Abstract
The general phenylpropanoid pathways generate a wide array of aromatic secondary metabolites that range from monolignols, which are ubiquitous in all plants, to sinapine, which is confined to crucifer seeds. The biosynthesis of these compounds involves hydroxylated and methoxylated cinnamyl acid, aldehyde, or alcohol intermediates. Of the three enzymes originally proposed to hydroxylate the 4-, 3-, and 5-positions of the aromatic ring, cinnamate 4-hydroxylase (C4H), which converts trans-cinnamic acid to p-coumaric acid, is the best characterized and is also the archetypal plant P450 monooxygenase. Ferulic acid 5-hydroxylase (F5H), a P450 that catalyzes 5-hydroxylation, has also been studied, but the presumptive 3-hydroxylase converting p-coumarate to caffeate has been elusive. We have found that Arabidopsis CYP98A3, also a P450, could hydroxylate p-coumaric acid to caffeic acid in vivo when expressed in yeast (Saccharomyces cerevisiae) cells, albeit very slowly. CYP98A3 transcript was found in Arabidopsis stem and silique, resembling both C4H and F5H in this respect. CYP98A3 showed further resemblance to C4H in being highly active in root, but differed from F5H in this regard. In transgenic Arabidopsis, the promoters of CYP98A3 and C4H showed wound inducibility and a comparable developmental regulation throughout the life cycle, except in seeds, where the CYP98A3 promoter construct was inactive while remaining active in silique walls. Within stem and root tissue, the gene product and the promoter activity of CYP98A3 were most abundant in lignifying cells. Collectively, these studies show involvement of CYP98A3 in the general phenylpropanoid metabolism, and suggest a downstream function for CYP98A3 relative to the broader and upstream role of C4H.
Plants synthesize thousands of secondary metabolites from offshoots of primary metabolism (Croteau et al., 2000). In most cases the biosynthetic routes are unknown and even in some of the well-studied pathways many aspects remain uncertain. Phenylpropanoid metabolism generates phenolic intermediates and end products that include lignin monomers, flavonoids, isoflavonoids, lignans, tannins, quinones, and sinapate esters (Strack, 1997; Dixon and Steele, 1999; Nair et al., 2000). Lignin constitutes approximately 15% to 30% of the dry weight in woody plants, and contributes about 30% of the organic carbon in plant biomass in general (Lewis and Yamamoto, 1990; Douglas, 1996; Boudet, 2000). Thus, lignin assembly places a huge demand on phenylpropanoid supply. Lignification is considered a biochemical adaptation to provide mechanical strength and “non-seeping” water transport channels as plants adopted terrestrial habitats. The biosynthetic pathways appear to have been further diversified and recruited to supply metabolites for a variety of other end uses such as attraction of pollinators for promoting sexual propagation, pest deterrence, pathogen resistance, UV radiation protection, and allelopathic exclusion of potentially competing plants (Dixon et al., 1996). The inherent inter- and intraplant variations in metabolite accumulation and environmentally modulated fluctuations make generalizations of the biosynthetic steps very tentative, but, nonetheless, composite views are useful to address specific aspects. Freudenberg and Neish (1968) laid out such a foundation for the pathways of general phenylpropanoid metabolism that led to the concept of l-Phe and l-Tyr entering secondary metabolism and undergoing various biochemical transformations.
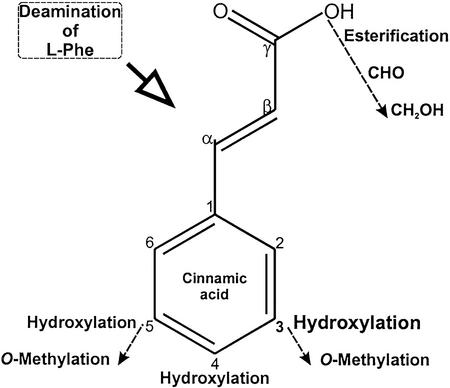
Deamination of l-Phe by phenylalanine ammonia lyase (PAL) is the first committed step in phenylpropanoid synthesis (Fig. 1). Cinnamic acid is then hydroxylated at the 4-position of the aromatic ring by cinnamate 4-hydroxylase (C4H) to generate p-coumarate. Subsequently, several independent or sequential transformations occur at the 3 and 5 positions of the ring (hydroxylation and methylation) and at the γ-carbon in the side chain. Recent studies have revealed that the order in which the ring substitutions and side chain modifications occur are not as clear-cut as previously depicted (Dixon et al., 2001; Humphreys and Chapple, 2002). The p-coumarate ring has been considered to undergo methoxylation successively at its 3 and 5 positions to generate 4-hydroxy-3,5-dimethoxycinnamic acid (sinapic acid) via the following intermediates in a linear pathway: 3,4-dihydroxycinnamic acid (caffeic acid), 3-methoxy-4-hydroxycinnamic acid (ferulic acid), and 3-methoxy 4,5-dihydroxycinnamic acid (5-hydroxy ferulic acid). Each of these intermediates are activated at their γ-carbon by 4-coumarate ligase (4CL) to generate corresponding CoA thioesters that in turn are successively reduced to aldehyde and alcohol forms. These linear pathways have been revised to a metabolic grid, and the latter is supported by the following observations: 5-hydroxylation occurs favorably on aldehyde and alcohol forms derived from feruloyl CoA rather than on ferulic acid (Humphreys et al., 1999; Osakabe et al., 1999); 0-methylation occurs on CoA esters of caffeate and 5-hydroxy ferulate, and on the respective aldehyde and alcohol forms (Zhong et al., 1998; Humphreys et al., 1999; Maury et al., 1999; Li et al., 2000; Parvathi et al., 2001).
Figure 1.
Biochemical transformations of the ring and side chain in cinnamic acid. It is the first phenolic acid in the general phenylpropanoid metabolism arising from deamination of l-Phe by PAL. 4-Hydroxylation of cinnamic acid by C4H generates p-coumaric acid, but further ring transformations can potentially occur when the γ-carbon is an acid, aldehyde, or ester. In the current depiction of the pathways, 3-hydroxylation occurs on p-coumaric acid or p-coumaroyl CoA, and the later 5-hydroxylation by ferulic acid 5-hydroxylase (F5H) occurs when the γ-carbon is in aldehyde or alcohol form. A fully methoxylated acid, i.e. 4-hydroxy,3,5-dimethoxycinnamic acid (sinapic acid) is an intermediate in sinapine and other sinapate ester synthesis. The gene encoding 3-hydroxylation has been elusive. Further details appear in the text.
Of the three hydroxylases implicated in ring substitutions, C4H was the first to be studied and the best characterized cytochrome P450 monooxygenase (CYP) from plants (Pierrel et al., 1994; Urban et al., 1994; Chapple, 1998; Blount et al., 2000). It is encoded by a single gene in Arabidopsis and is designated as CYP73A5 (http://drnelson.utmem.edu/CytochromeP450.html). F5H is also a P450 (Meyer et al., 1996; Chapple, 1998). Contrary to the historical nomenclature, it does not hydroxylate ferulic acid efficiently, but instead shows substrate preference for coniferaldehyde and coniferyl alcohol (Humphreys et al., 1999; Osakabe et al., 1999). Although C4H and F5H add single oxygen to the 4 and 5 positions of the phenolic ring, respectively, the situation with 3-hydroxylation has been unclear. Several types of enzymes have been implicated. Phenolases, also referred to as polyphenol oxidases, tyrosinases, and catechol oxidases, have been suggested (Freudenberg and Neish, 1968; Vaughan and Butt, 1970; Stafford and Dresler, 1972; Boniwell and Butt, 1986). These soluble enzymes differ from membrane-associated P450s in structural and mechanistic characteristics. Their poor substrate specificity and lack of coordinate gene expression with such genes as PAL and C4H have cast doubts on their involvement in the general phenylpropanoid pathway. Furthermore, chemical inhibition of phenolase activity in mung bean (Vigna mungo) seedlings does not prevent formation of caffeic acid derivatives (Duke and Vaughn, 1982).
A p-coumarate-specific hydroxylase responsible for caffeic acid formation has also been reported (Kojima and Takeuchi, 1989); however, there is no evidence for its widespread occurrence, a central characteristic for the general phenylpropanoid-related enzymes. Interestingly, Kneusel et al. (1989) have described a parsley (Petroselinum crispum) p-coumaroyl CoA hydroxylase, thus providing another entry point for 4-hydroxy coumaroyl CoA into the phenylpropanoid grid. An enzyme capable of hydroxylating p-coumaroyl Glc in sweet potato (Ipomoea batatas) has also been purified (Tanaka and Kojima, 1991). There are also reports that P450 enzyme(s) can catalyze the 3-hydroxylation of shikimate and quinate esters of p-coumarate to the corresponding caffeoyl esters (Heller and Kühnl, 1985; Kühnl et al., 1987). While this paper was in preparation, Schoch et al. (2001) reported that Arabidopsis CYP98A3 expressed in yeast (Saccharomyces cerevisiae) can catalyze 3-hydroxylation of p-coumaroyl shikimate and p-coumaroyl quinate, but not p-coumaric acid. This finding was surprising because p-coumaroyl shikimate and p-coumaroyl quinate were not generally considered as intermediates in the general phenylpropanoid metabolism.
The general phenylpropanoid network includes biosynthesis of seed-borne sinapine in crucifers (Regenbrecht and Strack, 1985). Sinapine is an antinutritional factor in canola (Brassica napus), an economically important oilseed crop, and consequently a target for elimination by breeding and biotechnology (Velasco and Möllers, 1998; Nair et al., 2000). Previously, we had characterized F5H genes from canola and showed that their transgenic suppression can result in up to 40% reduction in sinapine (Nair et al., 2000). We undertook the work presented here to investigate the elusive coumarate 3-hydroxylase (C3H) in Arabidopsis to gain an additional handle for metabolic engineering of sinapate ester synthesis in canola, a close relative of Arabidopsis.
RESULTS
CYP98A3 as a Gene Potentially Involved in the Phenylpropanoid Metabolic Network
As mentioned above, classical biochemical approaches have not led to the identification of a C3H gene. Therefore, we employed in silico and comparative gene expression analyses to identify the putative C3H gene. We set the following criteria for identifying potential candidate(s) for C3H: (a) Its product might have some structural resemblance to other hydroxylases of the phenylpropanoid network, namely C4H and F5H; (b) Expression of the candidate gene(s) should resemble that of C4H and F5H in being active in organs that have a high demand for phenylpropanoids (e.g. stem); (c) The promoter regions of the candidate gene(s) might have some features in common with C4H and F5H, suggesting similar gene regulation; (d) The gene product should be most evident in or very near lignifying cells so as to meet the huge demand for phenylpropanoids; and (e) Expression of the putative gene(s) in yeast might afford in vivo conversion of p-coumarate to caffeate.
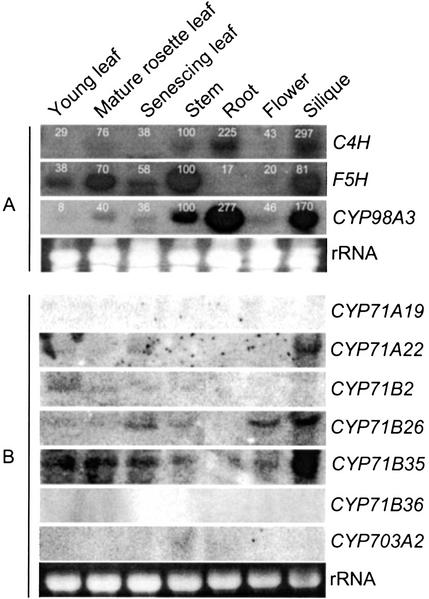
Initially, we used simple BLASTP searches of the Arabidopsis P450 database (http://www.Arabidopsis. org) with entire C4H and F5H amino acid sequences as the query. There were approximately 160 P450s in the database when this was done. The sequences identified as being related to both C4H and F5H were sorted out after visual examination of the identity and the scores. Those common to both C4H and F5H were accorded priority for further consideration. Among these, CYP98A3 was the highest scoring sequence, with an identity of 28% to C4H. Interestingly, CYP98A3-like expressed sequence tags had also been found among the cDNAs of xylem tissue from loblolly pine (Allona et al., 1998) and sweetgum (Osakabe et al., 1999). These observations encouraged us to investigate the Arabidopsis CYP98A3 gene further, and a few lower ranking candidates found in our screening were also included in the subsequent analysis. RNA from stems, roots, flowers, siliques, and leaves that were young, mature, or partially senescent was probed for transcripts of the following: C4H (CYP73A5) and F5H (CYP84), CYP98A3, CYP71A19, CYP71A22, CYP71B2, CYP71B26, CYP71B35, CYP71B36, and CYP703A2 (Fig. 2). CYP98A3 was the only one to show expression in stem, in common with C4H and F5H, and it was also the only one to resemble C4H in being highly active in root tissue (Bell-Lelong et al., 1997; Mizutani et al., 1997; Ruegger et al., 1999). However, there were some differences, such as low-level expression of F5H in roots.
Figure 2.
Expression patterns of some Arabidopsis CYP genes. PCR-amplified segments of the CYPs (http://drnelson.utmem.edu/CytochromeP450.html) were used for probing 15 μg of RNA. After initial analyses, C4H, F5H, and CYP98A3 expression was determined again for quantitation using a phosphor imager. A, The stem RNA signal was set as the reference (100%) for each probe. Expression of C4H, F5H, and CYP98A3 were analyzed successively in the indicated order after removal of the previous probe. B, Collectively, three membranes with identical RNA loading were used; a representative for rRNA loading is shown.
The promoters of the phenylpropanoid metabolism genes in many species contain conserved cis sequences that are referred to under various names (Douglas, 1996). MYB transcription factors are considered to regulate certain phenylpropanoid genes via interactions with MACCWAMC (M = A/C; W = A/T) elements present in them (Sablowski et al., 1994; Douglas, 1996). In the Arabidopsis C4H promoter region, Mizutani et al. (1997) have noted sequences resembling the “L box” (two; YCYYACCWACC; Y = C/T), and “P box” (four; YTYYMMCMAMCMMC); some of these overlap with or include the “H box”-like sequence and “Box 3”-like element found by Bell-Lelong et al. (1997). Some of the above P and L boxes comprise the MYB-binding consensus sequence (MACCWAMC; MYB element). We searched the 2-kb sequence upstream of the CYP98A3 open reading frame (ORF) and, for comparison, the 2-kb sequence upstream of Arabidopsis F5H ORF for perfectly conserved MYB elements. Five were found in CYP98A3 (−143 to −136, −1,116 to −1,109, −1,661 to −1,654, −1,674 to −1,667, and −1,779 to −1,772) and two in F5H (−90 to −83 and −1,126 to −1,119). In addition, A and H box-like sequences that were imperfectly conserved among C4H, F5H and CYP98A3 were also found. These provided additional impetus to characterize CYP98A3.
Expression of CYP98A3 in Yeast Affords 3-Hydroxylation of p-Coumaric Acid to Yield Caffeic Acid
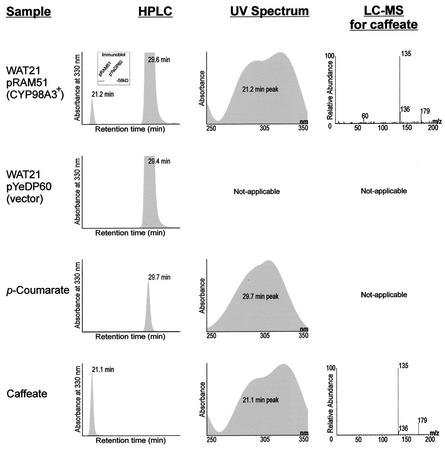
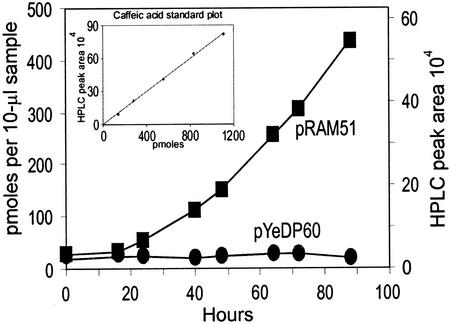
We used the P450 expression system of Pompon et al. (1996) to investigate CYP98A3 for hydroxylase activity. Plasmid pRAM51, a recombinant pYeDP60 with the CYP98A3 ORF, was introduced into the yeast WAT21 strain that produces an Arabidopsis P450 reductase, and the microsomal fractions of the pRAM51 and pYeDP60 strains were assayed for hydroxylation of cinnamic acid, p-coumaric acid, ferulic acid, coniferaldehyde, and coniferyl alcohol at substrate concentrations of 20 μm, 0.1 mm, and 1.0 mm. Even after 1 h of incubation at 30°C, no novel UV-absorbing products were detectable by HPLC. Assuming that this might be due to a slow reaction that escaped detection in the microsomal assays, we then assayed the yeast cultures supplemented with the above substrates for in vivo production of novel products. There was a new but very small peak in WAT21 (pRAM51) supplemented with p-coumarate but not with the other substrates. This product was found to be indistinguishable from caffeic acid according to three criteria (Fig. 3): (a) retention time in HPLC, (b) UV absorbance spectrum, and (c) analysis of the parent and daughter ions by liquid chromatography (LC)/negative ion electrospray mass spectrometry. The WAT21 (pYeDP60) control cultures did not produce this peak. Thus, the production of caffeic acid could be attributed to CYP98A3 in the recombinant yeast cells. The caffeic acid production analyzed over a period was linear (Fig. 4), and the apparent rate of caffeic acid production was 4.3 × 102 pmol h−1 for a unit of 2 × 108 cells and 1.2 × 103 pmol h−1 for 1-mg protein content of the cells. These results showed that CYP98A3 could hydroxylate p-coumaric acid in vivo.
Figure 3.
Hydroxylation of p-coumaric acid by yeast WAT21 (pRAM51) cells that produce Arabidopsis CYP98A3. Inset, Immunoblot of 15 μg of protein from the control strain containing the vector (pYeDP60) and the recombinant strain fractionated on SDS-PAGE. The new peak appearing in the CYP98A3+ strain matched an authentic caffeic acid standard. Negative ion electrospray tandem mass spectrometry identified a predominant daughter ion (m/z 135) from the parent ion (m/z 179).
Figure 4.
CYP98A3-catalyzed production of caffeic acid in yeast WAT 21 cells. The cells were grown in YPLA medium (Pompon et al., 1996) supplemented with 5 mm p-coumaric acid. Samples withdrawn at indicated times were analyzed for caffeic acid content by HPLC as described in “Materials and Methods.” Inset, Caffeic acid standard plot constructed with authentic sample.
Comparative Analysis of the Developmental Regulation of CYP98A3 and C4H
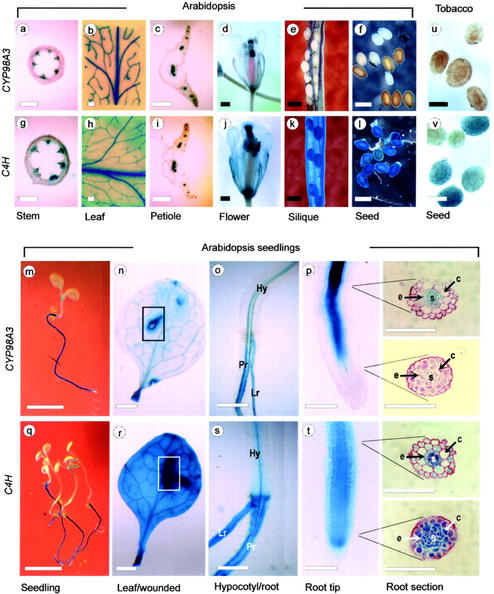
The results above and the commonality of CYP98A3 and C4H gene expression in various tissues (Fig. 2) were intriguing to warrant a comparison of the developmental regulation of the two genes in Arabidopsis. We reasoned that this comparative expression profiling would be informative with regard to the functional relationships of CYP98A3 with C4H. The 5′-upstream promoter sequences of the CYP98A3- and C4H-coding regions retrieved by PCR were fused to the ORF of the Escherichia coli β-glucuronidase (GUS) gene (Jefferson et al., 1986). At least 20 independent Arabidopsis transgenic lines were produced for each construct and analyzed initially for GUS expression. Five second generation transgenic lines were analyzed for detailed GUS activity in seedlings grown on Murashige and Skoog agar medium and in plants grown on soil (Fig. 5). The vascular bundles in stem, petiole, leaf, and silique wall showed evidence of promoter activity in both CYP98A3::GUS and C4H::GUS plants (Fig. 5, a–c, e, g–i, and k). The GUS staining was always more intense in the C4H::GUS plants than in the CYP98A3::GUS plants. The observed differences between these included an overall GUS staining of the flowers in the C4H::GUS plants (Fig. 5j), but in the case of the CYP98A3::GUS flowers (Fig. 5d), only in the vascular tissues of petal, sepal, anther, and stigma. The most contrasting characteristic between these two was the absence of GUS staining in the seeds of CYP98A3 lines, whereas it was very intense in C4H transgenic seeds (Fig. 5, e, f, k, and l). This difference was evident in transgenic tobacco (Nicotiana tabacum) seeds as well (Fig. 5, u and v), and the spatial expression of GUS in other parts of transgenic tobacco plants was comparable with that in Arabidopsis (data not shown). In Arabidopsis seedlings, both CYP98A3 and C4H promoters were most active in roots (Fig. 5, m and q). However, there were some zonal and tissue-specific differences: Unlike the C4H promoter (Fig. 5t), the CYP98A3 promoter was not active in the apical meristem of roots (Fig. 5p), and it was active only in stele and endodermis but not in epidermis or cortex, whereas the C4H promoter was active in all cells as shown in the cross sections of the upper and lower regions of the roots. Wounding induced both C4H and CYP98A3 promoters (Fig. 5, n and r). Thus, the two genes were generally comparable in spatial expression.
Figure 5.
Developmental regulation of CYP98A3 and C4H. Transgenic plants with promoter::GUS fusion constructs were analyzed for 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-gluc) staining. a, g, c, i, p, and t, Compound microscope; the remainder are from a dissecting microscope. p and t, Differential interference contrast microscopy. The material in a through l and u and v were from soil-grown plants, and the rest from seedlings germinated on Murashige and Skoog agar. The deliberately wounded parts of excised leaves are marked in boxes. Hy, Hypocotyl; Pr, primary root; Lr, lateral root. Cross sections: s, stele; e, endodermis; c, cortex. Bars: 100 μm for p and t; 400 μm for a through l, o, s, u, and v; 1 mm for n and r; and 4 mm for m and q.
CYP98A3 Protein Localizes to Lignifying Cells of Arabidopsis Stems and Roots
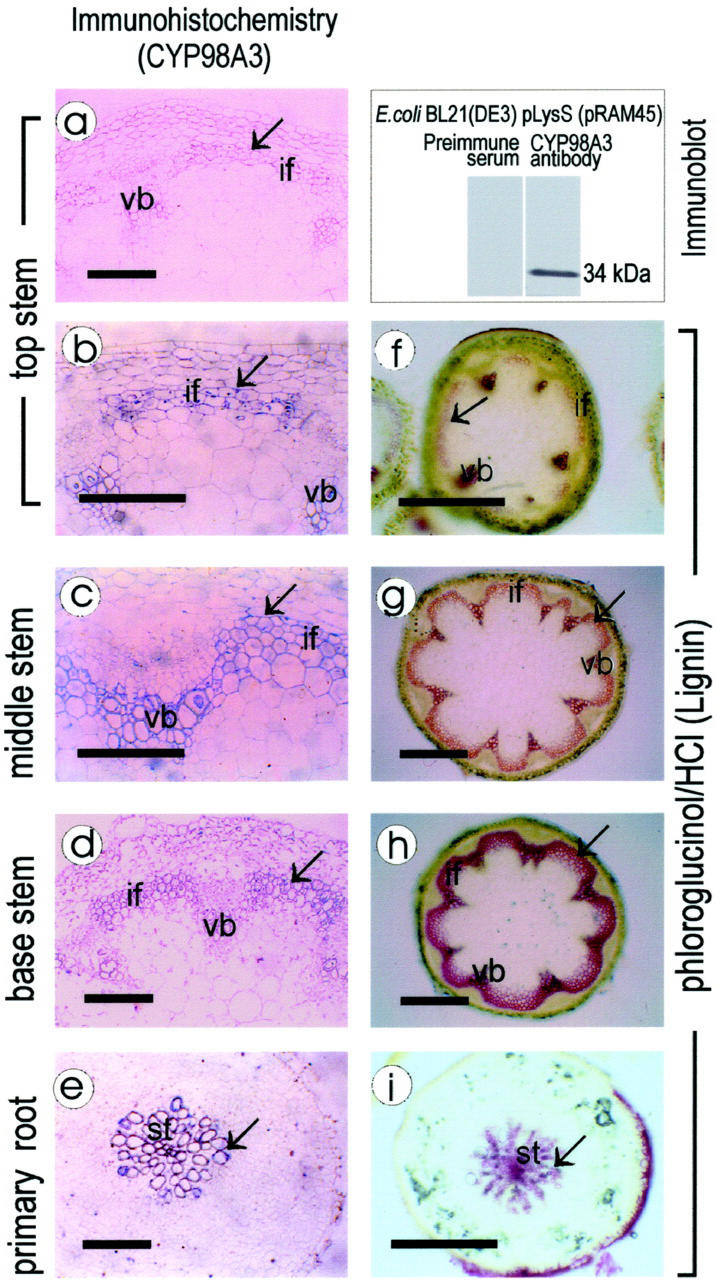
The common characteristics of CYP98A3 and C4H in the analyses described above suggested that CYP98A3 might also be produced in lignifying cells. This was found to be the case in immunolocalization by polyclonal antibodies raised against a truncated CYP98A3 expressed in recombinant E. coli. Although the pre-immune serum did not bind to lignifying tissue, the antiserum did. The meta- and protoxylem cells undergoing primary lignification in young stem showed a positive immunochemical reaction, and the lignified interfascicular fibers and xylem vessels in older stem showed a stronger reaction (Fig. 6). This correlated well with the lignin deposition patterns in Arabidopsis (Zhong et al., 2000). In root, the stele cells were reactive with the antiserum, providing further evidence that CYP98A3 was associated with lignification.
Figure 6.
CYP98A3 localization, and lignification. Immunochemistry with polyclonal antiserum raised against a truncated CYP98A3 produced in E. coli (inset). Paraffin sections of stem (a–d) and root (e) were probed with pre-immune serum (a) or antiserum (b–e). Hand sections of stem (f–h) or root (i) were stained with phloroglucinol for lignin. Bar = 100 μm in a through e and 400 μm in f through i. Arrows indicate tissues that are lignified. vb, Vascular bundles; if, interfascicular fiber; st, stele.
DISCUSSION
We have extensively characterized the developmental regulation of CYP98A3 and have shown that it resembles in many respects C4H that encodes the archetypal hydroxylase of phenylpropanoid metabolism. Taken together with the finding that CYP98A3 was most evident in lignifying cells and that it was capable of hydroxylating p-coumarate to caffeate, albeit at a very slow rate in yeast cells, it was attractive to consider CYP98A3 as a phenylpropanoid network enzyme. Historically, all such evidence has been used in assigning the role of a given gene to a specific function in phenylpropanoid biosynthesis, and, as discussed below, recent evidence shows that CYP98A3 is involved in general phenylpropanoid metabolism.
CYP98A3, the Elusive 3-Hydroxylase of the General Phenylpropanoid Metabolism
The independent in silico gene discovery approaches employed by Schoch et al. (2001) and us entailed assumptions of a potential 3-hydroxylase being structurally similar to the previously characterized hydroxylases of phenylpropanoid metabolism. These strategies were productive in the identification of CYP98A3, even with the following caveat that some P450s are enigmatic: Nearly identical P450s (in one example differing by just one amino acid) can mediate reactions on unrelated substrates and, conversely, dissimilar P450s can use a common substrate (Schuler, 1996). A 3-hydroxylase of the latter type, if present, would have escaped detection at the outset.
In recombinant yeast cells, CYP98A3 converted p-coumaric acid to caffeic acid, but so slowly that the reaction escaped detection in in vitro assays. The rate of caffeic acid production contrasts highly with that of the production of p-coumaric acid by C4H-expressing recombinant yeast cells; the latter is 1.6 × 106 pmol h−1 for 2 × 108 cells, as calculated from the data of Urban et al. (1994), and, thus, 3,700-fold greater than the caffeic acid production. Schoch et al. (2001) could not detect p-coumaric acid hydroxylation by yeast-produced CYP98A3 in in vitro assays. While our paper was in revision, Franke et al. (2002b) have also reported that the CYP98A3 activity with p-coumaric acid in vitro was too low to permit accurate measurement of the catalytic properties, even lower with p-coumaraldehyde, and undetectable with p-coumaryl alcohol. With regard to other potential substrates, p-coumaroyl CoA, p-coumaraldehyde, p-coumaryl alcohol, or 1-O-glucoside or 4-O-glucoside esters of p-coumaric acid are not hydroxylated in vitro (Schoch et al., 2001). However, CYP98A3 hydroxylates p-coumaroyl shikimate (Kcat/Km = 87 min−1 μm−1; Km = 7 μm) and, at a 4-fold less efficiency, p-coumaroyl quinate (Kcat/Km = 22 min−1 μm−1; Km = 18 μm) to the corresponding caffeoyl esters (Schoch et al., 2001). The apparent Km for the shikimate ester is similar to that reported for C4H toward cinnamic acid (4–10 μm; Urban et al., 1994, 1997) and for F5H toward coniferaldehyde (1 μm) and coniferyl alcohol (3 μm; Humphreys et al., 1999). The poor catalytic activity of CYP98A3 in p-coumaric acid hydroxylation, as surmised from our in vivo measurements, is reminiscent of the observations that Arabidopsis F5H and its corresponding enzyme from sweetgum hydroxylate coniferaldehyde at 150- to 1,000-fold greater efficiency in comparison with ferulic acid as the substrate (Humphreys et al., 1999; Osakabe et al., 1999). Although all these observations need to be considered in light of potential metabolic channeling that has been inferred for phenylpropanoid intermediates (Burbulis and Winkel-Shirley, 1999; Dixon et al., 2001), they do suggest that shikimate and quinate esters are likely the physiological substrates, as originally conceived by Heller and Kühnl (1985) and Kühnl et al. (1987), and that p-coumaric acid is not. The monooxygenase(s) present in carrot (Daucus carota; Heller and Kühnl, 1985) and parsley (Kühnl et al., 1987) may well be encoded by orthologs of the Arabidopsis CYP98A3.
How exactly caffeoyl shikimate and quinate esters fit into the general phenylpropanoid metabolism is unclear. Typical with the construction of the metabolic maps of phenylpropanoids, only a composite picture from multiple species is available. The shikimate and quinate esters are not methylated (Schoch et al., 2001), but caffeoyl CoA is (Dixon et al., 2001). Because caffeoyl shikimate or quinate do not accumulate in Arabidopsis, these esters might be acted upon by a reversible hydroxycinnamoyl CoA p-hydroxycinnamoyltransferases activity to generate caffeoyl CoA as proposed by Schoch et al. (2001). In parsley, caffeoyl CoA can be produced by a coumaroyl CoA 3-hydroxylase (Kneusel et al., 1989). In Arabidopsis, however, this route is either nonexistent or insignificant as deduced from the following observations: p-coumaroyl CoA is not hydroxylated by CYP98A3 (Schoch et al., 2001); the Arabidopsis ref8 mutant, with a chemically induced mutation in CYP98A3, accumulates p-coumaroyl esters, instead of sinapoyl esters, in its leaves and synthesizes lignins derived almost exclusively from p-coumaryl alcohol (Franke et al., 2002a). Thus, the genetic evidence not only confirms CYP98A3 mediating aromatic 3-hydroxylation but also shows that a distinct coumaroyl CoA 3-hydroxylase, if present in Arabidopsis, cannot appreciably substitute for CYP98A3. All of these studies necessitate a revision to the general phenylpropanoid metabolism to include an obligatory esterification at the γ-position before hydroxylation at the 3 position (Fig. 1), and furthermore portend additional revisions in regards to the formation of the 3-methoxy intermediate.
Developmental Regulation of CYP98A3. Commonality and Dissimilarity of CYP98A3 and C4H
CYP98A3 promoter activity, like C4H activity, was most abundant in vascular tissue. In addition, the CYP98A3 protein was most evident in lignifying cells in stem and root, which require a large supply of phenylpropanoids. Both promoters were also wound inducible, another characteristic of lignin biosynthesis-related genes (Ohl et al., 1990; Bell-Lelong et al., 1997; Mizutani et al., 1997; Ehlting et al., 1999). CYP98A3 gene expression was strong in roots (approximately 280% of stem; all comparisons are for a given probe, relative to the signal in stem sample as shown in Fig. 2) and resembled C4H (approximately 230%) in this respect. This differs from the results of Schoch et al. (2001) that show a high level expression of CYP98A3 in stem but much less in roots. In siliques, C4H expression was much more robust (approximately 300%) than in stems; CYP98A3 also showed this trend (approximately 170%).
The histochemical analyses of promoter function portrayed the commonality between CYP98A3 and C4H expression in general, and, in addition, showed some specific differences. Although C4H promoter activity was found in all cell layers of the root, CYP98A3 promoter function was localized to the stele and endodermis where the cells were lignified. Because C4H catalyzes the pivotal hydroxylation step that generates p-coumarate, which in turn is a common precursor for the biosynthetic pathways of monolignols, sinapates, and flavonoids, the broad gene expression is explicable on the basis of the upstream function for C4H. Notable in this regard is the in situ localization of chalcone synthase and chalcone isomerase to the epidermal and cortex cells of the primary root where flavonoids accumulate (Saslowsky and Winkel-Shirley, 2001). The absence of CYP98A3 promoter activity in these locations is consistent with a function of CYP98A3 occurring downstream of C4H, but not required for flavonoid biosynthesis. Most of the phenylpropanoid genes that exist as multigene families are differentially expressed. For example, the members within the Arabidopsis PAL, 4CL, and CCR gene families are differentially regulated (Liang et al., 1989; Ehlting et al., 1999; Lauvergeat et al., 2001). For single-copy genes, a higher basal level of gene expression might be necessary in those organs that potentially have a sudden and high demand for their activity. Considering this potential requirement, the deviations in the expression of CYP98A3 and C4H are likely to have some functional implications. The high transcript level of CYP98A3 in roots is intriguing. Given that roots are in intimate contact with other organisms in the biosphere, both CYP98A3 and C4H may also be involved in synthesizing phenylpropanoids pertaining to interactions with these organisms. For example, some phenylpropanoids have antimicrobial activity (Keen and Littlefield, 1979; Barber et al., 2000). Interestingly, F5H expression in roots was relatively low (17%), suggesting that the products further downstream of F5H action might not have such a function. The implications of the differential expression in roots remain to be unraveled, but these observations place C4H and CYP98A3—the relatively upstream hydroxylases—in one category and the downstream F5H in another.
The phenylpropanoid pathway in Arabidopsis generates leaf- and seed-associated sinapate esters such as sinapoylmalate and sinapine, respectively. Sinapine synthesis, a distinct hallmark of crucifers, occurs only in seed tissue, and is a subject of our interest (Nair et al., 2000). Although the CYP98A3 promoter was active in the silique walls of transgenic Arabidopsis, it was not active in seeds. The ref8 mutant is impaired in sinapine synthesis in Arabidopsis seeds (Franke et al., 2002a), showing that CYP98A3 in its native context does function in seeds. Together, our observation of the lack of GUS expression in seeds with the promoter construct suggests the presence of a hitherto uncharacterized seed-active cis element(s) in the CYP98A3 gene outside of the 2.2-kb promoter region included in our study. These element(s) might offer a means to differentially regulate this single-copy gene in seeds.
MATERIALS AND METHODS
Plant Growth Conditions and Chemicals
Arabidopsis (Columbia) was used throughout. Unless stated otherwise, plants were grown in pots containing RediEarth (Grace Horticultural Products, Ajax, ON, Canada) in a chamber under a 16-h-light/25°C and 8-h-dark/22°C cycle with a light intensity of 380 μmol m−2 s−1 photosynthetic photon flux density. Cinnamic acid, p-coumaric acid, caffeic acid, ferulic acid, coniferaldehyde, and coniferyl alcohol were from Sigma-Aldrich (Oakville, ON, Canada).
DNA and RNA Analysis
Young leaves, mature leaves, and roots were collected from 4-week-old plants. Main inflorescence stems, partly senescent leaves, siliques, and flowers, were collected from 8-week-old plants. Roots were washed with double-deionized water to remove the soil. All tissues were frozen in liquid nitrogen and stored at −80°C. All DNA and RNA extractions and analyses were as in Nair et al. (2000). A Molecular Dynamics PhosporImager:SI and ImageQuaNT program (Amersham, Sunnyvale, CA) were used for quantitating the RNA signal intensity according to the supplier's instructions. The CYP probe DNAs were amplified by PCR with the following primers: CYP98A3 (atgtcgtggtttctaatagc and aaggctagccgcgttatgttgt), CYP71A19 (tgtgcttaacaacgctcctt and cttcttgtaagaccggacca), CYP71A22 (gaagaaaagcaacacacctg and gttggtgaggagatggagga), CYP71B2 (cgatcttgctctgtttcttc and gatactagcggtgagggaga), CYP71B26 (tcgacgaataccatctcctc and tgactgcagagcttccttag), CYP71B35 (ggcttctgtcacttatcttc and gtcctgcagaccacactaac), CYP71B36 (ttgtattcttctagccgcct and tcgctcaagttaaccggagt), and CYP703A2 (atgattttcgtgctagcctc and cttgggcttcttttgggcta). GenBank entries are as follows: CYP98A3 sequence, locus At2g40890 and bacterial artificial chromosome AC002409; and F5H sequence, locus At4g36220 and bacterial artificial chromosome AL022141. The FINDPATTERNS program in the GCG software suite (Genetics Computer Group, Madison, WI; http://www.cbr.nrc.ca) was used for sequence analyses.
Reverse Transcriptase-PCR
First-strand cDNA was synthesized using 10 μg of total RNA as described in Nair et al. (2000). PCR was set in a 50-μL reaction volume containing 2.5 μL of the cDNA, 1× Pfu polymerase buffer (Stratagene, La Jolla, CA), 200 μm of each dNTP, and 50 pmol of each of the gene-specific primers (5′-gcggatccg-atgtcgtggtttctaatagcggtgg-3′ and 5′-gcgaattca-ttacatatcgtaaggcacgcgt-3′) to amplify the CYP98A3 ORF. After initial denaturation of DNA for 2 min, PCR was conducted for 25 cycles with 2.5 units of Pfu DNA polymerase (Stratagene) in a DNA Thermal Cycler (Perkin-Elmer Applied Biosystems, Foster City, CA) at a setting of 94°C for 45 s, 56°C for 1 min, and 72°C for 4 min for each cycle. The PCR product was purified, digested with BamHI and EcoRI (Life Technologies, Rockville, MD), and cloned into pBluescript SK− (Stratagene). The ORF sequence of CYP98A3 was confirmed by sequencing and the plasmid termed as pRAM48.
Genetic Transformation of Plants
The plant vector pRD420 (Datla et al., 1993) derivatives containing the promoter from Arabidopsis C4H (CYP73A5) or CYP98A3 as a HindIII-BamHI segment was fused to GUS ORF. These vectors were transformed into Agrobacterium tumefaciens GV3101 (pMP90). The promoters were originally retrieved from Arabidopsis genomic DNA by Pfu Turbo (Stratagene)-mediated PCR. 5′-gcgaagctt-tatttcctgcaaaagatgttataatg-3′ and 5′-gcggatcc-gaagttttgcttctatttttattttcgg-3′ were used for retrieving 2.2 kb immediately upstream of the CYP98A3 ORF, and 5′-gcaagctt-agaggagaaactgag-3′ and 5′-gcggatcc-tatagtttgtgtatccgcaatgatattg-3′ for 2.9 kb of the C4H promoter sequence. Arabidopsis plants with 3- to 5-cm stem bolts were transformed using the floral dip method of Clough and Bent (1998). Tobacco (Nicotiana tabacum) plants were transformed as described previously (Datla et al., 1993).
Production of CYP98A3 in Yeast (Saccharomyces cerevisiae), and Biochemical Analyses
The WAT21 strain, pYeDP60, microsome preparation and assays were as in Pompon et al. (1996) with the indicated modifications. The CYP98A3 ORF from pRAM48 was subcloned between the BamHI and EcoRI sites of pYeDP60 to generate pRAM51. The enzyme assays of microsomal preparations were done in a 500-μL reaction according to the method of Humphreys et al. (1999): 450 μL of 100 mm sodium phosphate buffer (pH 7.4), containing 1 mm NADP+, 10 mm Glc 6-phosphate, and 1 unit of Glc-6-phosphate dehydrogenase (Sigma-Aldrich), was pre-incubated at 30°C in the presence of one of the substrates (cinnamic acid, p-coumaric acid, ferulic acid, coniferaldehyde, or coniferyl alcohol) at three different concentrations of 20 μm, 0.1 mm, or 1.0 mm. The reaction was initiated by the addition of 5 or 50 μg of microsomes from the vector control or pRAM51 cells in 50 μL, allowed to proceed at 30°C for 15 or 60 min, and stopped by adding 50 μL of trifluoroacetic acid. The extract supernatant was analyzed by HPLC (Nucleosil C18 AB column, Alltech, Deerfield, IL) using an acetonitrile/phosphoric acid (1.5% [v/v]) gradient of 0% to 25% (v/v) acetonitrile over a 40-min period in a 60F multisolvent delivery system (Waters, Milford, MA) fitted with a Waters 600 controller. A330 was determined with a Waters 996 photodiode array detector.
The in vivo assays and the media compositions were as in Pompon et al. (1996), except for supplementation with the phenolics used here. A saturated culture of yeast grown at 28°C in 2 mL of N3AT medium was subcultured into 100 mL of YPGE medium for 24 h. Fifty milliliters of the culture was centrifuged and resuspended in 100 mL of YPLA medium, and was induced for 6 h with 2% (w/v) Gal. Ten milliliters of the induced culture was added to 100 mL of YPLA containing 2% (w/v) Gal and one of the following phenolics at 0.1, 1.0, or 5 mm: cinnamic acid, p-coumaric acid, ferulic acid, coniferaldehyde, and coniferyl alcohol. One-milliliter samples were then drawn at 0, 16, 24, 40, 48, 64, 72, and 88 h after incubation at 28°C, and 500 μL of the supernatant was added to 100 μL of trifluoroacetic acid, and 20 μL of the supernatant analyzed by HPLC as described above.
Negative ion electrospray tandem mass spectrometry analysis for caffeic acid was done by the Mass Spectrometry Unit with a Quattro-LC mass spectrometer (Micromass, Manchester, UK) following LC in an HP1100 HPLC (Agilent, Palo Alto, CA). The LC was done with a Genesis C18 reverse phase column (2.1 × 100 mm; Jones Chromatography, Lakewood, CO) and a mobile phase of aqueous (12 mm acetic acid, pH 3.3) solution with acetonitrile increasing from 0% to 25% (v/v) over 40 min. Multiple reaction monitoring was for caffeic acid (m/z of 179) and its most abundant daughter ion (m/z of 135) generated by argon collision.
Production of CYP98A3 Antisera, and Immunoblot
A partial CYP98A3 cDNA (bp 309–1,141 of the ORF) amplified using Pfu and the primers 5′-gaggatcc-tagccgcaacggtcag-3′ and 5′-gcaagctt-agcctccgatcttgacatct-3′ was ligated into the BamHI-HindIII sites of pRSET B vector (Invitrogen, Carlsbad, CA) to give pRAM45. Escherichia coli BL21(DE3) pLysS strain (Invitrogen) containing pRAM45 or pRSET B was grown overnight at 30°C in 2 mL of Luria-Bertani medium, subcultured in 50 mL of Luria-Bertani medium, and grown to A600 of 0.7. Isopropyl β-d-1-thiogalactopyranoside was added to 0.1 mm, and incubated for 3 h before harvesting and lysing in 6 m urea, pH 8.0. The protein was purified on nickel-nitrilotriacetic acid agarose columns (Qiagen, Valencia, CA) by the manufacturer's protocol. The 34-kD polypeptide, excised from an SDS-PAGE (10% acrylamide, w/v), was used to raise antibodies in rabbits at the facilities of Veterinary and Infectious Disease Organization (Saskatoon, SK, Canada). Western blotting was as described in Nair et al. (2000). Pre-immune serum was used as a control in all immunoblot and immunolocalization experiments.
GUS Activity in Situ, Lignin Staining, and Immunohistochemistry
Hand sections or explants were incubated in a solution of X-gluc [1 mm X-gluc in 50 mm phosphate buffer (pH 7.2) containing 0.5 mm K4Fe(CN)6.H2O, 0.5 mm K3Fe(CN)6, 10 mm EDTA, and 0.1% (v/v) Triton X-100] at 37°C overnight, destained, and rinsed in 70% (v/v) ethanol. Seedlings were stained for GUS according to Malamy and Benfey (1997). Whole root tips were mounted in 50% (v/v) glycerol. For cross sections, the GUS-stained root tips were fixed, dehydrated, and embedded in paraffin as described in Wan et al. (2002). Counterstaining was done with 0.01% (w/v) Safranin in deionized water for 15 min, and the sections were mounted with Cytoseal (Stephens Scientific, Kalamazoo, MI). Phloroglucinol stains lignin (Freudenberg and Neish, 1968) and hand-sectioned stem or root was stained with 1% (w/v) Phloroglucinol (Sigma-Aldrich) in 6 n HCl as described in Zhong et al. (2000). Immunolocalization methods for tissue preparation and detection were as in Wan et al. (2002), with CYP98A3 antiserum at 1:500 (v/v) dilution.
ACKNOWLEDGMENTS
We are grateful to Mr. Stephen Ambrose for mass spectrometry analyses; Ms. Yan Ge for tobacco transformation; Mr. Darrin Klassen, Mr. Barry Panchuk, and Ms. Inge Roewer for DNA sequencing; Ms. Deanna Ratzlaff and Mr. Don Schwab for oligonucleotide synthesis; Dr. Clint Chapple, Dr. John Balsevich, and the anonymous reviewers for their helpful suggestions; and Dr. Denis Pompon and Dr. Philippe Urban for plasmid pYeDP60 and WAT21 strain.
Footnotes
This is publication no. 45,239 of the National Research Council of Canada.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.008649.
LITERATURE CITED
- Allona I, Quinn M, Shoop E, Swope K, Cyr S, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R et al. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MS, McConnell VS, DeCaux BS. Antimicrobial intermediates of the general phenylpropanoid and lignin specific pathways. Phytochemistry. 2000;54:53–56. doi: 10.1016/s0031-9422(00)00038-8. [DOI] [PubMed] [Google Scholar]
- Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C. Cinnamate-4-hydroxylase expression in Arabidopsis: regulation in response to development and the environment. Plant Physiol. 1997;113:729–738. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA. Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 2000;122:107–116. doi: 10.1104/pp.122.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniwell JM, Butt VS. Flavin nucleotide-dependent 3-hydroxylation of 4-hydroxyphenylpropanoid carboxylic acids by particulate preparations from potato tubers. Z Naturforsch. 1986;41c:56–60. [Google Scholar]
- Boudet AM. Lignins and lignification: selected issues. Plant Physiol Biochem. 2000;38:81–96. [Google Scholar]
- Burbulis IE, Winkel-Shirley B. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc Natl Acad Sci USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:311–343. doi: 10.1146/annurev.arplant.49.1.311. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Croteau R, Kutchan TM, Lewis NG. Natural products (secondary metabolites) In: Buchanan B, Gruissem W, Jones R, editors. Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 1250–1318. [Google Scholar]
- Datla RSS, Bekkaoui F, Hammerlindl JK, Pilate G, Dunstan DI, Crosby WL. Improved high-level constitutive foreign gene expression in plants using an AMV RNA4 untranslated leader sequence. Plant Sci. 1993;94:139–149. [Google Scholar]
- Dixon RA, Chen F, Guo D, Parvathi K. The biosynthesis of monolignols: a “metabolic grid,” or independent pathways to guaiacyl and syringyl units? Phytochemistry. 2001;57:1069–1084. doi: 10.1016/s0031-9422(01)00092-9. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Lamb CJ, Masoud S, Sewalt VJH, Paiva NL. Metabolic engineering: prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses: a review. Gene. 1996;179:61–71. doi: 10.1016/s0378-1119(96)00327-7. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Steele CL. Flavonoids and isoflavonoids: a gold mine for metabolic engineering. Trends Plant Sci. 1999;4:394–400. doi: 10.1016/s1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- Douglas CJ. Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci. 1996;1:171–178. [Google Scholar]
- Duke SO, Vaughn KC. Lack of involvement of polyphenol oxidase in ortho-hydroxylation of phenolic compounds in mung bean seedlings. Physiol Plant. 1982;54:381–385. [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999;19:9–20. doi: 10.1046/j.1365-313x.1999.00491.x. [DOI] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J. 2002a;30:47–59. doi: 10.1046/j.1365-313x.2002.01267.x. [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002b;30:33–45. doi: 10.1046/j.1365-313x.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- Freudenberg K, Neish AC. Constitution and Biosynthesis of Lignin. New York: Springer-Verlag; 1968. [Google Scholar]
- Heller W, Kühnl T. Elicitor induction of a microsomal 5-O-(4-coumaroyl) shikimate 3′-hydroxylase in parsley cell suspension cultures. Arch Biochem Biophys. 1985;241:453–460. doi: 10.1016/0003-9861(85)90570-3. [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C. Rewriting the lignin roadmap. Curr Opin Plant Biol. 2002;5:224–229. doi: 10.1016/s1369-5266(02)00257-1. [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA. 1999;96:10045–10050. doi: 10.1073/pnas.96.18.10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsch D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Littlefield LJ. The possible association of phytoalexins with resistance gene expression in flax to Melamspora lini. Physiol Plant Pathol. 1979;14:265–280. [Google Scholar]
- Kneusel RE, Matern U, Nicolay K. Formation of trans-caffeoyl-CoA from trans-4-coumaroyl-CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elicitor-induced pH shift. Arch Biochem Biophys. 1989;269:455–462. doi: 10.1016/0003-9861(89)90129-x. [DOI] [PubMed] [Google Scholar]
- Kojima M, Takeuchi W. Detection and characterization of p-coumaric acid hydroxylase in mung bean, Vigna mungo, seedlings. J Biochem. 1989;105:265–270. doi: 10.1093/oxfordjournals.jbchem.a122651. [DOI] [PubMed] [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E. Chlorogenic acid biosynthesis: characterization of a light-induced microsomal 5–0-(4-coumaroyl)-d-quinate/shikimate 3′-hydroxylase from carrot (Daucus carota L.) cell suspension cultures. Arch Biochem Biophys. 1987;258:226–232. doi: 10.1016/0003-9861(87)90339-0. [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lassere E, Roby D, Grima-Pettenati J. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry. 2001;57:1187–1195. doi: 10.1016/s0031-9422(01)00053-x. [DOI] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- Li L, Popko JL, Umezawa T, Chiang VL. 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem. 2000;275:6537–6545. doi: 10.1074/jbc.275.9.6537. [DOI] [PubMed] [Google Scholar]
- Liang X, Dron M, Cramer CL, Dixon RA, Lamb CJ. Differential regulation of phenylalanine ammonia-lyase genes during plant development and by environmental cues. J Biol Chem. 1989;264:14486–14492. [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Maury S, Geoffroy P, Legrand M. Tobacco O-methyltransferases involved in phenylpropanoid metabolism: the different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 1999;121:215–223. doi: 10.1104/pp.121.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS. Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D, Sato R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997;113:755–763. doi: 10.1104/pp.113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RB, Joy RW, Kurylo E, Shi X, Schnaider J, Datla RSS, Keller WA, Selvaraj G. Identification of a CYP84 family of cytochrome P450-dependent monooxygenase genes in Brassica napus and perturbation of their expression for engineering sinapine reduction in the seeds. Plant Physiol. 2000;123:1623–1634. doi: 10.1104/pp.123.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl S, Hedrick SA, Chory J, Lamb CJ. Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell. 1990;2:837–848. doi: 10.1105/tpc.2.9.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL. Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci USA. 1999;96:8955–8960. doi: 10.1073/pnas.96.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo D, Blount JW, Dixon RA. Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 2001;25:193–202. doi: 10.1046/j.1365-313x.2001.00956.x. [DOI] [PubMed] [Google Scholar]
- Pierrel MA, Batrad Y, Kazmaier M, Mignotte-Vieux C, Durst F, Werck-Reichhart D. Catalytic properties of the plant cytochrome P450 CYP73 expressed in yeast. Substrate specificity of a cinnamate hydroxylase. Eur J Biochem. 1994;224:835–844. doi: 10.1111/j.1432-1033.1994.00835.x. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Regenbrecht J, Strack D. Distribution of 1-sinapoyl-glucose:choline sinapoyltransferase activity in the Brassicaceae. Phytochemistry. 1985;24:407–410. [Google Scholar]
- Ruegger M, Meyer K, Cusumano JC, Chapple C. Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol. 1999;119:101–110. doi: 10.1104/pp.119.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M. A flower-specific myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslowsky D, Winkel-Shirley B. Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 2001;27:37–48. doi: 10.1046/j.1365-313x.2001.01073.x. [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem. 2001;276:36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- Schuler MA. Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- Stafford HA, Dresler S. 4-hydroxycinnamic acid hydroxylase and polyphenolase activities in Sorghum vulgare. Plant Physiol. 1972;49:590–595. doi: 10.1104/pp.49.4.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack D. Phenolic metabolism. In: Dey PB, Harborne JB, editors. Plant Biochemistry. Toronto: Academic Press; 1997. pp. 387–416. [Google Scholar]
- Tanaka M, Kojima M. Purification and characterization of p-coumaroyl-d-glucose hydroxylase of sweet potato (Ipomoea batatus) roots. Arch Biochem Biophys. 1991;284:151–157. doi: 10.1016/0003-9861(91)90277-p. [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272:19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- Urban P, Werck-Reichhart D, Teutsch HG, Durst F, Regnier S, Kazmaier M, Pompon D. Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur J Biochem. 1994;222:843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]
- Vaughan PFT, Butt VS. The action of o-dihydric phenols in the hydroxylation of p-coumaric acid by a phenolase from leaves of spinach beet (Beta vulgaris L.) Biochem J. 1970;119:89–94. doi: 10.1042/bj1190089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco L, Möllers C. Nondestructive assessment of sinapic acid esters in Brassica species: II. Evaluation of germplasm and identification of phenotypes with reduced levels. Crop Sci. 1998;38:1650–1654. [Google Scholar]
- Wan L, Xia Q, Qiu X, Selvaraj G. Early stages of seed development in Brassica napus: a seed coat specific cysteine proteinase associated with programmed cell death of the inner integument. Plant J. 2002;30:1–10. doi: 10.1046/j.1365-313x.2002.01262.x. [DOI] [PubMed] [Google Scholar]
- Zhong R, Morrison WH, Negrel J, Ye Z-H. Dual methylation pathways in lignin biosynthesis. Plant Cell. 1998;10:2033–2045. doi: 10.1105/tpc.10.12.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ripperger A, Ye Z-H. Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 2000;123:59–69. doi: 10.1104/pp.123.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]