Abstract
Pyrrolizidine alkaloids (PAs) are constitutive plant defense compounds with a sporadic taxonomic occurrence. The first committed step in PA biosynthesis is catalyzed by homospermidine synthase (HSS). Recent evidence confirmed that HSS evolved by gene duplication from deoxyhypusine synthase (DHS), an enzyme involved in the posttranslational activation of the eukaryotic translation initiation factor 5A. To better understand the evolutionary relationship between these two enzymes, which are involved in completely different biological processes, we studied their tissue-specific expression. RNA-blot analysis, reverse transcriptase-PCR, and immunolocalization techniques demonstrated that DHS is constitutively expressed in shoots and roots of Senecio vernalis (Asteraceae), whereas HSS expression is root specific and restricted to distinct groups of endodermis and neighboring cortex cells located opposite to the phloem. All efforts to detect DHS by immunolocalization failed, but studies with promoter-β-glucuronidase fusions confirmed a general expression pattern, at least in young seedlings of tobacco (Nicotiana tabacum). The expression pattern for HSS differs completely from its ancestor DHS due to the adaptation of HSS to the specific requirements of PA biosynthesis.
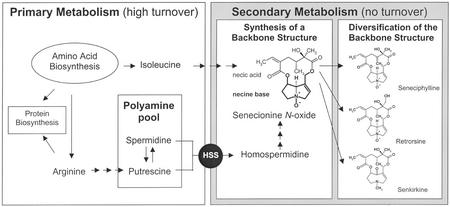
Homospermidine synthase (HSS) catalyzes the first pathway-specific step in the biosynthesis of pyrrolizidine alkaloids (PAs), a group of secondary compounds whose distribution is widely scattered among the angiosperms (Hartmann and Witte, 1995; Hartmann and Ober, 2000). PA-producing plants accumulate these compounds constitutively in all plant organs as chemical defenses against herbivores. In annual Senecio spp., the inflorescences are the major sites of PA accumulation (Hartmann and Zimmer, 1986). The function of PAs as powerful deterrents and toxins is supported by the impressive adaptations of certain insects that are specialized to feed on PA-containing plants and utilize plant-acquired PAs for their own protection (for review, see Hartmann and Witte, 1995; Hartmann, 1999; Hartmann and Ober, 2000).
The biosynthesis of PAs has been intensively studied, particularly in Senecio spp. (Asteraceae). Here, PAs are synthesized in the roots as N-oxides that are translocated into the shoot through the phloem (Hartmann et al., 1989; Witte et al., 1990). Specific carriers are involved in phloem loading and unloading of the polar PA N-oxides because species that do not produce PAs are unable to translocate them via the phloem (Hartmann et al., 1989). In all Senecio spp. studied so far, senecionine N-oxide is the primary product of biosynthesis. The backbone structure is modified by one- or two-step reactions (e.g. hydroxylations, dehydrogenations, epoxydations, O-acetyla-tions, etc.) that are species specific and lead to the unique PA pattern of a given plant population. In the living plant, PAs are spatially mobile but do not show any turnover or degradation (Hartmann and Dierich, 1998).
In PA biosynthesis, homospermidine is the first pathway-specific precursor of the necine base moiety of these ester alkaloids (Fig. 1). Homospermidine is a rare polyamine, which is not commonly present in plants. It is formed by HSS (spermidine specific; EC 2.5.1.45), which catalyzes the transfer of the aminobutyl moiety from spermidine to a putrescine molecule in an NAD+-dependent reaction. In contrast to its substrates putrescine and spermidine, which in Senecio spp. roots show a dynamic turnover, homospermidine does not exhibit any metabolic activity apart from its incorporation into PAs (Boettcher et al., 1993). In Senecio spp. roots, free homospermidine is only detectable in the presence of β-hydroxyethylhy-drazine, a diamine oxidase inhibitor, which efficiently blocks the subsequent step in PA biosynthesis. If the inhibition is released, PA biosynthesis starts again at the expense of accumulated homospermidine.
Figure 1.
Biosynthesis of PAs in the roots of S. vernalis. Putrescine and spermidine from primary metabolism are used as substrates for HSS to catalyze the formation of homospermidine. This is the first committed step in PA biosynthesis. The resulting homospermidine is exclusively incorporated into the necine base moiety of senecionine N-oxide. During translocation of this parent PA to the aerial parts of the plant, its structure is chemically modified to provide the PA derivatives found in S. vernalis.
Recently obtained molecular data about HSS from Senecio vernalis provided conclusive evidence for its close phylogenetic relation to deoxyhypusine synthase (DHS; EC 2.5.1.46), an enzyme involved in the posttranslational activation of the eukaryotic initiation factor 5A (eIF5A; Ober and Hartmann, 1999b). DHS catalyzes the first of the two enzymatic reactions leading to one of the most specific posttranslational modifications known (Krishna and Wold, 1993) to produce activated eIF5A. Although inhibition of hypusine formation stops cell growth at the G1/S boundary (Hanauke-Abel et al., 1995), the function of eIF5A remains elusive (for review, see Park et al., 1997). Recently, eIF5A was localized at the nuclear pore complex (Rosorius et al., 1999), where it is efficiently exported by the transport receptor exportin 4 (Lipowsky et al., 2000). It probably functions as a carrier for the export of specific RNAs from the nucleus to the cytosol. In plants, these mRNAs may be required for programmed cell death (Wang et al., 2001). DHS and eIF5A seem to be conserved among eukaryotes (Gordon et al., 1987) and archaebacteria (Bartig et al., 1990). Recently, the same mechanism of activation was confirmed in plants by cloning and functional expression of DHS and eIF5A from tobacco (Nicotiana tabacum; Ober and Hartmann, 1999a) and S. vernalis (Ober and Hartmann, 1999b).
The recruitment of HSS from DHS has been interpreted as evolution by change of function (Ober and Hartmann, 2000) because HSS was recruited for PA biosynthesis, a totally different function in plant secondary metabolism, and maintained under the selection pressure of herbivory (Ober and Hartmann, 2000). Mechanistically, HSS and DHS catalyze analog reactions, both transferring an aminobutyl moiety of spermidine to their substrates, i.e. putrescine in the case of HSS and to a specific protein-bound Lys residue in the case of DHS. Interestingly, purified DHS also catalyzes as a side reaction the aminobutylation of putrescine with the same kinetic properties as HSS (Ober and Hartmann, 1999b; D. Ober, R. Harms, L. Witte, and T. Hartmann, unpublished data). This suggests that HSS could have evolved from DHS by simply losing its ability to bind the eIF5A protein. Because HSS controls the substrate flow into the alkaloid pathway, it occupies a central position in the regulation of PA biosynthesis. HSS and DHS are cytosolic enzymes and the activities of both enzymes seem to be correlated with cell growth (Ober and Hartmann, 2000).
The intention of this study was to elucidate the spatial localization of HSS in roots of S. vernalis and to understand the biochemical aspects of the compartmentation discussed above. Using polyclonal antibodies against HSS, we studied the tissue-specific and subcellular localization of HSS. The identification of groups of distinctive cells close to the phloem as sites of homospermidine formation indicate a highly cell-specific expression of alkaloid biosynthesis with possible symplastic connection to the phloem. In addition, RNA gel-blot analysis and semiquantitative PCR were performed to compare the tissue-specific expression pattern of the closely related HSS and DHS genes in S. vernalis. For comparison, tobacco, a plant that is devoid of PAs, was included in these studies. Due to difficulties in detecting DHS by immunolocalization, promoter-β-glucuronidase (GUS)/green fluorescent protein (GFP) fusions were constructed to analyze the tissue specificity of the promoter of tobacco DHS.
RESULTS
Analysis of hss and dhs Gene Expression in S. vernalis and Tobacco
To compare hss and dhs gene expression in different organs of S. vernalis and tobacco, we employed two strategies. First, northern-blot analysis was performed with complete cDNAs encoding HSS and DHS as probes. Second, because both genes are closely related and the respective enzymes share identities of 83% and 79% on nucleic acid and amino acid level, respectively (Ober and Hartmann, 1999b), we performed semiquantitative reverse transcriptase (RT)-PCR to prove the specificity of the probes in the northern blot. In these experiments primers were applied, which are highly specific for the cDNAs of HSS and DHS, respectively. However, this technique allows only demonstration of the presence or absence of gene expression in a tissue without rigorous quantitative comparison.
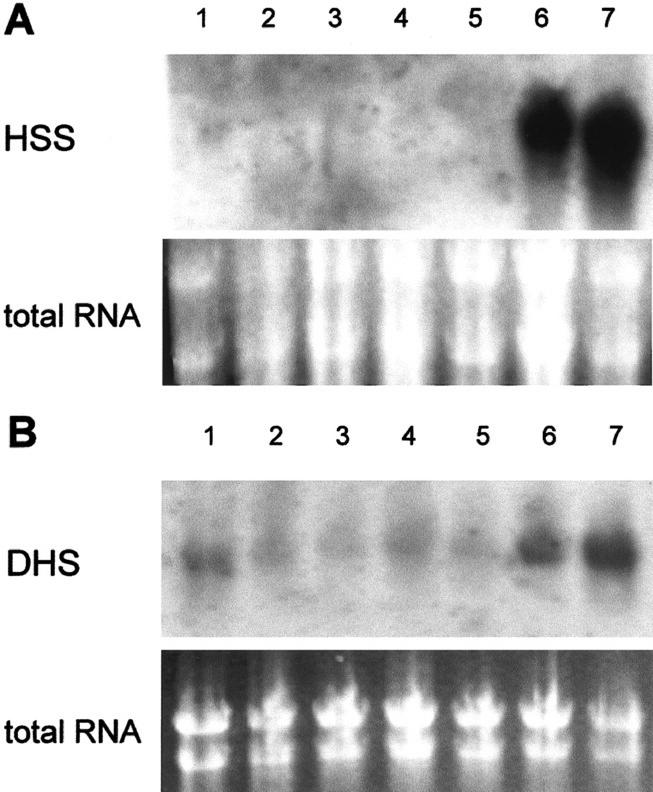
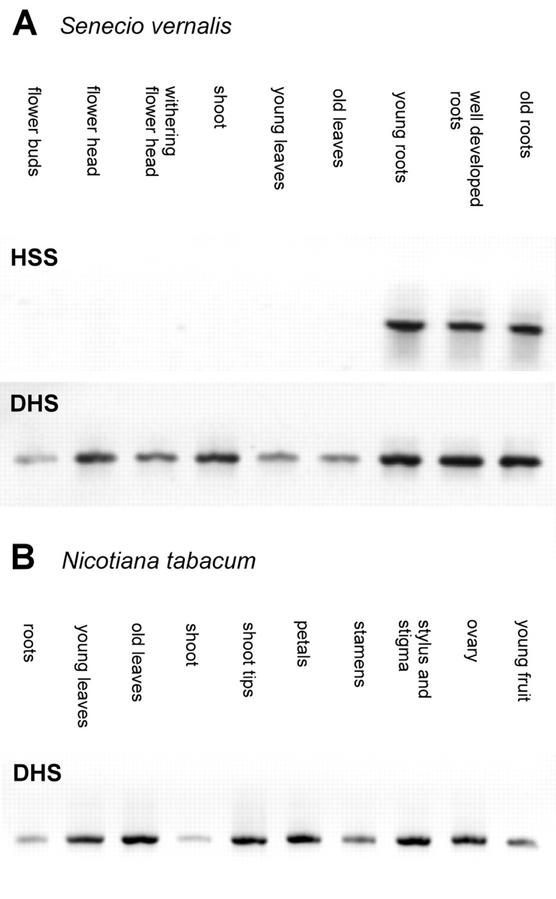
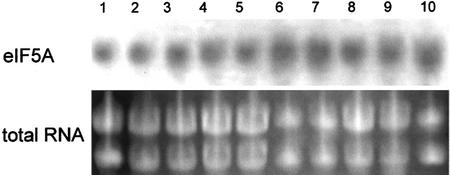
Figure 2 shows the results of the northern-blot analysis. The hss gene is expressed at high levels in the roots of S. vernalis independently of the root age, but is not expressed in the aerial parts of the plant like the buds, flower heads, leaves, or stems. This result supports the biochemical evidence that roots of Senecio spp. are the exclusive site of PA biosynthesis, as was shown with in vitro root cultures and detached plant organs (Hartmann and Toppel, 1987; Toppel et al., 1987; Hartmann et al., 1989). Western-blot analysis of six successive 1-cm segments beginning with the root tip showed expression of HSS in all tested root segments except the undifferentiated first centimeter of the root tip. The hypocotyl was devoid of any HSS expression (data not shown). In contrast to the selective expression of the hss gene, the dhs gene was expressed at low levels in all tested organs of S. vernalis, with higher levels in the roots (Fig. 2B). Semiquantitative RT-PCR (Fig. 3) confirmed that the dhs gene is expressed in all tested tissues of S. vernalis as well as of tobacco, whereas the hss gene is expressed selectively only in the roots of S. vernalis. The test of whether the expression of the dhs gene correlates with expression of the gene encoding the substrate of DHS, eIF5A, also showed for the eIF5A an expression at constant level in all tested tissues (Fig. 4), which is well in accord with the data for eIF5A from maize (Zea mays; Dresselhaus et al., 1999). Also, ovaries of different developmental stages (of young flower bud through developed fruit-containing seeds) showed a constant level of dhs gene expression (data not shown).
Figure 2.
Northern-blot analysis and RNA loading control of different tissues of S. vernalis. RNA of S. vernalis tissues was hybridized with digoxigenin (DIG)-labeled HSS probe (A) and DHS probe (B) of S. vernalis (buds, lane 1; young leaves, lane 2; young stem, lane 3; flower heads, lane 4; old stem, lane 5; young root, lane 6; and old root, lane 7).
Figure 3.
Semiquantitative RT-PCR. Reverse transcription and PCR were performed with RNA of different tissues of S. vernalis with primers specific for HSS and DHS of S. vernalis (A) and with RNA of different tissues of tobacco with primers specific for DHS of tobacco (B).
Figure 4.
Northern-blot analysis and RNA loading control of different plant organs of tobacco with eIF5A probe. RNA of different tissues was hybridized with DIG-labeled probe of eIF5A (roots, lane 1; shoots, lane 2; old leaves [about 20 cm in length], lane 3; young leaf [about 2 cm in length], lane 4; stamina of open flower, lane 5; stamina of closed flower bud, lane 6; petals of open flower, lane 7; petals of closed flower bud, lane 8; ovary of open flower, lane 9; and ovary of closed flower bud, lane 10).
Immunolocalization of HSS in Root Sections of S. vernalis
The tissue-specific localization of HSS was studied with roots of field- or greenhouse-grown S. vernalis. Although in vitro-cultured roots proved to be an ideal system for biochemical studies of PA biosynthesis (Hartmann et al., 1988), they turned out to be unsuitable for histological studies because of incomplete tissue differentiation in the central cylinder.
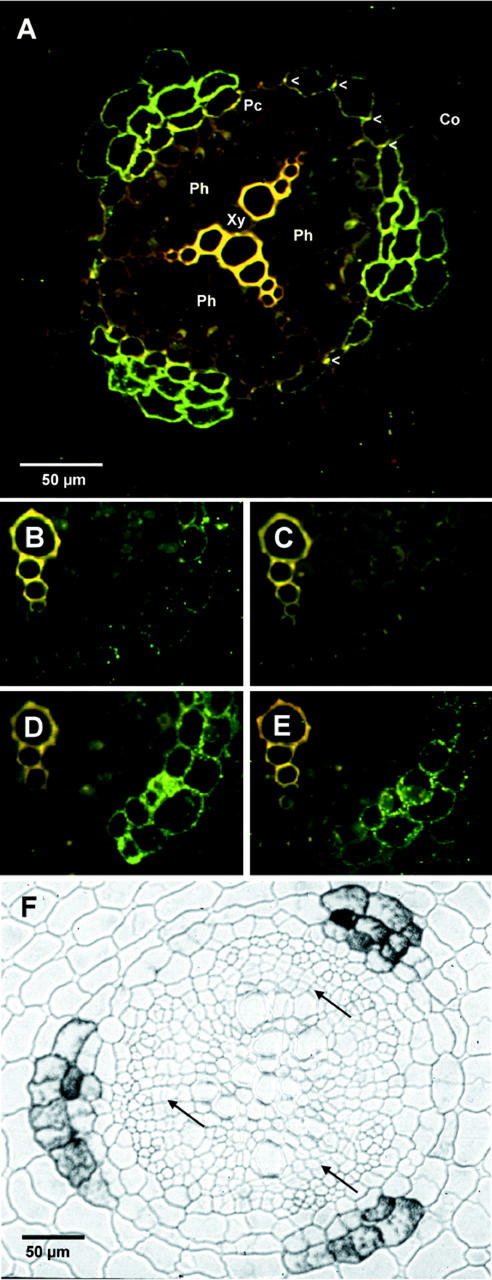
The immunostaining of HSS was performed with polyclonal antibodies from rabbits raised against S. vernalis HSS. Before use, the antibodies were purified by affinity chromatography with immobilized recombinant HSS. Figure 5A shows a fluorescence photograph of FITC-labeled root sections. The xylem vessels of the triarch vascular bundle show intensive yellow autofluorescence, due to lignification. Yellow autofluorescence is also visible in the radial cell walls of the endodermis cells caused by suberin (casparian strip, marked with arrowheads). A strong antibody labeling is detected in endodermis cells and the adjacent parenchyma cells of the root cortex, which are located opposite of the phloem tissue. The phloem is located in between the radially arranged ridges of the xylem, separated from the endodermis by the pericycle, the outer cell layer of the central cylinder. The pericycle cells are devoid of any labeling.
Figure 5.
Immunolabeling of HSS in root sections of S. vernalis. A, Specific labeling using fluorescein isothiocyanate (FITC) detection of specialized cells within the endodermis and the adjacent parenchyma cells of the root cortex with affinity-purified anti-HSS antibody (Co, parenchyma cells of the root cortex; En, endodermis; Ph, phloem; Xy, xylem; and the casparian strips in the radial cell walls of the endodermis are marked by arrowheads). B through E, Specific labeling as in A, but in the presence of purified HSS (B and C) and in the presence of purified DHS (D and E) in a molar ratio of antibody:added protein of 10:1 and 1:3, respectively. F, Immunogold labeling enhanced with silver of a root section with beginning secondary growth (cells introduced by cambium activity are marked by arrows). The HSS-specific label is still detectable.
To confirm the specificity of this labeling, various control experiments were performed. Incubation of the section with either pre-immune serum or the secondary FITC-labeled antibody omitting the primary antibody gave no labeling (data not shown). In protein gel-blot analysis, the antibody raised against the HSS showed weak but detectable cross-reactivity with DHS of S. vernalis, even after affinity purification (data not shown). Because DHS was shown to be expressed in roots (Figs. 2B and 3A), any influence on HSS immunolocalization by cross-reactivity with DHS needed to be excluded. Therefore, the labeling experiment was repeated by addition of either purified HSS or purified DHS. In case of a specific labeling of HSS, the label should be reduced by pre-incubation of the antibody with added HSS protein, but not with DHS protein. The pre-incubations were performed at room temperature 10 min before the mixture was applied to the root section. Two different protein concentrations were used, resulting in a molar ratio of antibody to added protein of 10:1 and 1:3. Figure 6, B through E, show that only the addition of soluble HSS reduces (10:1 ratio) and even completely blocks (1:3 ratio) the specific labeling, whereas addition of the DHS protein is ineffective.
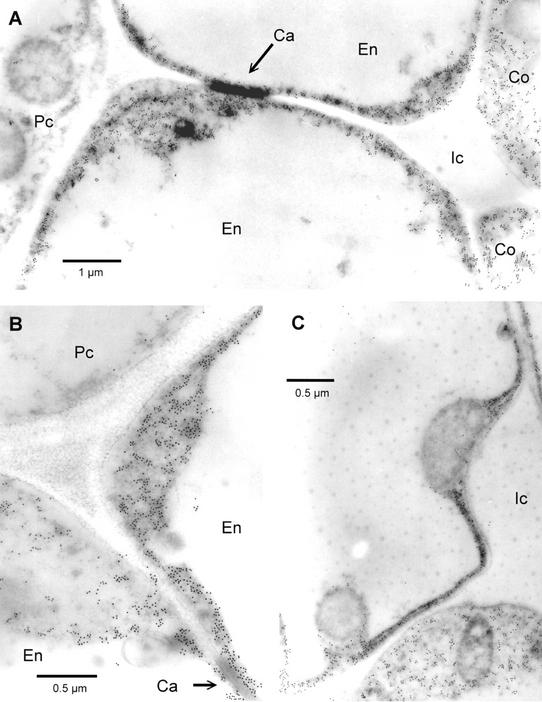
Figure 6.
Electron micrographs of in situ immunogold-labeled HSS in roots of S. vernalis. A, Two cells of the endodermis (En) with the casparian strip (Ca) as incrustation in the radial cell wall. The endodermis cells are accompanied by cells of the pericycle (Pc) and the cortex parenchyma (Co). Gold label is only found in the endodermis and the adjacent cortex cells, but no label is detectable in the pericycle. B, Cell junction of two endodermis cells (En; casparian strip at bottom right) with a cell of the pericycle (Pc). C, Detail of labeled cells that show the label exclusively in the cytoplasm. All detectable organelles are devoid of any label (at the right is an intercellular space, Ic).
Localization of HSS by immunogold labeling coupled with silver enhancement under the light microscope shows the same labeling pattern in a root that just started with secondary growth. Between the well-differentiated metaxylem and the phloem, rows of cells parallel to the radii of the axes are formed by the periclinal divisions of the cambium (Fig. 5F).
Intracellular Localization of HSS in S. vernalis
Figure 6 shows electron micrographs of cross sections of S. vernalis roots labeled with 18-nm gold particles after incubation with the affinity-purified antibody against HSS. In the center of the picture, the casparian strip is visible as a dark incrustation in the wall between two endodermis cells. These are joined at left by a cell of the pericycle. The gold particles are localized exclusively in the cytoplasm of the endodermis cells and in the adjoining cells of the cortex. There is no label associated with any cell organelle. The pericycle cells are also devoid of any label. These results clearly support the biochemical observation that HSS is a soluble cytosolic protein (Boettcher et al., 1993). Control incubations with preserum instead of the anti-HSS antibody did not show any label (data not shown).
Tissue-Specific Expression of Tobacco dhs-Promoter Fusions
In contrast to HSS, all efforts to localize DHS by immunolabeling have failed so far. Two reasons may be given to explain these negative results. First, the expression level of DHS is comparatively low. We were never able to demonstrate DHS activity in plant extracts, whereas HSS activity is easily detectable in crude root extracts of S. vernalis. Second, the localization of HSS in specific root cells suggests high local enzyme concentrations that should be detected more easily than the suspected more dispersed tissue distribution of DHS. To analyze DHS expression in more detail, we used the promoter-GUS/GFP fusion technique. From the eukaryotic genome projects (i.e. human, Arabidopsis, Drosophila melanogaster, and yeast [Saccharomyces cerevisiae]), it is known that DHS is represented by a single gene copy.
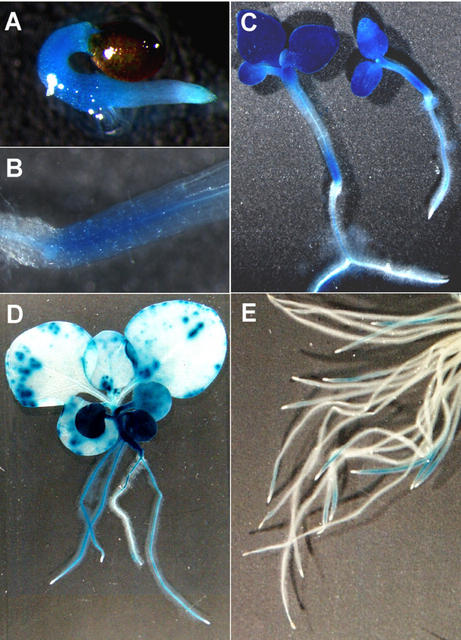
In tobacco, a 2.0-kb fragment upstream of the dhs gene was identified that should contain the regulatory elements of the dhs promoter. To analyze the tissue-specific expression of the dhs gene, we used this fragment to construct a promoter GFP/GUS fusion and transformed it into tobacco plants using Agrobacterium tumefaciens. GUS staining was performed with seedlings and young plantlets of the F1 generation of multiple independent transgenic lines. A significant staining was already detectable after 6 h of incubation, and longer incubation times did not alter the staining patterns. Figure 8, A through D, show strong dhs promoter activity in young tobacco seedlings. The radicle and the hypocotyl just emerging from the seed already showed intensive blue coloration (Fig. 7A). The promoter activity in the hypocotyl remains stable in older seedlings with expanded cotyledons, but only in the lower part, directly above the root-shoot connection (Fig. 7, B showing a detail of C). Intensive dhs promoter activity is also detectable in the cotyledons and the two primary leaves (Fig. 7C), whereas the following leaves show only a slight staining, indicating low expression (Fig. 7D). Roots of young seedlings also exhibit promoter activity, with the exception of the still undifferentiated growing zone of the root tip, which is generally devoid of any blue coloration (Fig. 7, C and D). If adventitious roots of the F0 generation are stained, some show promoter activity behind the root tips, with the exception of the tip itself in a 0.5- to 1.0-cm-long region, whereas others are devoid of any label (Fig. 7E). No success was achieved in trying to visualize GFP expression due to autofluorescence in the tissues.
Figure 7.
GUS expression in tobacco plants transformed with the dhs-promoter GUS fusion. A, Young seedling with stained root and hypocotyl. B, Detail of the transition between root and shoot of a seedling shown in C. C, Seedlings with developed cotyledons, one with two primary leaves. D, Young plant with GUS expression in the cotyledons, successive leaves, the hypocotyl, and some of the roots. Older leaves only show weak GUS expression. E, Adventitious roots of an F0 plant, of which only some show GUS expression in a section of approximately 0.5 to 1.0 cm in length directly behind the root tips.
DISCUSSION
Plant secondary metabolism is characterized by an immense diversity of chemical structures of restricted taxonomic distribution that are produced with high specificity and under stringent genetic control. Genes encoding enzymes mediating the high specificity of these pathways must have been recruited during evolution from primary metabolism by gene duplication and modified under selection pressure. Their integration into a secondary pathway implies not only the adaptation of enzymatic activities to new functions but also a proper regulatory integration of the gene, including its expression level and tissue specificity.
Comparison of hss and dhs Expression Patterns
The expression patterns of the hss and dhs genes revealed that they are differentially regulated. The dhs gene is expressed in all plant organs analyzed, with slightly higher levels occurring in roots, whereas the hss gene is expressed exclusively in roots (Figs. 2 and 3A). These results suggest that recruitment of the hss gene from dhs was accompanied by changes in the regulation of this gene to accommodate its new and unique role in PA biosynthesis. The root-specific expression of HSS corroborates earlier physiological studies that proved that the biosynthesis of PAs is restricted to the roots in Senecio spp. It is interesting to note that on a whole-root basis in S. vernalis, the mRNA coding for HSS is expressed in much higher concentration than the mRNA coding for DHS. This becomes even more remarkable if we consider that the HSS expression is restricted to certain root cells. Obviously, the regulatory elements are also modulated in regard to the expression level of the gene product. Pichersky and Gang (2000) postulate that the increased expression level of an enzyme involved in the biosynthesis of defense compounds in comparison with its ancestor from primary metabolism could increase the fitness of an organism. This is probably the case for the HSS-DHS system because the low turnover number of DHS is well known (Ober and Hartmann, 1999a), although in plants accumulating defense compounds, low enzyme activities may suffice to establish an efficient defense system.
Nothing is known about the localization and function of DHS in plants. In fact, tobacco and S. vernalis are the first plants for which the occurrence of DHS and its function in eIF5A activation was confirmed (Ober and Hartmann, 1999a, 1999b). Recently, Wang et al. (2001) isolated the cDNAs for DHS and eIF5A from a senescence-induced cDNA library of tomato (Lycopersicon esculentum). Only few data are available concerning plant eIF5A. Pay et al. (1991) succeeded in cloning and sequencing of the first plant eIF5A cDNA from Medicago sativa. Two cDNA sequences for eIF5A were isolated from Nicotinana plumbaginifolia. One of the corresponding genes is constitutively expressed in all tested tissues, whereas the second gene seems to be expressed mainly in photosynthetically active parts of the plant (Chamot and Kuhlemeier, 1992). In maize, eIF5A is also constitutively expressed in all tissues without any correlation of gene expression with cell division activity (Dresselhaus et al., 1999). Our results suggest that in plants, neither the expression patterns of eif5A nor DHS appear to be correlated with cell growth because root and leaf tissues of various developmental stages do not show noticeable differences in their expression levels (Figs. 2–4). Thus, in plants, the function of activated eIF5A may be different from that described for other eukaryotic systems, where eIF5A appears to be essential for cell proliferation (Kang and Hershey, 1994). In addition to the results of Wang et al. (2001), the dhs promoter-GUS fusions we expressed in tobacco also indicate the involvement of activated eIF5A in processes other than plant senescence. An essential role was postulated for eIF5A in RNA export from the nucleus (see above). Such a mechanism may be required in physiological processes such as those involved in senescence or early development of seedlings.
Cell-Specific Localization of HSS in Root Sections of S. vernalis
HSS expression was localized to specialized endodermis and the neighboring parenchyma cells just opposite the phloem. Cells of the endodermis are coated with a lipid (suberin)-containing casparian strip in the radial and transverse walls that produce a physical barrier for apoplastic transport of solutes from the cortex to the central cylinder. We assume that these specialized endodermis and adjacent cortex cells are not only the specific sites of HSS expression but are the intrinsic sites of the biosynthesis of senecionine N-oxide, the backbone structure of the Senecio spp. PAs. This view is corroborated by experimental evidence from earlier physiological and biochemical studies. Thus, in Senecio spp., the roots are not only the specific site of HSS expression but also the exclusive site of de novo PA synthesis (Hartmann and Toppel, 1987; Toppel et al., 1987). The findings of Boettcher et al. (1993) that endogenously formed homospermidine is exclusively incorporated into PAs and is not a substrate for degrading polyamine oxidases indicate that the formation of homospermidine and the subsequent steps of its utilization for alkaloid biosynthesis should share the same cellular compartment. Our data prove that cell organelles are not involved in compartmentation of homospermidine formation.
The arrangement of the HSS-expressing cells vis-à-vis the phloem matches with the phloem-specific root-to-shoot translocation of senecionine N-oxide (Hartmann et al., 1989). To enter a sieve tube, newly synthesized senecionine N-oxide has only to cross the single cell layer of the pericycle (see Fig. 5). The mechanism of the root-specific phloem loading of alkaloids is not known, but the polar salt-like senecionine N-oxide was found to be only phloem mobile in PA-synthesizing Senecio spp. (Hartmann et al., 1989). The specific phloem loading with senecionine N-oxide may be either apoplastic or symplastic. We know that PAs are spatially mobile (Hartmann and Dierich, 1998). In whole plants, PAs circulate slowly between plant organs and accumulate transiently at the preferential storage sites (i.e. inflorescences and peripheral stem tissue). Even in Senecio spp. root cultures, there is a continuous translocation of PAs between old and new tissues (Sander and Hartmann, 1989). In plants and cultured roots, there is no significant loss of PAs over prolonged periods (i.e. >4 weeks with plants and >2 weeks with root cultures), indicating that during their spatial mobility, the PAs are well protected from apoplastic degradation. In root cultures, not even traces of PA N-oxides are lost into the medium. Thus, a PA transport via the apoplast seems to be unlikely.
The expression of other alkaloid-specific enzymes in particular plant organs, defined cells, or in cell organelles has been described (for review, see De Luca and St-Pierre, 2000). In this respect, PAs and nicotine as well as the tropane alkaloids, such as the anticholinergic drugs l-hyoscyamine and scopolamine, found in certain species of the Solanaceae, share some striking similarities. Like PAs, they are exclusively synthesized in the roots and translocated into the shoot. However, in contrast to PAs, they are translocated via the transpiration stream in the xylem (Neumann, 1985). Putrescine N-methyltransferase, the first enzyme of scopolamine biosynthesis, and hyoscyamine 6β-hydroxylase, the last enzyme of scopolamine biosynthesis, are only expressed in the pericycle of young roots of Hyoscyamus niger and Atropa belladonna (Hashimoto et al., 1991; Kanegae et al., 1994; Suzuki et al., 1999a). The transformation of H. niger and A. belladonna plants with a GUS fusion construct containing the promoter of hyoscyamine 6β-hydroxylase revealed GUS expression mainly in those cells of the pericycle, which are adjacent to the primary xylem poles (Kanegae et al., 1994; Suzuki et al., 1999b). Thus, like PA biosynthesis, tropane alkaloid formation appears to take place spatially adjacent to the translocation path of the alkaloids. However, in contrast to the pericycle-specific expression of these two enzymes of tropane alkaloid biosynthesis, tropinone reductase I, another specific enzyme of this pathway, is expressed in the endodermis and outer cortex. The differential compartmentation of the biosynthetic enzymes indicates that intermediates must be shuttled between the pericycle and the endodermis to complete the pathway leading to the formation of scopolamine (Nakajima and Hashimoto, 1999).
Complex intercellular compartmentation has also been found in the biosynthesis of the monoterpenoid indole alkaloids in Catharanthus roseus. Specific enzymes of this pathway were localized in at least two different cell types, also requiring intercellular translocation of pathway intermediates (St-Pierre et al., 1999). The hypothesis of De Luca and St-Pierre (2000) that already compartmentalized reactions were recruited from primary metabolism to participate in alkaloid biosynthesis cannot be the case for PA biosynthesis. HSS exhibits a tissue-specific expression that is clearly different from the expression pattern of DHS, its evolutionary ancestor.
The high specificity of expression of enzymes involved in alkaloid biosynthesis may be an important requirement for the establishment of new secondary pathways in evolution. Although we have good evidence that the HSS-expressing cells are the specific sites of PA biosynthesis, a similar intercellular compartmentation of the biosynthetic enzymes, as suggested for tropane and monoterpenoid indole alkaloid biosynthesis, cannot be excluded. Further enzymes involved in PA biosynthesis need to be identified and localized.
MATERIALS AND METHODS
Polyclonal Antibody Preparation and Affinity Purification
Polyclonal sera were raised in rabbits against HSS purified from roots of Senecio vernalis Waldst. & Kitaibel and against purified recombinant DHS protein (Ober and Hartmann, 1999b), provided by Eurogentec (Seraing, Belgium) and Bioscience (Göttingen, Germany), respectively. For affinity purification of the sera, purified recombinant HSS and DHS of S. vernalis were coupled to activated Sepharose 4B (Amersham Biosciences, Freiburg, Germany) according to the manufacturer's instructions. These affinity matrices were incubated overnight at room temperature with the respective sera, washed with 0.1 m sodium acetate (pH 4.5) containing 0.5 m NaCl, and eluted with 0.2 m sodium acetate (pH 2.7) containing 0.5 m NaCl. The eluting purified antibodies were rebuffered to phosphate buffered saline (PBS), concentrated, and stored at −20°C until further use.
Protein Gel-Blot Analysis
Proteins were separated on 12% (w/v) SDS-PAGE gels using a discontinuous buffer system (Laemmli, 1970) at 200 V of constant voltage. Gels not used for immunoblotting were stained with Coomassie Blue. Protein gels were electroblotted onto polyvinylidene fluoride membrane (Immobilon P, Millipore, Bedford, MA) with a current density of 2.5 mA cm2. The blots were then blocked with Tris-buffered saline supplemented with 0.1% (v/v) Tween 20 (TBS-T) containing 10% (v/v) fetal calf serum (Sigma, St. Louis) for 1 h at room temperature. With the affinity-purified polyclonal antibody (diluted 1:20,000 [v/v] in blocking solution), the blot was incubated for 1 h at room temperature, followed by successive washing steps (each 2 × 5 min) in TBS-T, TBS-T + 0.5 m NaCl, TBS-T + 0.5% (v/v) Triton X-100, and once in TBS-T. After incubation with a goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (diluted 1:5,000 [v/v], Dianova, Hamburg, Germany), the washing steps were repeated before chemoluminescence detection was performed with the ECL Western Blotting System (Amersham Biosciences) and documented on XAR5 x-ray film (Eastman-Kodak, Rochester, NY).
RNA Isolation and Gel-Blot Analysis
Tissue samples were collected, frozen in liquid nitrogen, and stored at −80°C. Total RNA was extracted with RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Ten micrograms per sample was separated on a formaldehyde-agarose gel and transferred onto positively charged nylon membranes (Roche Diagnostics, Mannheim, Germany) by capillary blotting. RNA gel blots were hybridized overnight in DIG Easy Hyb buffer or High-SDS hybridization buffer at 42°C with HSS and DHS probes and at 39°C with the eIF5A probe, all of which were digoxigenin labeled by using the PCR DIG Probe Synthesis Kit (Roche Diagnostics). Chemoluminescent detection was performed with CSPD (Roche Diagnostics) according to the manufacturer's instructions. Exposure times were 2 h for the HSS- and DHS-specific probes and 1 h for the eIF5A-specific probe.
Semiquantitative RT-PCR
Per sample, 2 μg of total RNA was used as template for oligo(T) cDNA synthesis with an oligo(dT)17 primer (0.1 μm, 5′-dGTCGACTCGAGA-ATTC(T)17-3′, MWG-Biotech, Ebersberg, Germany) using Superscript II RT (Invitrogen, Carlsbad, CA) in a total volume of 50 μL according to the manufacturer's instructions. PCR was performed with specific primers for HSS and DHS, previously used for amplification of the full-length cDNAs from S. vernalis (Ober and Hartmann, 1999b) and tobacco (Nicotiana tabacum; Ober and Hartmann, 1999a), Taq DNA polymerase (Invitrogen), and the following temperature program: 5 min at 95°C initial denaturation, 40 cycles with 95°C for 45 s, 57°C for 1 min, and 72°C for 2 min. Aliquots of the reaction were taken after cycle 26, 28, 30, 32, 34, and 40 and analyzed by agarose gel electrophoresis.
Tissue Preparation for Immunohistochemistry
Roots and other plant organs of S. vernalis and tobacco grown in the greenhouse were cut into small segments (approximately 0.5–1.0 cm) and immediately fixed for 1 h under reduced pressure in 4% (v/v) formaldehyde (freshly prepared from paraformaldehyde) and 0.2% (v/v) glutaraldehyde in sample buffer (0.05 m potassium phosphate buffer, pH 7.2). Afterward, the samples were washed twice for 10 min in sample buffer, dehydrated in a graded ethanol series, and embedded in Technovit 7100 resin (Heraeus-Kulzer, Hanau, Germany) according to the manufacturer's instructions. Sections (3–5 μm) were cut with a microtome and mounted on glass slides coated with Teflon (Roth, Karlsruhe, Germany).
Immunocytochemical Analysis by UV and Light Microscopy
Sections were blocked at room temperature for 30 min with 0.15 m Gly followed by 30 min with PBS supplemented with 10% (w/v) bovine serum albumin and 0.1% (w/v) fish gelatin. After washing with PBS, the sections were incubated with either pre-immune serum (without dilution) or affinity-purified primary antibody (1:500 dilution [v/v]) diluted with 1% (w/v) bovine serum albumin in PBS for 1 h at 37°C in a humid chamber. After five 1-min washings with PBS, the sections were incubated for 1 h at room temperature with the secondary goat anti-rabbit antibody. For immunogold labeling, the secondary antibody was coupled to 18-nm gold particles (diluted 1:20 [v/v], Dianova) and for fluorescence detection, coupled with FITC (1:100 [v/v], Sigma). For visualization in a light microscope (Photomicroscope III, Zeiss, Jena, Germany), gold particle-labeled sections were exposed to a silver enhancement reagent according to the manufacturer's instructions (Amersham Biosciences). FITC-labeled sections were excitated by UV light of 450 to 490 nm and recorded using a Photomicroscope III or Axioskop 2 (Zeiss).
Immunocytochemical Analysis by Transmission Electron Microscopy
Sections for transmission electron microscopy were fixed and dehydrated as described above, and then were embedded in Unicryl resin (Plano, Wetzlar, Germany) according to the manufacturer's instructions. Sections of 80 nm were cut with an ultramicrotome and mounted onto nickel grids (300 mesh, Plano) coated with Butvar B-98 (Sigma) according to Handley and Olsen (1979). Blocking and antibody incubation (goat anti-rabbit with 18-nm gold particles as secondary antibody) was performed as described for light microscopy. After post-staining with 2% (w/v) aqueous uranyl acetate for 20 min, the sections were analyzed using a transmission electron microscope (300 EM, Philips, Eindhoven, The Netherlands).
Construction of Promoter-GUS Fusions and Plant Transformation
Genomic DNA was isolated from field-grown tobacco leaves using the hexadecyl-trimethyl-ammonium bromide protocol (Doyle and Doyle, 1990). To identify the promoter region, a gene walking method (Siebert et al., 1995) was used with primers specific to the known cDNA sequence of tobacco DHS (Ober and Hartmann, 1999a). The 2.0-kb fragment of the genomic DNA directly upstream of the open reading frame was amplified by PCR using the Advantage Genomic PCR Kit (BD Biosciences Clontech, Palo Alto, CA) and two specific primers, with the forward primer including a native HindIII restriction site 1,871 pb upstream of the start-ATG of the DHS gene and the reverse primer encompassing a SpeI site directly behind the start-ATG of the open reading frame. The HindIII-/SpeI-digested PCR product was inserted into pCAMBIA1304, replacing the cauliflower mosaic virus 35S promoter directly in front of an mGFP5*/gusA fusion. The hygromycin resistance cassette of this plasmid was replaced by the ampicillin resistance cassette of pCAMBIA2300. This construct was propagated in Escherichia coli XL1-blue and transformed into competent Agrobacterium tumefaciens strain C58C1:pGV2260 to transform tobacco cv SNN as described by Rosahl et al. (1987). The nucleotide sequences for the promoter region of the dhs gene of tobacco has been submitted to the EMBL Nucleotide Sequence Database (accession no. AJ428400).
GUS Reporter Analysis
For histochemical GUS analysis, seedlings and young plantlets of the F1 generation of tobacco plants transformed with the dhs promoter-GFP/GUS fusion were immersed in the GUS reaction buffer according to Urao et al. (1999) followed by a slight vacuum infiltration for 20 min. Tissues were incubated at 37°C for 6 to 18 h and afterward cleared in 70% (v/v) ethanol. Photos were taken using a stereomicroscope (M8, Wild Heerbrugg, Heerbrugg, Switzerland) with an AxioCam HRc (Zeiss).
ACKNOWLEDGMENTS
We thank Benoit St-Pierre for valuable advice in establishing our method for immunolabeling, and Bettina Hause for many helpful discussions and for the possibility to use the microscopic equipment at Institute of Plant Biochemistry (Halle, Germany). We thank Wolfgang Lein and Andrea Knospe for their help in learning tobacco transformation and Jonathan Gershenzon for discussion of the manuscript. Anita Backenköhler is thanked for her excellent technical assistance.
Footnotes
The work was supported by the Deutsche Forschungsgemeinschaft (grant to T.H. and D.O.) and by the Fonds der Chemischen Industrie (grant to T.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004259.
LITERATURE CITED
- Bartig D, Schümann H, Klink F. Posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the archaebacterial kingdom. Syst Appl Microbiol. 1990;13:112–116. [Google Scholar]
- Boettcher F, Adolph RD, Hartmann T. Homospermidine synthase, the first pathway-specific enzyme in pyrrolizidine alkaloid biosynthesis. Phytochemistry. 1993;32:679–689. [Google Scholar]
- Chamot D, Kuhlemeier C. Differential expression of genes encoding the hypusine-containing translation initiation factor, eIF-5A, in tobacco. Nucleic Acids Res. 1992;20:665–669. doi: 10.1093/nar/20.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, St-Pierre B. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 2000;5:168–173. doi: 10.1016/s1360-1385(00)01575-2. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dresselhaus T, Cordts S, Loerz H. A transcript encoding translation initiation factor eIF-5A is stored in unfertilized egg cells of maize. Plant Mol Biol. 1999;39:1063–1071. doi: 10.1023/a:1006176819213. [DOI] [PubMed] [Google Scholar]
- Gordon ED, Mora R, Meredith SC, Lee C, Lindquist SL. Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved among eukaryotes. J Biol Chem. 1987;262:16585–16589. [PubMed] [Google Scholar]
- Hanauke-Abel HM, Slowinska B, Zagulska S, Wilson RC, Staiano CL, Hanauske AR, McCaffrey T, Szabo P. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary: proposal of a role for eIF-5A in onset of DNA replication. FEBS Lett. 1995;366:92–98. doi: 10.1016/0014-5793(95)00493-s. [DOI] [PubMed] [Google Scholar]
- Handley DA, Olsen BR. Butvar B-98 as a thin support film. Ultramicroscopy. 1979;4:479–480. doi: 10.1016/s0304-3991(79)80025-x. [DOI] [PubMed] [Google Scholar]
- Hartmann T. Chemical ecology of pyrrolizidine alkaloids. Planta. 1999;207:483–495. [Google Scholar]
- Hartmann T, Dierich B. Chemical diversity and variation of pyrrolizidine alkaloids of the senecionine type: biological need or coincidence? Planta. 1998;206:443–451. [Google Scholar]
- Hartmann T, Ehmke A, Eilert U, von Borstel K, Theuring C. Sites of synthesis, translocation and accumulation of pyrrolizidine alkaloid N-oxides in Senecio vulgaris L. Planta. 1989;177:98–107. doi: 10.1007/BF00392159. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Ober D. Topics in Current Chemistry. Vol. 209. Berlin: Springer; 2000. Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores; pp. 207–244. [Google Scholar]
- Hartmann T, Sander H, Adolph RD, Toppel G. Metabolic links between the biosynthesis of pyrrolizidine alkaloids and polyamines in root cultures of Senecio vulgaris. Planta. 1988;175:82–90. doi: 10.1007/BF00402884. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Toppel G. Senecionine N-oxide, the primary product of pyrrolizidine alkaloid biosynthesis in root cultures of Senecio vulgaris. Phytochemistry. 1987;26:1639–1644. [Google Scholar]
- Hartmann T, Witte L. Pyrrolizidine alkaloids: chemical, biological and chemoecological aspects. In: Pelletier SW, editor. Alkaloids: Chemical and Biological Perspectives. Vol. 9. Oxford: Pergamon Press; 1995. pp. 155–233. [Google Scholar]
- Hartmann T, Zimmer M. Organ-specific distribution and accumulation of pyrrolizidine alkaloids during the life history of two annual Senecio species. J Plant Physiol. 1986;122:67–80. [Google Scholar]
- Hashimoto T, Hayashi A, Amano Y, Kohno J, Iwanari H, Usuda S, Yamada Y. Hyoscyamine 6 β-hydroxylase, an enzyme involved in tropane alkaloid biosynthesis, is localized at the pericycle of the root. J Biol Chem. 1991;266:4648–4653. [PubMed] [Google Scholar]
- Kanegae T, Kajiya H, Amano Y, Hashimoto T, Yamada Y. Species-dependent expression of the hyoscyamine 6-β-hydroxylase gene in the pericycle. Plant Physiol. 1994;105:483–490. doi: 10.1104/pp.105.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HA, Hershey JWB. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- Krishna RG, Wold F. Post-translational modification of proteins. Adv Enzymol Rel Areas Mol Biol. 1993;67:265–298. doi: 10.1002/9780470123133.ch3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Gorlich D. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Hashimoto T. Two tropinone reductases, that catalyze opposite stereospecific reductions in tropane alkaloid biosynthesis, are localized in plant root with different cell-specific patterns. Plant Cell Physiol. 1999;40:1099–1107. doi: 10.1093/oxfordjournals.pcp.a029494. [DOI] [PubMed] [Google Scholar]
- Neumann D. Storage of alkaloids. In: Mothes K, Schütte HR, Luckner M, editors. Biochemistry of Alkaloids. Weilheim, Germany: VCH; 1985. pp. 49–55. [Google Scholar]
- Ober D, Hartmann T. Deoxyhypusine synthase from tobacco: cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J Biol Chem. 1999a;274:32040–32047. doi: 10.1074/jbc.274.45.32040. [DOI] [PubMed] [Google Scholar]
- Ober D, Hartmann T. Homospermidine synthase, the first pathway-specific enzyme of pyrrolizidine alkaloid biosynthesis, evolved from deoxyhypusine synthase. Proc Natl Acad Sci USA. 1999b;96:14777–14782. doi: 10.1073/pnas.96.26.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober D, Hartmann T. Phylogenetic origin of a secondary pathway: the case of pyrrolizidine alkaloids. Plant Mol Biol. 2000;44:445–450. doi: 10.1023/a:1026597621646. [DOI] [PubMed] [Google Scholar]
- Park MH, Lee YB, Joe YA. Hypusine is essential for eukaryotic cell proliferation. Biol Sign. 1997;6:115–123. doi: 10.1159/000109117. [DOI] [PubMed] [Google Scholar]
- Pay A, Heberle-Bors E, Hirt H. Isolation and sequence determination of the plant homologue of the eukaryotic initiation factor 4D cDNA from alfalfa, Medicago sativa. Plant Mol Biol. 1991;17:927–929. doi: 10.1007/BF00037075. [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- Rosahl S, Schell J, Willmitzer L. Expression of a tuber-specific storage protein in transgenic tobacco plants: demonstration of an esterase activity. EMBO J. 1987;6:1155–1159. doi: 10.1002/j.1460-2075.1987.tb02348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosorius O, Reichart B, Kraetzer F, Heger P, Dabauvalle MC, Hauber J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci. 1999;112:2369–2380. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- Sander H, Hartmann T. Site of synthesis, metabolism and translocation of senecionine N-oxide in cultured roots of Senecio erucifolius. Plant Cell Tissue Org Cult. 1989;18:19–32. [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, Vazquez-Flota FA, De Luca V. Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell. 1999;11:887–900. doi: 10.1105/tpc.11.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Ki, Yamada Y, Hashimoto T. Expression of Atropa belladonna putrescine N-methyltransferase gene in root pericycle. Plant Cell Physiol. 1999a;40:289–297. doi: 10.1093/oxfordjournals.pcp.a029540. [DOI] [PubMed] [Google Scholar]
- Suzuki Ki, Yun DJ, Chen XY, Yamada Y, Hashimoto T. An Atropa belladonna hyoscyamine 6β-hydroxylase gene is differentially expressed in the root pericycle and anthers. Plant Mol Biol. 1999b;40:141–152. doi: 10.1023/a:1026465518112. [DOI] [PubMed] [Google Scholar]
- Toppel G, Witte L, Riebesehl B, von Borstel K, Hartmann T. Alkaloid patterns and biosynthetic capacity of root cultures from some pyrrolizidine alkaloid producing Senecio spp. Plant Cell Rep. 1987;6:466–469. doi: 10.1007/BF00272784. [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi SK, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TW, Lu L, Wang D, Thompson JE. Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato. J Biol Chem. 2001;276:17541–17549. doi: 10.1074/jbc.M008544200. [DOI] [PubMed] [Google Scholar]
- Witte L, Ehmke A, Hartmann T. Interspecific flow of pyrrolizidine alkaloids; from plants via aphids to ladybirds. Naturwissenschaften. 1990;77:540–543. [Google Scholar]