboratory. We asked the question: If Phi is interfering with signal transduction pathway, what happens to the coordinated expression of genes regulated by the Pi starvation response mechanism in plants? Here, we provide molecular evidence for suppression of coordinated expression of several Pi starvation-induced genes by Phi.
RESULTS
Phi Severely Affects Plant Growth and Development by Interfering with Pi Starvation- Induced Responses
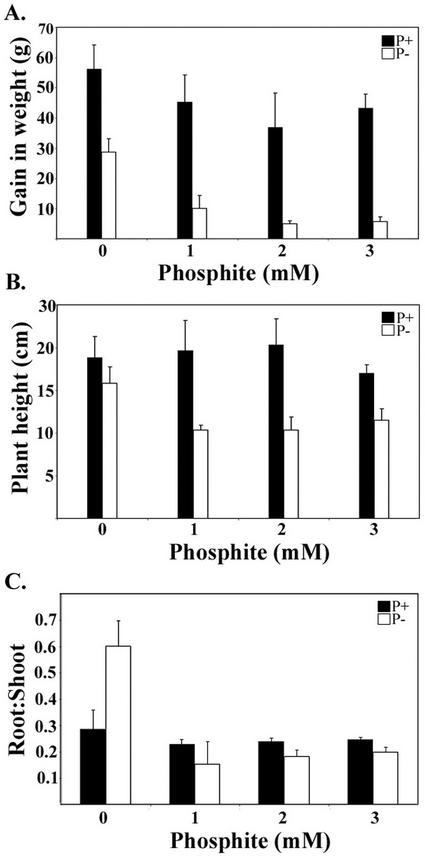
Effect of Phi on tomato (Lycopersicon esculentum) growth and development was analyzed using hydroponic plants grown in the presence or absence of Pi. Increasing concentrations of Phi in Pi-deficient media had significant negative effects on plant growth and development. A 2-fold difference in fresh weight between Pi-sufficient and -deficient seedlings was observed (Fig. 1A). However, in the presence of 1 mm Phi, gain in fresh weight of Pi-deficient plants was less than 30% of the control. Further suppression of growth was observed with increasing concentrations (2 and 3 mm) of Phi under Pi starvation. There were no obvious effects of Phi on Pi-sufficient plants. The pattern of accumulation of anthocyanin in leaves and stem, a hallmark of Pi deficiency, was distinctly different in Phi-treated plants. Pi-starved plants exhibited relatively uniform purple coloration in leaves and stem, whereas Phi-treated plants in the absence of Pi accumulated anthocyanin in patches, primarily on the lower surface of older leaves. Leaves of Phi-treated plants under Pi deficiency were epinastic, which is an uncharacteristic response to Pi deficiency. In addition, yellowing of lamina of young leaves was also observed in Phi-treated, Pi-deficient plants.
Figure 1.
Phi inhibits growth of Pi-starved tomato. Three-week-old tomato plants were transferred to one-half-strength Hoagland solution. After 7 d of growth, plants were treated with different concentrations of Phi (0–3 mm) in the presence or absence of Pi. A, Difference in total fresh weight (g) measured at the beginning and the end of 5 d of Phi treatment was expressed as gain in weight. Error bars indicate ± sd. B, Plant height (cm) was measured from the crown region to the apex of plants at the end of 5 d of Phi treatment. Error bars indicate ± sd. C, Root to shoot ratio of hydroponically grown tomato plants treated with different concentrations of Phi. Error bars indicate ±sd.
Reduction in plant growth in the presence of Phi could be attributed to two major morphological changes in plants. One distinguishing feature of Phi-treated, Pi-deficient plants was reduced internodal length. A significant reduction in plant height was noticed by addition of Phi to Pi-deficient medium (Fig. 1B), whereas Pi-sufficient plants appeared normal even in the presence of higher concentration of Phi (3 mm). The second major factor was a pronounced suppression of root growth in Pi-deficient plants treated with Phi (Fig. 1C). Deleterious effects of Phi on root growth in Pi-starved plants were quite obvious at 1 mm concentration, which caused a 3-fold reduction in root to shoot ratio. Even lower concentrations (0.1 and 0.3 mm) of Phi were sufficient to cause a decline in root to shoot ratio under Pi-deficient conditions (data not shown). In contrast, the root to shoot ratio of Pi-sufficient plants supplied with Phi showed minimal changes. These morphological differences indicated that none of the typical Pi starvation-induced symptoms were obvious in Pi-deficient plants grown in the presence of increasing concentrations of Phi.
Phi Specifically Inhibits the Root Elongation under Pi Stress
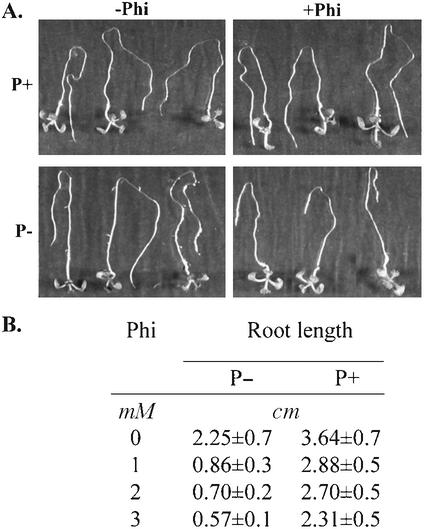
Effect of Phi on root growth was analyzed further by root bending assay of Arabidopsis. Root bending assay is an effective way to measure root elongation in response to a treatment. Phi strongly suppressed root growth of Pi-starved plants (Fig. 2A). One millimolar Phi resulted in nearly a 3-fold reduction in root growth (Fig. 2B). Reduction in root elongation paralleled with a decrease in root mass. Root weight of Pi-starved plants (0.51 ± 0.0001 mg plant−1, n = 300) decreased by 50% (0.24 ± 0.0001 mg plant−1, n = 300) in the presence of 3 mm Phi. There were no major differences in number of root hairs of Pi-deficient plants grown in the presence or absence of Phi for 5 d. However, the number of lateral roots initiated under Pi deficiency was suppressed by Phi (data not shown). In addition, leaves of Pi-deficient Arabidopsis plants were dark green, whereas newly emerging leaves of Pi-deficient plants grown in the presence of Phi showed signs of bleaching. This response was very similar to that observed in Pi-deficient tomato plants treated with Phi. These changes are a clear reflection of strong inhibitory effects of Phi on root growth under Pi deficiency conditions.
Figure 2.
Phi inhibits root growth of Pi-starved Arabidopsis seedlings. Arabidopsis seedlings were grown for 5 d on membranes placed on one-half-strength Murashige and Skoog agar medium. Plates were inclined vertically to allow the roots to grow down. A, The membranes along with seedlings were transferred to plates containing Phi in the presence (P+) or absence (P−) of Pi and allowed to grow for 5 more d. Plates were inverted such that the roots were pointing up to monitor root growth and bending. B, The membranes along with seedlings were transferred to plates containing different concentrations of Phi in the presence (P+) or absence (P−) of Pi. After 5 d of transfer, root length of plants was measured. The table represents the mean ± sd of root lengths of 20 seedlings.
Phi Interferes with the Expression of Pi Starvation-Induced Gene Expression
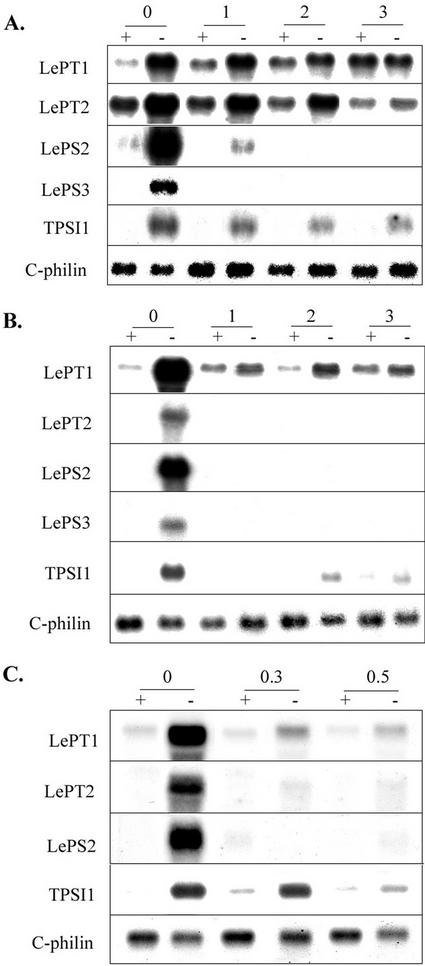
Effect of Phi on expression of several Pi starvation-induced genes was analyzed by northern blots. In general, Phi suppressed the expression of all Pi starvation-induced genes analyzed. Transcript levels of the high-affinity Pi transporters increased under Pi deficiency. Addition of Phi to Pi-deficient medium suppressed transcript accumulation of the transporters. The LePT1 and TPSI1 expression appeared to be less sensitive to Phi as compared with that of other Pi starvation-induced genes. Decrease in the level of transcripts was associated with increasing concentrations of Phi. Expression of LePS2, an acid phosphatase gene isolated from tomato (Baldwin et al., 2001), and two other novel Pi starvation-induced genes LePS3 and TPSI1 (Liu et al., 1998; Baldwin et al., 1999), was also suppressed in Pi-deficient, Phi-treated plants (Fig. 3A). A strong suppression of LePS2 and LePS3 transcript accumulation indicates that their expression is more sensitive to Phi. Effect of Phi was less pronounced in tomato plants grown under Pi sufficiency conditions (Fig. 3A). In addition, Phi treatment did not suppress the constitutive expression of tomato cyclophilin gene either in the presence or absence of Pi, indicating that inhibitory effects are specific to Pi stress-induced genes.
Figure 3.
Phi suppresses Pi starvation-induced gene expression in tomato. A, Three-week-old tomato seedlings were transferred to one-half-strength Hoagland nutrient solution. After 7 d, they were treated with Phi (0, 1, 2, and 3 mm) in the presence (+) or absence (−) of Pi for 5 d. Total RNA extracted from the roots was used for northern-blot analysis of Pi starvation-induced genes. cDNAs of genes indicated on the left of the figure were used as probes. Tomato cyclophilin (C-philin) served as a control. B, Tomato cell cultures grown in full-strength Murashige and Skoog medium were transferred to medium containing Phi (0, 1, 2, and 3 mm) with (+) or without (−) Pi. Cells were harvested after 48 h for RNA isolation and northern-blot analysis. C, Tomato cells grown in full-strength Murashige and Skoog medium were transferred to medium containing relatively low concentrations of Phi (0, 0.3, and 0.5 mm) in the presence (+) or absence (−) of Pi. Cells were harvested after 48 h for RNA isolation and northern-blot analysis.
The effect of Phi on Pi starvation-induced gene expression was more pronounced in cell cultures (Fig. 3, B and C). Pi starvation-induced expression of LePT2, LePS2, and LePS3 was highly suppressed in the presence of Phi. Expression of TPSI1 in tomato cells was reduced with increasing concentrations of Phi under Pi limitation. Negative effect of Phi on gene expression was evident even at lower concentrations. Application of 0.3 mm Phi had a significant effect on the expression of LePT1, LePT2, and LePS2 under Pi deficiency. Addition of 0.5 mm Phi was sufficient to suppress the expression of TPSI1. These data support the notion that Pi starvation-induced gene expression is one of the major targets of Phi action.
Phi Interferes with the Transcription of Pi Starvation-Induced Reporter Gene Expression
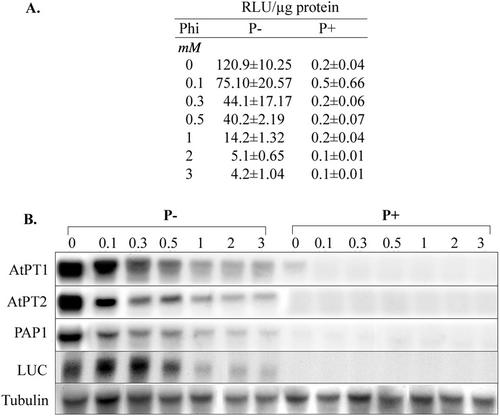
Analysis of reporter gene expression driven by a specific promoter in response to a treatment is an effective way to study gene expression. Plants expressing the luciferase reporter gene driven by the promoter of Arabidopsis Pi transporter (AtPT2:LUC), which is specifically induced under Pi starvation (Karthikeyan et al., 2000), were used to analyze the effect of Phi on transcription of Pi starvation-induced genes. Luciferase (Luc) activity served as an indicator of the ability of Phi and Pi to alter the transcription of Pi starvation-induced genes. Pi-sufficient (1.25 mm) plants showed relatively low levels of luminescence both in the presence or absence of Phi (Fig. 4A). However, suppression of Pi starvation-induced Luc activity, even at lower concentrations (0.1, 0.3, and 0.5 mm) of Phi, was evident in transgenic plants. Nearly a 30-fold inhibition in the Pi starvation-induced Luc activity was observed in plants supplied with 3 mm Phi (Fig. 4A). Interestingly, the level of suppression of Luc activity with increasing concentration of Phi was correlated with levels of LUC and AtPT2 transcripts (Fig. 4B). Similar repression of AtPT1 and PAP1 (purple acid phosphatase, a gift from Dr. Thomas McKnight, Texas A&M University, College Station, TX) genes by Phi was also observed under Pi stress. The reporter gene expression studies provided the strongest evidence that Phi interferes with Pi starvation-induced responses at the level of transcription.
Figure 4.
Suppression of Pi starvation-induced gene expression in Arabidopsis by Phi. A, AtPT2-LUC transgenic Arabidopsis were grown in one-half-strength Murashige and Skoog liquid medium for 6 d. The media was replaced with fresh solution containing indicated concentrations of Phi either in the presence (P+) or absence (P−) of Pi. After 5 d of Phi treatment plants were analyzed for Luc activity. Luc activity was expressed as relative luminescence units per microgram of protein. Values represent the mean of three replicates ± sd. B, Six-day-old Arabidopsis seedlings grown in one-half-strength Murashige and Skoog liquid medium were transferred to medium containing increasing concentrations of Phi (0–3 mm) in the presence (P+) or absence (P−) of Pi. After 5 d of transfer, plants were harvested for total RNA isolation and northern-blot analysis. cDNAs of genes representing AtPT1, AtPT2, PAP1, and LUC were used as probes. Arabidopsis tubulin gene probe served as a control.
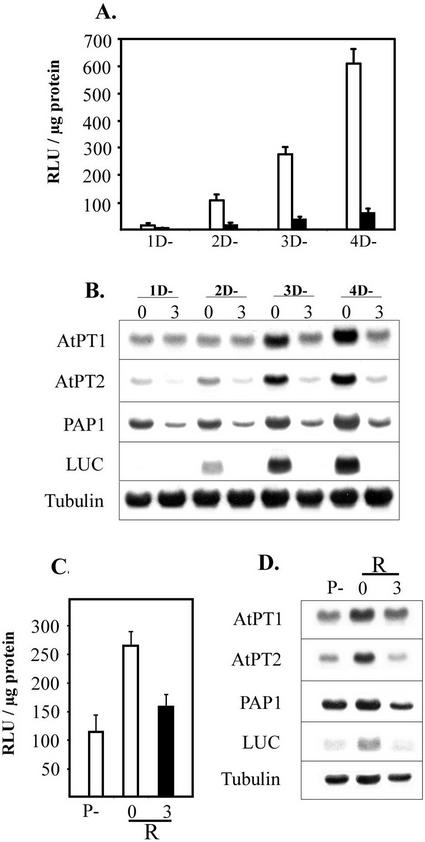
Phi effectively suppressed the expression of Pi stress responsive genes before any noticeable changes in plant growth (Fig. 5B). One day of Phi treatment was sufficient to suppress AtPT2 and PAP1 expression, whereas LUC transcript was reduced within 2 d, and by the 3rd d a distinct suppression was noticed in expression of all the examined Pi stress response genes. Suppression of Luc enzyme activity mimicked that of gene expression (Fig. 5A). Increasing duration of Pi deficiency increased the activity of Luc, whereas Phi suppressed the Pi starvation-induced activity of the enzyme. A nearly 3-fold decrease in LUC activity after 1 d of Phi treatment further supports the notion that repression of Pi starvation-induced gene expression is likely to be a primary mode of action of Phi. The rapidity of suppression of gene expression by Phi was examined further by transferring Pi-starved Arabidopsis to Pi-deficient media containing 3 mm or no Phi for 24 h. Interestingly, there was a drastic decrease in the Pi starvation-induced reporter gene activity in the presence of Phi (Fig. 5D). Similar decreases in transcript level of LUC, AtPT1, AtPT2, and PAP1 were also noticed in the presence of Phi (Fig. 5C). In all these experiments, the expression of tubulin, a constitutively expressed gene, remained relatively unaltered. Taken together, these data suggest that Phi specifically inhibits Pi starvation-induced gene expression before any visible changes in plant morphology.
Figure 5.
Inhibition of Pi starvation-induced genes is an early response to Phi treatment. Arabidopsis seedlings were grown in one-half-strength Murashige and Skoog liquid media for 6 d and transferred to P− media without (0) Phi (white bars) or with 3 mm Phi (black bars). Replicated samples were harvested at indicated time (1–4 d) after Phi treatment for measuring LUC activity (A), and isolating RNA for northern-blot analysis (B). In a separate experiment, 6-d-old Arabidopsis seedlings were transferred to Pi-deficient medium for 2 d (P−) to activate Pi starvation-induced gene expression. Sets of Pi-starved plants received (R) an additional day of Pi stress in the absence (0) or presence of 3 mm of Phi. After 24 h of treatment, plants were harvested for measuring LUC activity (C) and isolating RNA for northern-blot analysis (D). The northern blots were probed with 32P-labeled cDNAs of AtPT1, AtPT2, PAP1, LUC, and tubulin.
DISCUSSION
Plants have evolved several biochemical mechanisms to utilize both inorganic and organic Pi complexes in the rhizosphere (Plaxton and Carswell, 1999; Raghothama, 1999). Despite all these biochemical adaptations, there are no data demonstrating that plants can convert Phi to Pi (MacIntire et al., 1950; Guest and Grant, 1991). 31P NMR studies provided evidence that Phi could not be converted to other Pi-containing compounds in plants, indicating that Phi is unable to serve as a substrate for Pi-dependent enzymes (Carswell et al., 1996). However, structural similarity between Pi and Phi allow both to be taken up via Pi transporters. Uptake kinetics revealed a rapid acquisition and translocation of Phi in plants (d'Arcy-Lameta and Bompeix, 1991). The similarity between Pi and Phi appears to end at the level of translocation. Because Phi is not converted into Pi in plants, it fails to enter the biochemical pathways. This may be one reason for observed changes in morphology of both tomato and Arabidopsis. Interestingly, the negative effect of Phi on plant growth was amplified in the absence of Pi. Root growth was severely inhibited, young leaves were partially bleached, and internodal length was reduced in Phi-treated, Pi-deficient plants. These changes are similar to the deleterious effects of Phi on growth of onion, tomato, B. nigra, and Arabidopsis (Sukarno et al., 1993; Carswell et al., 1996; Forster et al., 1998; Ticconi et al., 2001). Phi treatment led to a reduction in root to shoot ratio of Pi-starved tomato and Arabidopsis. In addition, suppression of both root hair initiation and elongation in Pi-starved Arabidopsis has been reported (Ticconi et al., 2001). Proteoid root formation in white lupin (Lupinus albus), a typical response to Pi deficiency, was also suppressed by Phi treatment (Gilbert al., 2000). Unlike in whole plants, fresh weight of Pi-starved tomato cell suspensions did not change much during 2 d of Phi treatment. The cells were in early lag phase of growth during these 2 d; hence, weight gain was particularly minimal in Pi-deficient cultures.
The negative effects of Phi were suppressed in plants provided with sufficient Pi. This is likely to be due to a combination of reduced uptake of Phi or its inability to interfere with biochemical reactions in the presence of Pi. The competition between Pi and Phi for the common transport system is well documented in fungi and plant cells (Barchietto et al., 1989; Griffith et al., 1989; Grant et al., 1992; Carswell et al., 1997). In the presence of both Pi and Phi, organisms preferentially acquire Pi. An inverse relationship between fungal sensitivity to Phi and Pi concentration has been established (Grant et al., 1992; Griffith et al., 1993). In B. nigra, the concentration of Phi in roots and leaves was lower in Pi-sufficient than -deficient plants (Carswell et al., 1996). These data strongly point to an antagonistic interaction between Phi and Pi at the level of uptake. This interaction may in part explain the observed differences in growth of tomato and Arabidopsis in the presence or absence of Pi. Even if Phi is able to overcome the competitive effect, and enter the plant, it may still be incapable of interfering with Pi-dependent metabolism in the presence of Pi. This may be particularly due to ability of enzymes to discriminate Phi in the presence of Pi (Plaxton, 1998) and account for the lack of deleterious effects of Phi on plants grown under adequate Pi nutrition. Reasons for inhibitory effects of Phi on plant growth are less well understood compared with extensive literature describing its effects on fungi. Phi acts as a systemic fungicide against several species of fungi (Guest and Grant, 1991). Its toxicity appears to be due to a combination of factors, including decrease in total adenylate pool, accumulation of poly- and pyrophosphates, and inhibition of several enzymes involved in the glycolytic and pentose Pi pathways (Griffith et al., 1990; Niere et al., 1990, 1994; Guest and Grant, 1991; Barchietto et al., 1992; Stehmann and Grant, 2000). Furthermore, addition of Phi to Pi-starved yeast resulted in reduced levels of sugar Pis, pyrophosphates, and polyphosphates, and it also inhibited Pi stress-repressible acid phosphatase (McDonald et al., 2001b). In contrast to this extensive documentation of antifungal action of Phi, its influence on plant metabolism is not well understood. In Brassica napus cell cultures, Phi greatly suppressed the induction of several Pi stress marker enzymes (Carswell et al., 1996, 1997). Phi has also been shown to disrupt protein phosphorylation during Pi stress, thus affecting key events common to multiple Pi stress-induced mechanisms. In Arabidopsis, Phi suppressed the activity of nucleolytic enzymes and expression of acid phosphatase and Pi transporter genes (Ticconi et al., 2001).
In this study, we provide molecular evidence that Phi suppresses expression of several Pi starvation-induced genes in plants. The suppressed genes include high-affinity Pi transporters, phosphatases, a homolog of glycerol-3-Pi permease (LePS3), and a novel gene (TPSI1) induced under Pi starvation. In general, the effect of Phi on gene expression in tomato cell culture was much more pronounced than that in plants. This may be due differences in rate of uptake and accumulation of Phi in cell culture and plants. Expression of TPSI1 appeared to be relatively less sensitive to Phi as compared with that of other Pi starvation-induced genes in both plants and cell culture. In addition, accumulation of high-affinity Pi transporter protein (LePT1) was also suppressed in Pi-starved plants by Phi (data not shown). Similar observations in expression of several Pi starvation-induced genes in Arabidopsis further substantiated its effect on gene expression. Taken together, these results indicate that one of the primary mechanisms by which Phi affects plant growth is by interfering with Pi starvation-induced gene expression. Furthermore, suppression of Luc reporter gene expression driven by the AtPT2 promoter in Arabidopsis provided the strongest evidence for inhibition of Pi stress responses at the level of transcription. This seems to be an early response, as suggested by a rapid decrease in transcript levels of Pi starvation-induced genes and the reporter gene activity within 24 h of Phi treatment. The suppressive effect of Phi appears to be specific for Pi starvation-induced gene expression as indicated by relatively constitutive levels of tomato cyclophilin and Arabidopsis tubulin transcripts. Similar observations were also made with respect to the expression of auxin-induced genes in Arabidopsis (Ticconi et al., 2001). Furthermore, concentrations of Phi used in these experiments were less than the concentration (5 mm) suggested to be toxic to Arabidopsis. The intriguing question is how can Phi synchronously suppress multiple responses of plants to Pi starvation.
Suppression of multiple Pi starvation responses by Phi may be due to inhibition of primary Pi starvation response mechanism(s). The target of Phi appears to be signal transduction during Pi starvation (Carswell et al., 1997). It is likely that the signal transduction pathway may be recognizing Phi as Pi and thus coordinately suppressing the expression of multiple genes involved in the response mechanism. Under these conditions, the plants will be severely starved for Pi, whereas the Pi-sensing mechanism is thoroughly compromised by Phi, creating a “pseudo Pi sufficiency” condition. Many of the physiological, biochemical, and molecular responses of plants to Phi substantiate this assumption. Irrespective of the utility of Phi as a readily available source of Pi, it could serve as a powerful research tool in dissecting the Pi starvation-induced molecular changes. There is growing evidence suggesting that Pi stress response-disrupting properties of Phi can be exploited to study Pi signaling in plants (Carswell et al., 1997; Ticconi et al., 2001). Its use in elucidating the Pi stress signal transduction pathway requires further investigation.
MATERIALS AND METHODS
Hydroponic Culture of Tomato (Lycopersicon esculentum L. VFNT “Cherry”) Plants
Tomato var. OS4 seeds were germinated in seedling trays filled with Metro mix 360 (Scotts Co,. Marysville, OH). Four leaf stage seedlings were removed from the media and roots were washed free of media and transferred to aerated, one-half-strength Hoagland nutrient solution. Hydroponic culture of tomato was carried out in a green house (23.5°C day/21°C night, 230 μm m2 s−1 light). After 7 d, the seedlings were weighed and transferred to one-half-strength Hoagland nutrient solution containing different concentrations of Phi (0, 1, 2, or 3 mm) in the presence of 250 μm (P+) or absence (P−) of Pi. A stock solution of 250 mm of Phi was prepared by dissolving phosphorus acid (Aldrich, Milwaukee, WI) in distilled water and pH adjusted to 6.5 with 10 n NaOH. The stock solution was added to the media to obtain required concentrations of Phi. After 5 d of Phi treatment, plant growth parameters such as plant height, fresh weight, internodal length, gain in fresh weight, and root to shoot ratio were recorded. Representative samples were collected for RNA isolation.
Determination of Arabidopsis Root Growth in the Presence of Phi
Growth of Arabidopsis ecotype Columbia roots in the presence of Phi was determined by root bending assay. Seeds were surface sterilized by rinsing in 70% (v/v) ethanol and treating with 50% (v/v) bleach for 10 min, followed by several washes of sterile water. After stratification for 2 d at 4°C, seeds were sown in three rows on cellophane membranes (Bio-Rad Laboratories, Hercules, CA) layered over one-half-strength Murashige and Skoog solid medium in square petri plates. Plates were placed in vertical position to allow the roots to grow down for 5 d under 16-h-light (34.3 μm m−2 s−1)/8-h-dark cycle at 25°C. Membranes along with the seedlings were transferred to fresh plates containing 1.25 mm (P+) or no (P−) Pi and different concentrations of Phi (0–3 mm). The orientation of plates was reversed such that roots were pointing up to monitor root bending and growth. Length of roots from the point of bending was recorded after 5 d of transfer. Twenty seedlings were used in replicated experiments to analyze the effect of Phi on root growth.
Analysis of Effects of Phi on Short-Term Growth of Arabidopsis
Surface sterilized Arabidopsis seeds were stratified for 2 d. Approximately 40 seeds were germinated in one-half-strength Murashige and Skoog liquid medium in 16-h-light (23.3 μm m−2 s−1)/8-h-dark cycle at 25°C in a gyratory shaker set at 100 rpm. After 6 d of germination, plants were rinsed with water and transferred to P+ (1.25 mm Pi) or P− medium with different concentrations of Phi (0–3 mm) for 1 to 5 d. In a separate experiment effect of Phi on Pi starvation-induced gene expression was further analyzed by transferring Pi-starved plants to Phi containing medium. In this study, 6-d-old transgenic Arabidopsis seedlings were starved for Pi for 2 d to induce gene expression. Starved plants were transferred to fresh Pi-deficient medium with (3 mm) or without Phi for 24 h. The plants were harvested, rinsed in distilled water, frozen in liquid nitrogen, and stored at −80°C.
Treatment of Tomato Cell Culture with Phi
Tomato cell suspension culture was maintained as described earlier (Bressan et al., 1981). Seven days after subculturing, cells were collected on Miracloth (Calbiochem-Novabiochem Corporation, La Jolla, CA), washed with Pi-deficient (0 μm, P−) or -sufficient (1.25 mm, P+) media. Filter-sterilized 250 mm stock solution of Phi was added to 150 mL of the same media used for washing to obtain final concentrations of 0, 0.3, 0.5, 1, 2, and 3 mm of Phi. Then, 2.25 g of appropriately washed cells was transferred to the media. Cells were cultured for 2 d in the presence or absence of Phi, collected on Miracloth, frozen in liquid nitrogen, and stored at −80°C.
RNA Extraction and Northern Analysis
Total RNA was isolated from tomato and Arabidopsis by hot-phenol extraction and lithium chloride precipitation (Pawlowski et al., 1994). Ten micrograms of total RNA was electrophoretically separated on 1.2% (w/v) denaturing formaldehyde agarose gels and blotted onto a nitrocellulose membrane (Schleicher & Schull, Keene, NH) according to Sambrook et al. (1989). The nitrocellulose membranes were hybridized overnight with a 32P-labeled probe (106 cpm mL−1) in a solution containing 50% (v/v) formamide, 5× Denhardt's solution, 0.1% (w/v) SDS, 6× SSPE, and 100 μg mL−1 denatured salmon sperm DNA at 42°C. Probes represented the high-affinity Pi transporters (LePT1 and 2), phosphatase (LePS2), novel Pi starvation-induced genes (TPSI1 and LePS3), cyclophilin of tomato, and Pi transporters (AtPT1 and 2), purple acid phosphatase (PAP1), and LUC and tubulin from Arabidopsis. Hybridization and washing conditions were same as those described earlier (Liu et al., 1998).
Measurement of Reporter Gene Activity in Phi-Treated Transgenic Arabidopsis Plants
Arabidopsis ecotype Columbia plants carrying the LUC reporter gene driven by the promoter of the high-affinity Pi transporter AtPT2 were used in these studies. Reporter gene activity was observed in transgenic plants grown under Pi-deficient conditions. Approximately 40 stratified seeds of Arabidopsis were transferred to flasks containing 25 mL of Murashige and Skoog liquid media. The seedlings were grown for 6 d under 16-h-light/8-h-dark cycle at 25°C in a gyratory shaker set at 100 rpm. Seedlings were rinsed in water and transferred to one-half-strength Murashige and Skoog media with (1.25 mm) and without Pi, and different concentrations of Phi (0, 0.1, 0.3, 0.5, 1, 2, and 3 mm). The seedlings grown for 5 d in the presence of Phi were rinsed with distilled water, frozen in liquid nitrogen, and stored at −80°C. Frozen samples were ground to a fine power in a pestle and mortar and extracted with a buffer containing 0.1 m KH2PO4 (pH 7.8), 1 mm EDTA, 10 mm dithiothreitol, and 0.25% (v/v) glycerol. Cell debris was removed by centrifugation at 12,000 rpm for 5 min. The supernatant was diluted 10-fold with extraction buffer before determining the Luc activity. Luc activity was measured using the assay buffer containing 50 mm HEPES, 20 mm MgCl2, 10 mm ATP (prepared in 0.2 m KH2PO4, pH 7.8), and 0.5 mg mL−1 bovine serum albumin. The cuvette containing the assay buffer and crude enzyme extract was placed in the Luminometer (TD-20/20, Turner Designs, Sunnyvale, CA). Enzyme reaction was initiated by injecting 50 μL of 1 mm luciferin (pH 7). The instrument automatically records and computes the light produced by the luciferin-Luc reactions. Measurements were taken on three replicated samples representing three independent treatments. Total protein concentration of samples was determined by dye binding assay (Bradford, 1976). The Luc activity was expressed as relative luminescence units per microgram of soluble protein.
ACKNOWLEDGMENTS
We thank Drs. Angus Murphy and Peter Goldsbrough for critical reading of the manuscript.
Footnotes
This work was supported in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 590 1165–2614 to K.G.R.). This is journal paper no. 16,793 of the Purdue University Agriculture Research Program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010835.
LITERATURE CITED
- Baldwin JC, Karthikeyan AS, Raghothama KG. Annual American Society of Plant Physiologists Meeting, July 24–28, Baltimore. Rockville, MD: American Society of Plant Physiologists; 1999. LePS3, a novel phosphate starvation induced gene in tomato (abstract no. 937) p. 190. [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG. LePS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiol. 2001;125:728–737. doi: 10.1104/pp.125.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchietto T, Saindrenan P, Bompeix G. Characterization of phosphonate uptake in two Phytophthora spp. and its inhibition by phosphate. Arch Microbiol. 1989;151:54–58. [Google Scholar]
- Barchietto T, Saindrenan P, Bompeix G. Physiological responses of Phytophthora citrophthora to a sub-inhibitory concentration of phosphonate. Pestic Biochem Physiol. 1992;42:151–166. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bressan RA, Hasegawa PM, Handa AK. Resistance of cultured higher plant cells to polyethylene glycol-induced water stress. Plant Sci Lett. 1981;21:23–30. [Google Scholar]
- Carswell MC, Grant BR, Plaxton WC. Disruption of the phosphate-starvation response of oilseed rape suspension cells by the fungicide phosphonate. Planta. 1997;203:67–74. doi: 10.1007/s00050166. [DOI] [PubMed] [Google Scholar]
- Carswell C, Grant BR, Theodorou ME, Harris J, Niere JO, Plaxton WC. The fungicide phosphonate disrupts the phosphate starvation response in Brassica nigra seedlings. Plant Physiol. 1996;110:105–110. doi: 10.1104/pp.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey MD, Joseph MC. Effects of phosphorous acid and fosetyl-Al on the life cycle of Phytophthora cinnamomi and P. citricola. Phytopathology. 1985;75:1042–1046. [Google Scholar]
- d'Arcy-Lameta A, Bompeix G. Systemic transport of tritiated phosphonate in tomato plantlets (Lycopersicon esculentum Mill) Pestic Sci. 1991;32:7–14. [Google Scholar]
- El-Hamalawi ZA, Menge JA, Adams CJ. Methods of fosetyl-Al application and phosphonate levels in avocado tissue needed to control stem canker caused by Phytophthora citricola. Plant Dis. 1995;79:770–778. [Google Scholar]
- Forster H, Adaskaveg JE, Kim DH, Stanghellini ME. Effect of phosphite on tomato and pepper plants and on susceptibility of pepper to Phytophthora root and crown rot in hydroponic culture. Plant Dis. 1998;82:1165–1170. doi: 10.1094/PDIS.1998.82.10.1165. [DOI] [PubMed] [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. Proteoid root development of phosphorus deficient lupin is mimicked by auxin and phosphonate. Ann Bot. 2000;85:921–928. [Google Scholar]
- Grant BR, Grant JH, Harris J. Inhibition of growth of Phytophthora infestans by phosphate and phosphonate in defined media. Exp Mycol. 1992;16:240–244. [Google Scholar]
- Griffith JM, Akins LA, Grant BR. Properties of the phosphate and phosphite transport systems of Phytophthora palmivora. Arch Microbiol. 1989;152:430–436. [Google Scholar]
- Griffith JM, Coffey MD, Grant BR. Phosphonate inhibition as a function of phosphate concentration in isolates of Phytophthora palmivora. J Gen Microbiol. 1993;139:2109–2116. [Google Scholar]
- Griffith JM, Smillie RH, Grant BR. Alterations in nucleotide and pyrophosphate levels in Phytophthora palmivora following exposure to the antifungal agent potassium phosphonate (phosphite) J Gen Microbiol. 1990;136:1285–1291. [Google Scholar]
- Guest D, Grant BR. The complex action of phosphonates as antifungal agents. Biol Rev. 1991;66:159–187. [Google Scholar]
- Karthikeyan AS, Uthappa MT, Varadarajan D, D'Urzo PM, Raghothama KG. Eleventh International Conference of Arabidopsis Research, June 24–28, Madison: University of Wisconsin; 2000. Transcriptional regulation of the high affinity phosphate transporters (abstract no. 440) [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntire WH, Winterberg SH, Hardin LJ, Sterges AJ, Clements LB. Fertilizer evaluation of certain phosphorus, phosphorous and phosphoric materials by means of pot cultures. Agron J. 1950;42:543–549. [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. Ed 2. San Diego: Academic Press; 1995. [Google Scholar]
- McDonald AE, Grant BR, Plaxton WC. Phosphite (phosphorous acid): its relevance in the environment and agriculture and influence on plant phosphate starvation response. J Plant Nutr. 2001a;24:1505–1519. [Google Scholar]
- McDonald AE, Niere JO, Plaxton WC. Phosphite disrupts the acclimation of Saccharomyces cerevisiae to phosphate starvation. Can J Microbiol. 2001b;47:969–978. doi: 10.1139/w01-099. [DOI] [PubMed] [Google Scholar]
- Niere JO, DeAngelis G, Grant BR. The effect of phosphonate on the acid-soluble phosphorus components in the genus Phytophthora. Microbiology. 1994;140:1661–1670. [Google Scholar]
- Niere JO, Griffith JM, Grant BR. 31P NMR studies on the effect of phosphite on Phytophthora palmivora. J Gen Microbiol. 1990;136:147–156. doi: 10.1099/00221287-136-1-147. [DOI] [PubMed] [Google Scholar]
- Ohtake H, Wu H, Imazu K, Anbe Y, Kato J, Kuroda A. Bacterial phosphonate degradation, phosphite oxidation and polyphosphate accumulation. Res Cons Recy. 1996;18:125–134. [Google Scholar]
- Ouimette DG, Coffey MD. Phosphonate levels in Avocado (Persea americana) seedlings and soil following treatment with fosetyl-Al or potassium phosphonate. Plant Dis. 1989;73:212–215. [Google Scholar]
- Ouimette DG, Coffey MD. Symplastic entry and phloem translocation of phosphonate. Pestic Biochem Physiol. 1990;38:18–25. [Google Scholar]
- Pawlowski K, Kunze R, deVries S, Bisseling T. Isolation of total, poly (A) and polysomal RNA from plant tissue. In: Gelvin SB, Shiperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–13. [Google Scholar]
- Plaxton WC. Metabolic aspects of phosphate starvation in plants. In: Lynch JP, Deikman J, editors. Phosphorus in Plant Biology: Regulatory Roles in Molecular, Cellular, Organismic, and Ecosystem Processes. Rockville, MD: American Society of Plant Physiology; 1998. pp. 229–241. [Google Scholar]
- Plaxton WC, Carswell MC. Lerner HR, ed, Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganization. NY: Marcel Dekker; 1999. Metabolic aspects of the phosphate starvation response in plants; pp. 349–372. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate transport and signaling. Curr Opin Plant Biol. 2000;3:182–187. [PubMed] [Google Scholar]
- Rickard DA. Review of phosphorus acid and its salts as fertilizer materials. J Plant Nutr. 2000;23:161–180. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- Stehmann C, Grant BR. Inhibition of enzymes of the glycolytic pathway and hexose monophosphate bypass by phosphonate. Pestic Biochem Physiol. 2000;67:13–24. [Google Scholar]
- Sukarno N, Smith SE, Scott ES. The effect of fungicides on vesiculararbuscular mycorrhizal symbiosis: I. The effects on vesicular-arbuscular mycorrhizal fungi and plant growth. New Phytol. 1993;25:139–147. doi: 10.1111/j.1469-8137.1993.tb03872.x. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 2001;127:963–972. [PMC free article] [PubMed] [Google Scholar]
- Ullrich-Eberius CI, Novacky A, Fischer E, Luttge U. Relationship between energy-dependent phosphate uptake and the electrical membrane potential in Lemna gibba G1. Plant Physiol. 1981;67:797–801. doi: 10.1104/pp.67.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CJ, Shearer BL, Jackson TJ, St J, Hardy GE. Variation in sensitivity of Western Australian isolates of Phytophthora cinnamomi to phosphite in vitro. Plant Pathol. 2001;50:83–89. [Google Scholar]