Abstract
Phosphorus deficiency is one of the major abiotic stresses affecting plant growth. Plants respond to the persistent deficiency of phosphate (Pi) by coordinating the expression of genes involved in alleviation of the stress. The high-affinity Pi transporters are among the major molecular determinants that are activated during Pi stress. In this study, using three reporter genes (green fluorescent protein, luciferase, and β-glucuronidase) regulated by two Pi transporter promoters, we have carried out an extensive analysis of transcriptional and spatial regulation of gene expression. Activation of the genes was rapid, repressible, and specific in response to changes in Pi availability. The phytohormones auxin and cytokinin suppressed the expression of the reporter gene driven by the AtPT1 promoter, and that of the native gene, suggesting that hormones may be involved in regulation of some component(s) of Pi starvation response pathway. These studies also provide molecular evidence for a potential role of high-affinity Pi transporters in mobilizing Pi into reproductive organs. The results suggest that members of the Pi transporter family may have similar but nonredundant functions in plants.
Phosphorus is one of the major, but least available, essential nutrients acquired by plants. The evolution of plants in environments where phosphate (Pi) availability is low in the rhizosphere has led to numerous adaptations required for the survival of plants (Raghothama, 1999). These include the capacity to enhance Pi uptake during nutrient deficiency (McPharlin and Bieleski, 1987; Furihata et al., 1992). This increase has been correlated with an increased number of high-affinity Pi transporters assembled in the plasma membrane (Drew and Saker, 1984; Shimogawara and Usuda, 1995; Muchhal and Raghothama, 1999).
The high-affinity Pi transporters, encoded by a small family of genes, translocate Pi from the external media containing very low levels of the nutrient into the cytoplasm (Raghothama, 2000a, 2000b). Nine genes with a high degree of similarity to high-affinity Pi transporters have been identified in the Arabidopsis genome. Two members of this family have been shown to function as the high-affinity Pi transporters (Muchhal et al., 1996; Mitsukawa et al., 1997). Most of the cloned Pi transporters are expressed preferentially in roots under Pi starvation, consistent with their role in the nutrient acquisition (Muchhal et al., 1996; Leggewie et al., 1997; Smith et al., 1997; Liu et al., 1998a, 1998b). In addition, Pi transporters are also involved in in planta translocation of the nutrient. It is generally assumed that low-affinity Pi transporters are responsible for mobilization of Pi within the plant (Daram et al., 1999). However, expression of some of the high-affinity Pi transporters in plant parts other than roots, including stem, leaf, tuber, and flowers, points to their potential involvement in internal translocation of Pi (Leggewie et al., 1997; Daram et al., 1998; Liu et al., 1998a).
Ion uptake kinetic studies have shown that high-affinity Pi transport activity is inducible during Pi starvation, whereas the low-affinity transport remains constitutive (Furihata et al., 1992). This observation was further substantiated by the constitutive expression of the low-affinity Pi transporters in Arabidopsis (Daram et al., 1999). Furthermore, the transcript levels of high-affinity Pi transporters increased rapidly as a specific response to Pi starvation (Muchhal et al., 1996; Liu et al., 1998a). There is growing evidence supporting the transcriptional activation of high-affinity Pi transporters during Pi deficiency (Muchhal and Raghothama, 1999).
An effective approach to study transcriptional regulation is to monitor the activity of reporter genes driven by the specific gene promoter. Expression of reporter genes such as luciferase (LUC), green fluorescent protein (GFP), and β-glucuronidase (GUS) has been successfully used to understand transcriptional regulation in plants (Ow et al., 1986; Jefferson et al., 1987; Haseloff et al., 1997). Although GUS is an ideal reporter for histochemical analysis, the destructive nature of the staining procedure makes it unsuitable for sequential analysis of effects of multiple inducers. Perhaps the most useful aspect of using LUC as a reporter is its noninvasive assay procedure. This unique feature makes it possible to study the effects of multiple factors influencing gene expression by sequentially imposing different stimuli. However, both GUS and LUC reporter systems require the addition of substrates to detect their activity. In contrast, the detection of GFP is noninvasive and does not require any substrate. Illumination of GFP expressing plants with long-wave UV (395 nm) or blue light (475 nm) results in bright-green fluorescence (510 nm). To take advantage of the usefulness of each of these reporter systems, we have generated transgenic plants expressing reporter genes under the regulation of AtPT1 and AtPT2 promoters.
Plants exhibit an array of physiological changes, as well as morphological and architectural modifications in roots, in response to altered availability of phosphorus (Lynch, 1997). Some of these responses, particularly changes in root morphology and architecture, are very similar to those resulting from phytohormone treatments. Auxin and ethylene are known to have profound effects on root growth, root hair initiation, and elongation (Lynch and Brown, 1997). Ethylene is involved in the lateral root development, root hair initiation, and elongation (Lynch and Brown, 1997; Dolan, 2001; Ma et al., 2001). Interestingly, Pi deficiency that leads to similar morphological changes also enhances ethylene production in plants (Borch et al., 1999). It has been suggested that Pi starvation-induced changes in root morphology may involve both ethylene-dependent and -independent pathways (Schmidt, 2001). Application of auxin to Pi-sufficient white lupin (Lupinus albus) resulted in the formation of proteoid roots, a response commonly observed under Pi deficiency (Gilbert et al., 2000; Neumann et al., 2000). In addition, a decrease in the level of cytokinins during Pi deficiency has been correlated with altered root morphology (Kupier et al., 1988). Although these findings revealed the involvement of phytohormones in Pi deficiency responses, not much is known about their role in the Pi starvation-induced signaling pathways or in gene expression. Inhibitors of auxin transport and ethylene synthesis have been shown to influence the expression of genes during Pi deficiency in the proteoid roots of white lupin (Gilbert et al., 2000). A recent report showing the suppression of Pi starvation-induced gene expression by cytokinin is quite interesting (Martin et al., 2000). The cytokinin effects are presumed to be due to altered long distance signaling during Pi starvation.
In this study, we have carried out an extensive analysis of transcriptional regulation and tissue-specific expression of three reporter genes driven by the promoters of two high-affinity Pi transporters of Arabidopsis. The role of hormones such as auxin, ethylene, and cytokinin in the regulation of Pi starvation-induced gene expression is also examined. Results show that the transporters are transcriptionally regulated by Pi in a rapid and reversible manner. This study also showed that auxin and cytokinin specifically suppressed expression of the high-affinity Pi transporter AtPT1. This supports the notion that high-affinity Pi transporters are involved in nutrient remobilization during Pi starvation.
RESULTS
Generation of Transgenic Arabidopsis Plants
To understand the transcriptionally regulated tissue specific expression of Pi transporters in Arabidopsis, we have generated transgenic Arabidopsis plants expressing reporter genes under the regulation of the AtPT1 and AtPT2 promoters. About 25 independent transgenic lines from each construct (AtPT1-GUS/GFP, AtPT2-GUS, and AtPT2-LUC) were selected and evaluated for the expression of reporter genes in response to Pi starvation. One transgenic line from each category with representative expression was chosen for detailed analysis of expression. Because AtPT2 gene is specifically induced under Pi starvation, transgenic plants carrying the AtPT2 promoter were subjected to detailed analysis of reporter gene activity. This analysis provided evidence that the reporter genes are under the transcriptional regulation of Pi transporter promoters (Fig. 1).
Figure 1.
Pi starvation-induced expression of reporter genes. A, Transgenic Arabidopsis expressing the reporter genes GUS (1 and 2) and GFP (3 and 4) under the regulation of AtPT1 promoter are shown. 1 and 3, Expression of reporter genes under Pi sufficiency conditions. 2 and 4, Expression under Pi deficiency conditions. B, Transgenic Arabidopsis expressing the reporter gene GUS (1 and 2) or LUC (5 and 6) under the regulation of AtPT2 promoter are shown. 1 and 5, Absence of expression of GUS and LUC in plants grown under Pi sufficiency. Expression of GUS and LUC under Pi-starved conditions is shown in 2 and 6, respectively. 3 and 4, Pi-sufficient and -deficient plants used for monitoring LUC expression in roots.
Pi Deficiency Is the Underlying Factor in Gene Expression
One of the important features of high-affinity Pi transporters is their inducibility by low Pi concentrations (Muchhal et al., 1996). To better understand this response mechanism, a detailed analysis of the effect of different Pi concentrations was carried out. Seven-day-old seedlings of transgenic Arabidopsis lines grown in one-half-strength Murashige and Skoog medium were transferred to Murashige and Skoog medium supplemented with different concentrations of Pi (0–1.25 mm) for 5 d. LUC activity in plants (AtPT2-LUC) grown in the presence of 1.25 mm Pi was barely detectable (Fig. 2A). A dramatic increase in the reporter gene activity was noticed with decreasing Pi concentration, which reached nearly 85-fold at 25 μm. Further lowering of Pi concentration did not significantly alter the level of expression of the reporter gene (Fig. 2A). Analysis of plants expressing GUS under the regulation of AtPT2 promoter also showed a similar trend in the reporter gene activity (Fig. 2B). The GUS activity was quite obvious in transgenic plants carrying the AtPT1 promoter, even under Pi sufficiency conditions. Transferring these plants to Pi-deficient conditions increased the activity by approximately 2-fold (Fig. 3A).
Figure 2.
Effect of Pi concentration on gene expression. A, Seven-day-old seedlings of AtPT2-LUC plants were transferred to Murashige and Skoog media containing varying concentrations of Pi (0, 5, 10, 25, 50, 125, 250, 500, and 1,250 μm) for 5 d. Plants were harvested for measuring the LUC activity and RNA isolation. B, Seven-day-old seedlings of AtPT2-GUS plants were grown in the presence of different concentrations of Pi as indicated for 5 d and utilized for GUS reporter expression analysis. C, Northern analysis of total RNA from AtPT2-LUC plants supplemented with different concentrations of Pi. Ten micrograms of total RNA was electrophoretically separated on denaturing formaldehyde agarose gels and blotted on to nylon membranes. The membranes were probed with 32P-labeled AtPT1 and AtPT2 cDNAs. The ethidium bromide-stained gel picture shows uniform loading of RNA samples.
Figure 3.
Activity of AtPT1 promoter in response to Pi starvation. Seven-day-old AtPT1-GUS/GFP expressing plants were transferred to liquid Murashige and Skoog medium with (1,250 μm) and without (0 μm) Pi for 5 d. Soluble protein was extracted from the Pi-treated plants to determine GUS activity. The enzyme activity was measured using 4-methyumbelliferyl-β-d GlcUA as the substrate. Error bars show sd. B, Northern-blot analysis of total RNA isolated from Arabidopsis plants grown in the presence (P+) and absence (P−) of Pi for 5 d. The blot was probed with 32P-labeled AtPT1. The ethidium bromide-stained gel shows uniform loading and integrity of RNA.
Interestingly, the expression of the native genes (AtPT1 and AtPT2) parallels the expression and activity of the reporter genes. A strong correlation between decreasing Pi concentration and increasing transporter-specific message accumulation was observed (Fig. 2C). When the Pi in the medium was sufficient (1.25 mm), the expression of Pi transporters either remained at basal level (AtPT1) or was barely detectable (AtPT2). A significant increase in the abundance of these transcripts was observed at 500 μm Pi and it reached a maximum in plants grown in the medium lacking Pi. These results suggest that reporter gene activity could be used as an indicator of transcriptional activation of native gene expression under Pi starvation.
Transcriptional Activation of Gene Expression Is an Early and Reversible Response to Pi Availability
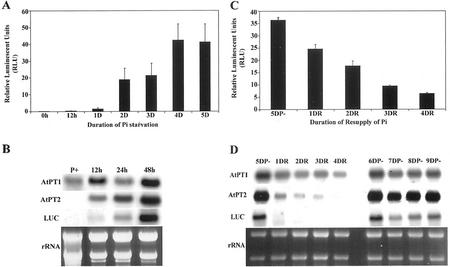
The inducible and repressible mode of Pi uptake based on internal Pi requirement is well documented in plants (Liu et al., 1998a; Muchhal and Raghothama, 1999). Accumulation of mRNA and appearance of Pi transporter protein within 12 to 24 h after removal of Pi have been observed in tomato (Lycopersicon esculentum; Liu et al., 1998a; Muchhal and Raghothama, 1999). In this study, the rapidity of gene induction and repression by Pi was examined. Seven-day-old seedlings (AtPT2-LUC) were transferred to Murashige and Skoog medium with and without Pi. Samples were collected at different time intervals after the initiation of treatment and assayed for the expression of LUC. A small but reproducible increase in LUC activity was observed as early as 12 h after transferring plants to Pi-deficient medium. A 60-fold increase in the LUC activity was recorded after 2 d of starvation (Fig. 4A). The activity reached the highest level of nearly 100-fold by 4 d of starvation and stabilized thereafter. Northern analysis of total RNA from these plants indicates the accumulation of LUC-specific mRNA as early as 12 h. The transcript levels increased with increasing duration of Pi starvation. This increase in the LUC transcripts appears to serve as a prelude to enzyme activity during Pi starvation (Fig. 4B).
Figure 4.
Temporal expression of Pi transporters. A, Rapid induction of AtPT2 promoter-driven expression of LUC during Pi deficiency was monitored in the transgenic plants. Seven-day-old seedlings of AtPT2-LUC plants were transferred to Murashige and Skoog medium without Pi. The seedlings were removed at different times after Pi starvation to measure reporter gene activity and for northern analysis of transcripts. B, Total RNA from the plants harvested at different times of Pi deficiency was separated on denaturing formaldehyde agarose gels and blotted onto nylon membranes. The membranes were probed with 32P-labeled cDNA fragments of AtPT1, AtPT2, and LUC. C, Reversibility of induction of genes was studied by resupplying Pi to Pi-starved plants. The LUC expressing plants (AtPT2-LUC) were starved for Pi for 5 d and then transferred to Murashige and Skoog medium containing sufficient Pi (1,250 μm). Other samples of seedlings were grown continuously under Pi deficiency. Plants were harvested at different time periods (12 h, and 1, 2, 3, and 4 d) after transfer, and analyzed for reporter gene activity and isolation of RNA. DR, Days after replenishment with Pi; DP, days plants continued to be grown in the absence of Pi. D, Northern blot showing the expression of AtPT1, AtPT2, and LUC in plants subjected to Pi replenishment experiments. The nylon membranes containing the RNA were probed with 32P-labeled cDNAs.
Expression of native Pi transporters, especially AtPT2, correlated with the expression of LUC and the enzyme activity. These results confirm a link between transcriptional activation of Pi transporters and Pi uptake during the nutrient starvation. Expression of reporter gene was not only inducible by Pi starvation but also repressible by replenishment of the nutrient (Fig. 4C). A 3-fold reduction in LUC activity was apparent within 24 h of Pi replenishment. The activity progressively decreased to one-sixth of the induced level within 4 d of Pi resupply, whereas the LUC activity remained at a high level in plants that continued to grow in Pi-deficient medium. Northern analysis of total RNA from these plants showed that repression of Pi transporter and the reporter gene is a rapid response to Pi supplementation. In the case of AtPT1, the reversal of induction was gradual, which is consistent with a relatively constitutive nature of its expression (Fig. 4D). There was a noticeable difference between the relatively rapid decrease in the AtPT2-LUC transcript abundance and the LUC enzyme activity in Pi-replenished plants.
Pi Transporters Are Expressed in Specific Tissues under Pi Starvation
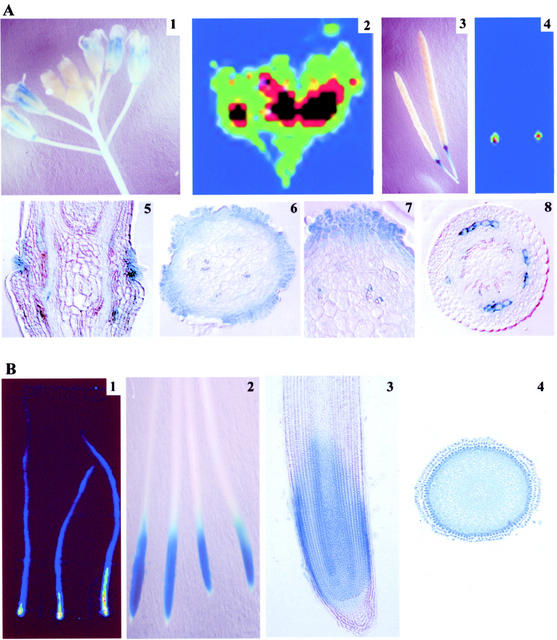
One of the effective ways to analyze tissue-specific expression of genes is by localizing the reporter gene activity in transgenic plants. There were distinct differences in the patterns of expression of reporter genes driven by the AtPT1 and AtPT2 promoters in roots of Pi-starved plants. AtPT1 promoter-driven reporter gene activity was lacking in root tips; in contrast, the AtPT2 promoter-driven reporter gene expression was observed in all cells of undifferentiated segments of the root, including the tip (Fig. 5A). Furthermore, in AtPT1 transgenic plants, GUS and GFP activity was quite high in the newly formed secondary root branches. In the case of AtPT2-GUS plants, the pattern of expression of the reporter gene remained the same irrespective of development of the secondary roots. The expression of GUS in the root hairs in both AtPT1-GUS and AtPT2-GUS plants was quite evident under Pi starvation (Fig. 5A). The root hairs and emerging secondary roots of AtPT1 transgenic plants showed highest levels of GFP expression. The histochemical analysis further confirmed the lack of expression of the reporter gene driven by AtPT1 promoter in root tips. The tissue sections also revealed that AtPT1 is strongly expressed in epidermal and endodermal layers. The GUS activity was detected in all the cell types in the meristamatic region of AtPT2-GUS roots. In the mature region, the activity of GUS was observed in the epidermis, cortex, and particularly in the stellar region (Fig. 5B). These studies show that different Pi transporters may have tissue specific role in acquiring and transferring Pi within the roots. A relatively strong expression of reporter genes in epidermal layers provides further evidence for the role of epidermally localized Pi transporters in Pi acquisition.
Figure 5.
Pi transporter promoter-mediated expression of reporter genes in Arabidopsis root. A, AtPT1 promoter-mediated expression of GUS (1–3) and GFP (4–6) in Arabidopsis roots and root hairs (3 and 6) are shown. Stronger GUS activity can be observed in newly formed branches, whereas the intensity of GUS staining decreased in the primary root (2). Lack of GUS expression in the tips of primary and lateral roots of AtPT1-GUS plants is clearly depicted (1 and 2). AtPT2 promoter-driven expression of GUS in all parts of the roots, including root tips and root hairs, is shown in 7 through 9. B, Expression of the reporter gene activity was examined in thin sections of roots of transgenic plants. The Pi-deficient plant roots expressing GUS were allowed to develop blue color in the presence of X-Gluc. The root segments were fixed and embedded in Technovit resin and 8-μm sections were cut and placed on a slide. The sections were photographed under a microscope (Olympus Corporation, Lake Success, NY). Top, GUS staining of the transverse sections of roots; bottom, cross-sectional view. 1 through 4, Expression of the AtPT1-GUS in the root tip (1 and 3) and the differentiated region of a root (2 and 4). Similarly, the AtPT2-GUS expression can be seen in the root tip (5 and 7) and the differentiated region (6 and 8).
The High-Affinity Transporters May Play a Role in Internal Pi Mobilization
High-affinity Pi transporters have been shown to express preferentially in roots during Pi starvation. However, reporter gene expression studies revealed a very interesting pattern of gene expression in mature plants subjected to Pi starvation. In this analysis, 1-month-old transgenic plants with young flowers and fruits, expressing LUC or GUS under the control of the AtPT2 promoter, were transferred to a hydroponic nutrient solution with or without Pi for 2 weeks. Under Pi deficiency conditions, a strong expression of LUC was observed in flowers and at the junction between silique and peduncle (Fig. 6A). In addition, some of the senescing leaves of Pi-starved plants also showed the expression of AtPT2 promoter-driven reporter genes (data not shown). These results suggest a possible role for AtPT2 in the mobilization Pi during the nutrient starvation.
Figure 6.
A, AtPT2 promoter-mediated expression of reporter genes in flowers and fruits of Pi-starved Arabidopsis. Expression of GUS (1 and 3) and LUC (2 and 4) in flowers (1 and 2) and fruits (3 and 4) was examined in Pi-starved plants. The reporter gene expression is confined to the silique and stalk junctions in fruits (3 and 4). Transverse (5) and cross-sectional (6 and 7) views of the silique and stalk junction clearly show the GUS expression. Strong expression of reporter gene is observed in outer cell layers (reminiscent of nectaries) and some cells in tracheary elements and vascular tissues. Cross-sectional view of the fruit stalk (8) showing the expression of GUS in some cells of cortical region. B, Heterologus expression of reporter genes in tobacco (Nicotiana tabacum). Expression of LUC (1) and GUS (2) under the regulation of AtPT2 promoter in tobacco roots. Longitudinal (3) and cross-sectional (4) view of the root showing the GUS expression.
AtPT2 Promoter Drives the Expression of Reporter Genes in a Heterologous System
Transgenic tobacco plants expressing LUC or GUS under the regulation of the Arabidopsis Pi transporter promoter (AtPT2) were generated by Agrobacterium tumefaciens-mediated leaf disc transformation method. A number of independent transgenic lines were examined for the reporter gene activity. In all the lines examined, reporter gene expression was induced only under Pi starvation and no detectable activity was observed in the presence of Pi. Expression of reporter genes was quite pronounced in the region behind the root cap (Fig. 6B). Similarly, the tips of newly emerging secondary roots of Pi-starved plants exhibited strong reporter gene activity. In addition, GUS expression was also observed in root hairs of Pi-starved tobacco (data not shown). These data indicate that the conserved cis-elements present in the AtPT2 promoter are sufficient to drive the Pi starvation-mediated expression of reporter genes in a heterologous plant.
Auxins and Cytokinins Suppress Expression of the AtPT1 Gene
Because many morphological responses of plants to phytohormones resemble those of Pi deficiency, we examined the effect of auxin, ethylene, or cytokinin on Pi starvation-induced gene expression. Transgenic plants expressing reporter genes under the regulation of AtPT1 (AtPT1-GUS/GFP) and AtPT2 (AtPT2-GUS) promoters were used in these studies. GUS activity under regulation of the AtPT1 promoter was suppressed by auxin and cytokinins (Fig. 7A). Suppression due to auxin or cytokinin was obvious both under Pi sufficiency and deficiency conditions. Increasing differences in the reporter gene activity were observed with increasing concentrations of Ki. Interestingly, there were no significant differences in the reporter gene activity in AtPT2-GUS plants either in the presence or absence of hormones (Fig. 7B). The suppression of reporter gene activity by hormones paralleled the suppression of AtPT1 expression (Fig. 7C). In contrast, expression of AtPT2 and other Pi starvation-induced genes such as RNase2, At4, purple acid phosphatase, and a constitutively expressed tubulin were comparable in hormone-treated and -untreated plants. Furthermore, there was no major difference in the transcript levels of Pi starvation-induced genes, including the transporters in ACC-treated plants. Some of the differences in gene expression observed in this study and an earlier study by Martin et al. (2000) may be due to differences in culture conditions and duration of hormone treatments.
Figure 7.
Effect of hormones or hormone inhibitors on GUS activity and expression of Pi starvation-responsive genes. Seven-day-old seedlings of AtPT1-GUS/GFP and AtPT2-GUS were transferred to Murashige and Skoog medium with (P+) and without (P−) Pi supplemented with 2,4-dichlorophenoxyacetic acid (2,4-D; 0.5 μm), naphthalene acetic acid (0.5 μm), kinetin (Ki; 0.1, 1.0 and 10 μm), 6-benzyleaminopurine (1.0 μm), 1-aminocyclopropane-1-carboxylic acid (ACC; 50 μm), 2,3,5-triiodobenzoic acid (1.0 μm), naphthalmic acid (1.0 μm), or α-2-aminoethoxyvinyl Gly (AVG; 10 μm). Seedlings were harvested after 48 h of treatment for enzyme assay and RNA isolation. Effect of hormones and inhibitors of hormones on AtPT1 (A and D) and AtPT2 (B and E) promoter-driven expression of GUS under Pi-sufficient (white bars) and -deficient (black bars) conditions. Error bars represent sd. Northern analysis of total RNA from AtPT2-GUS plants treated with different hormones and hormone inhibitors (C and F). Fifteen micrograms of total RNA was transferred to nylon membranes. The membranes were hybridized with 32P-labeled AtPT1, AtPT2, At4, PAP, RNase2, and tubulin cDNA fragments.
The effect of auxin and ethylene on gene expression was further analyzed by treating plants with auxin transport and ethylene biosynthesis inhibitors. The reporter gene activity of plants treated with inhibitors was similar to that of control plants (Fig. 7, D and E). There was no obvious change in the transcript levels of Pi starvation-induced genes in the inhibitor-treated or -untreated plants (Fig. 7F). These studies point to the complex nature of hormone interaction during Pi starvation and highlight the need for further experimentation.
DISCUSSION
Pi acquisition is a highly regulated process in plants, and the ability of plants to acquire the nutrient increases during Pi starvation (Drew and Saker, 1984; Furihata et al., 1992; Shimogawara and Usuda, 1995). Interestingly, kinetic analysis of nutrient uptake pointed to an increase in the Vmax of the high-affinity Pi transporters without much change in the Km. Based on these results, it was concluded that during Pi starvation the number of high-affinity Pi transporters increases, whereas the low-affinity Pi transporter's activity remains unchanged (Furihata et al., 1992). A clear link between increased transcription, translation, and assembly of Pi transporters into the plasma membrane during Pi starvation was obtained using gene-specific probes and antibodies for the transporter (Muchhal and Raghothama, 1999). In this study, we report an extensive analysis of transcriptional regulation, and tissue-/organ-specific expression of transporter genes under Pi deficiency. The data obtained with three different reporter genes under the regulation of two high-affinity Pi transporter gene promoters provide compelling evidence for transcriptional regulation of Pi uptake in plants. These promoters represent two highly expressed Pi transporters (AtPT1 and AtPT2) of Arabidopsis.
The similarity between the expression of reporter genes and native genes suggests that the promoters used in these studies are likely to contain all the conserved elements. The sequences responsible for root preferential expression and Pi starvation responsiveness appear to be located in the promoters used to drive the expression of these reporter genes. At present, the specific DNA regions associated with these responses are not completely elucidated. A conserved MYB transcription factor (PHR1) binding sequence has been identified in many of the Pi starvation-induced genes (Rubio et al., 2001). The PHR1 protein interacts with the conserved cis-element present in Pi starvation-induced AtIPS1 gene of Arabidopsis. Interestingly, mutation in PHR1 affects several Pi starvation-induced responses of Arabidopsis (Rubio et al., 2001). DNA-protein interaction studies have shown that two regions in the AtPT2 promoter specifically interact with nuclear protein factors that are present under Pi sufficiency but either disappear or are modified during Pi deficiency (Mukatira et al., 2001). It is likely that interaction of trans-acting factors with cis-elements located on the promoters is responsible at least in part for root preferential and Pi deficiency-induced gene expression.
The reporter gene activity increased as a direct consequence of changes in Pi concentration in the medium. This response is reminiscent of earlier Pi uptake studies carried out in plants that were subjected to Pi starvation (Clarkson et al., 1978; Drew and Saker, 1984). Expression of the AtPT1-reporter gene, which was obvious even under Pi sufficiency condition, increased by 2-fold during Pi deficiency. In contrast, the AtPT2 promoter-driven reporter gene activity was observed only under Pi deficiency conditions. These findings point to the existence of two types of high-affinity Pi transport systems operating under low Pi conditions, one that responds rapidly and specifically to Pi deficiency, and the expression of the other increases during the deficiency. The differential expression of two Pi transporters is a reflection of the ability of plants to adjust uptake rates based on internal Pi demand. Furthermore, induction of the reporter gene activity was not only rapid but also reversible upon resupply of the nutrient to Pi-deficient plants. The reversible induction of transporters responsible for regulated uptake of nutrients is crucial for maintaining cellular ion homeostasis. This is particularly relevant for Pi, where excess uptake results in imbalance of ions such as Zn and leads to physiological disorders (Clarkson and Scattergood, 1982).
One distinct advantage of using reporter genes is the relative ease of analyzing tissue- and organ-specific expression of genes. Both promoters activated the expression of reporters primarily in roots. Expression of the AtPT2 promoter-driven reporter gene was observed all along the roots, confirming the notion that the entire root system retains the ability to acquire Pi during the nutrient deficiency (Clarkson et al., 1978; Muchhal and Raghothama, 1999). The newly emerging secondary roots had noticeably higher levels of AtPT1 promoter-driven reporter gene activity. Both AtPT1 and AtPT2 promoters strongly activated reporter gene expression in root hairs. The elongating root hairs are considered to be an important component of Pi acquisition. Because of their large surface area and ability to access narrow pores in soil, they are geometrically placed in a better position to obtain Pi. It has been shown that nearly 63% of Pi may be acquired by root hairs and their role becomes more important under Pi deficiency conditions (Gahoonia and Neilsen, 1998). The data presented here provide molecular evidence that under Pi deficiency, root hairs not only increase surface area but also have enhanced Pi uptake ability. The integration of changes in root morphology, biochemistry, and gene expression under Pi deficiency has been studied more extensively in the proteoid roots of white lupin (Tadano and Sakai, 1991; Johnson et al., 1996). Under Pi deficiency, these roots increase surface area and act as organs of organic acid biosynthesis and secretion. Furthermore, they also serve as sites for the production of phosphatases and exhibit enhanced Pi uptake. Taken together, these studies suggest that roots and root hairs produced in response to Pi starvation are morphologically and biochemically distinct from those of Pi-sufficient plants.
Histochemical analysis of sections of roots further confirmed the distinct patterns of expression of AtPT1 and AtPT2. Although AtPT2-GUS activity was observed in all tissue types in the undifferentiated root tip and elongation zone, strong activity was evident in the vascular tissues of differentiated region. This indicated that high-affinity Pi transporters are not only involved in acquiring Pi from soil but they may also be involved in its transfer to the vascular tissues. In contrast, the AtPT1-GUS activity was clearly absent in the root tips and root caps. Another distinguishing feature is that the level of AtPT1-GUS expression decreased in main branches upon formation of secondary roots. Higher expression of reporter genes in newly emerging roots, root hairs, and epidermis resemble that of in situ transcript localization of Pi transporter in tomato (Daram et al., 1998; Liu et al., 1998).
It is generally assumed that high-affinity Pi transporters are involved in Pi uptake by roots, whereas low-affinity transporters regulate movement of Pi within the plant. This assumption was supported by the preferential expression of a number of cloned high-affinity Pi transporters in roots (Muchhal et al., 1996; Smith et al., 1997; Liu et al., 1998a, 1998b). However, analysis of reporter gene activity driven by the AtPT2 promoter in Pi-starved mature plants revealed the expression of reporter genes in floral organs. Interestingly, the majority of reporter gene activity was found in the thalamus region of the flower and at the junction of the silique and fruit stalk. Based on these patterns of gene expression, it is tempting to suggest that AtPT2 may be involved in loading of Pi to reproductive organs during Pi deficiency. The reporter gene expression studies also revealed that the AtPT1 promoter specified a different expression pattern compared with the AtPT2 promoter. It will be interesting to analyze the tissue-specific expression and Pi starvation responsiveness of the other seven members of the high-affinity Pi transporter family in Arabidopsis. Expression of reporter genes in roots of Pi-deficient tobacco points to the conservation of the components of Pi starvation-induced signal transduction pathway and response mechanisms across plant species. Because plants are constantly interacting with Pi-limited ecosystems, they may have developed highly conserved Pi starvation sensing (monitoring) and signaling mechanisms.
Despite extensive information on gene expression during Pi deficiency, little is known about signal transduction during Pi starvation. Similarities between Pi starvation-induced responses and that of auxin and ethylene has led to the hypothesis that these hormones play a role in Pi starvation (Bates and Lynch, 1996; Lynch and Brown, 1997; Gilbert et al., 2000). In addition, cytokinins also have been shown to be involved in suppression of Pi starvation-induced gene expression (Martin et al., 2000). In this study, auxin, particularly 2,4-D, and cytokinin suppressed the activity of a reporter gene driven by the AtPT1 promoter. In addition, hormones also suppressed accumulation of AtPT1 transcripts. This effect appears to be specific to AtPT1 expression because there was no marked difference in the induction of other Pi starvation-associated genes or the reporter gene activity driven by the AtPT2 promoter. Similarly, ethylene precursor ACC or its biosynthesis inhibitor AVG had no obvious effect on Pi starvation-induced gene expression. These results suggest that some components of the Pi starvation response may be directly affected by auxin and cytokinin and this response may not involve ethylene.
There is growing evidence supporting the differential effect of hormones on Pi starvation-induced responses (Gilbert et al., 2000; Martin et al., 2000). This has been clearly demonstrated in the proteoid roots of white lupin, wherein auxin transport inhibitors decreased the levels of Pi starvation-induced phosphoenolpyruvate carboxylase and malate dehydrogenase, but not the secretion of acid phosphatase (Gilbert et al., 2000). Similarly, cytokinins have been shown to suppress the expression of some Pi starvation-induced genes, including AtPT1, but not the modifications of root growth (Martin et al., 2000). These observations support the notion that both hormone signaling-dependent and -independent pathways are involved in Pi starvation response. The tissue-specific expression of reporter genes in newly emerging secondary roots and root hairs under Pi starvation point to a fine coordination of hormone action and Pi starvation-induced gene expression.
The data presented here provide strong evidence for the transcriptional regulation of high-affinity Pi transporters and their potential involvement in internal Pi mobilization. Different members of the high-affinity Pi transporter family are likely to be involved in Pi uptake and mobilization in the plant, along with the low-affinity transporters. These studies also show that morphological and physiological changes occurring during Pi starvation are highly coordinated with the molecular changes. The phytohormones may play a dual role in Pi starvation response by enhancing root biomass and specifically altering the expression of genes.
MATERIALS AND METHODS
Generation of Expression Vectors
A 2.1-kb 5′-flanking sequence of the AtPT1 gene representing the promoter region was PCR amplified from Arabidopsis genomic DNA using specific 5′ (CGGGATCCGATGGATGAGTCTACGTAC) and 3′ (GGGAAGCTTCCTAGAGCTCTATAATC) primers. The amplified fragment was cloned into pGEM5 vector (Promega, Madison, WI) and the authenticity of the PCR product was confirmed by restriction enzyme digestion and DNA sequencing. The promoter fragment was released from pGEM5 by restriction enzyme digestion and transcriptionally fused to GUS/GFP reporter genes in the binary vector pCAMBIA1303 (CAMBIA, Canberra, Australia). Similarly, the AtPT2 promoter (2.3 kb) was also amplified from genomic DNA using the specific 5′ (TCCCATTCGTGAATGAAAAC) and 3′ (TCTTCTTCTCCTCTGC) primers and cloned into pBSIIKS+. The amplified product contains a 1.1-kb intron located in the untranslated 5′ region of the gene. The promoter fragment was released from the pBSIIKS+ and cloned into binary vectors pLPTV-BAR and pGPTV-BAR (Becker et al., 1992), resulting in the transcriptional fusion of the promoter to the reporter genes firefly LUC and GUS, respectively. The binary vector pLPTV-BAR was constructed by replacing the HindIII-SacI fragment of pGPTV-BAR representing the coding sequence of GUS with an HindIII-SacI fragment harboring the firefly LUC coding sequence from RD29A-LUC (Ishitani et al., 1997). The binary vectors were transferred to the Agrobacterium tumefaciens (PGV3101) and used for transformation.
Plant Material and Transformation
Transgenic Arabidopsis (ecotype Columbia) plants expressing reporter genes were generated by the A. tumefaciens floral dip method as described by Clough and Bent (1998). Seeds of transformed plants (T0) were germinated in the Scotts Metro-mix-360 with coir medium (Scotts-Sierra Company, Marysville, OH) or petri plates with sterile agar media to select transgenic plants resistant to herbicide or antibiotics. Homozygous transgenic plants expressing reporter gene were selected in the following generations. These plants were used in all the molecular and biochemical analysis of Pi starvation response. One-month-old transgenic seedlings were also examined for the expression of reporter genes in different parts of the plant. The seedlings grown in the Scotts Metro-mix-360 were removed and roots were carefully washed free of the medium and transferred to modified Hoagland nutrient solution containing 250 μm Pi or no Pi (Liu et al., 1998a). After 2 weeks of the transfer, different parts of plants were analyzed for the reporter gene expression. Transgenic tobacco (Nicotiana tabacum) plants were generated by A. tumefaciens-mediated leaf disc transformation method. Seeds from transgenic tobacco plants were germinated on the Scotts Metro-mix-360 and seedlings were selected for herbicide resistance. Four-week-old seedlings were transferred to hydroponics solutions containing 250 μm or no Pi for 2 weeks before harvesting for gene expression analysis.
Liquid Culture of Arabidopsis Seedlings
Seeds from homozygous Arabidopsis plants expressing reporter genes were treated for a minute in 70% (v/v) ethanol and rinsed twice with sterile water. The seeds were then treated in 50% (v/v) commercial bleach with 0.1% (v/v) Tween 20 for 10 min and rinsed thoroughly with sterile water to remove the residual bleach. Surface sterilized seeds were subject to stratification at 4°C for 2 d before dispensing in conical flasks containing sterile Murashige and Skoog (Murashige and Skoog, 1962) liquid medium. The seedlings were grown under a 16-h-light/8-h-dark cycle at 25°C with constant shaking (85 rpm). Seedlings grown in liquid culture were used for all biochemical and molecular analysis unless otherwise indicated. All the experiments were repeated for a minimum of three times with at least three replications.
Analysis of Temporal- and Pi Concentration-Mediated Expression of Genes
Transgenic Arabidopsis plants expressing GUS or LUC under the regulation of AtPT1 and AtPT2 were used in this study. Seven-day-old seedlings grown in one-half-strength Murashige and Skoog medium were rinsed once with sterile distilled water followed by a rinse with sterile Murashige and Skoog medium without Pi and transferred to sterile Murashige and Skoog medium supplemented with different concentrations of Pi (0, 5, 10, 25, 50, 125, 250, 500, and 1,250 μm). The plants were harvested after 5 d and used for gene expression analysis. To study temporal induction of Pi transporters and the reporter gene in response to Pi starvation, 7-d-old transgenic seedlings (AtPT2-LUC) grown in liquid medium were rinsed once with sterile water, followed by a rinse with sterile Murashige and Skoog medium deficient in Pi, and transferred to fresh medium without Pi. Plants were collected at different time periods (12, 24, 48 h, 3, 4, and 5 d) after the transfer and used for isolating total RNA and measuring the reporter gene expression.
Evaluation of the Effect of Hormones on Gene Expression
Seven-day-old transgenic seedlings (AtPT1-GUS/GFP and AtPT2-GUS) grown in one-half-strength sterile Murashige and Skoog liquid medium were rinsed once with sterile distilled water followed by P+ or P− Murashige and Skoog medium and transferred to P+ or P− Murashige and Skoog medium supplemented with auxins 2,4-D or naphthalene acetic acid, the cytokinins Ki or 6-benzyleaminopurine, and the ethylene biosynthesis precursor ACC. Similarly, seedlings were also treated with auxin transport inhibitors 2,3,5-triiodobenzoic acid or naphthalmic acid and the ethylene biosynthesis inhibitor AVG. After 48 h of treatment, seedlings were collected for GUS analysis and RNA isolation.
RNA Isolation and Northern Hybridization Analysis
Total RNA was isolated by hot phenol and lithium chloride precipitation method (Pawlowski et al., 1994). Ten micrograms of total RNA was electrophoretically separated in a denaturing formaldehyde agarose gel and blotted onto nylon membranes. The nylon filters were hybridized overnight with 32P-labeled DNA probes in a hybridization solution containing 50% (v/v) formamide, 5× Denhardt's solution, 0.1% (w/v) SDS, 6× SSPE, and 100 μg mL−1 denatured salmon sperm DNA at 42°C. The membranes were washed three times with 2× SSC/2% (v/v) SDS for 15 min each at 55°C before autoradiography.
LUC Imaging
Imaging of LUC activity in transgenic Arabidopsis and tobacco plants was done with a high-performance CCD camera (Princeton Instruments, Trenton, NJ). Seedlings or the parts of the plants to be imaged were sprayed uniformly with 100 mm luciferin (dissolved in 0.1% Triton X-100) and kept in the dark for 5 min. Then the seedlings were transferred to a dark chamber equipped with a CCD camera and the images were acquired for 5 min and analyzed with the software “Winview” (Princeton Instruments).
Quantitative Analysis of LUC Activity
LUC activity was recorded with a luminometer TD-20/20 (Turner Designs, Sunnyvale, CA) following the manufacturer's instructions. The tissue was ground to a fine power in liquid nitrogen and about 200 mg of the powder was transferred to a microfuge tube containing 500 μL of LUC extraction buffer (100 mm K2HPO4, pH 7.8; 1 mm EDTA; 10 mm dithiothreitol; and 0.25% [v/v] glycerol). The contents were mixed thoroughly on a vortex mixer and centrifuged at 15,000 rpm for 5 min at 4°C. Supernatant was transferred to a fresh tube and used as a source of the enzyme. The enzyme activity was determined within 2 h of extraction. The reaction mixture consisted of 100 μL of LUC assay buffer (50 mm HEPES; 20 mm MgCl2; 10 mm ATP in 0.2 m potassium Pi buffer, pH 7.0; and 0.05% [w/v] bovine serum albumin) and 50 μL of enzyme extract. The assay buffer and enzyme extracts were mixed in a polypropylene cuvette and placed inside the luminometer chamber. The reaction was initiated by injecting 50 μL of 1 mm luciferin (Promega). The instrument automatically measures and computes the light produced by the luciferin-LUC reaction. The luminescence values were expressed as relative luminescence units per milligram total protein. Total protein in the crude enzyme extract was determined according to Bradford (1976).
Quantitative Analysis of GUS Activity
Fluorometric quantification of GUS activity was done as described by Jefferson and Wilson (1991). About 200 mg of plant tissue powder was transferred to a microfuge tube containing 500 μL of GUS extraction buffer (50 mm NaHPO4, pH 7.0; 10 mm β-mercaptoethanol; 10 mm EDTA; 0.1% [v/v] Triton X-100; and 0.1% [w/v] sodium lauryl sarcosyl) and thoroughly mixed. The contents were centrifuged (15,000 rpm) for 5 min at 4°C. The supernatant was transferred to a fresh tube and used as the source of enzyme. The reactions were started by mixing 450 μL of prewarmed (37°C) GUS assay buffer (2 mm 4-methyumbelliferyl-β-d GlcUA in GUS extraction buffer) with 50 μL of enzyme extract in a microfuge tube and incubating at 37°C for 30 min. The reaction was terminated by transferring 100 μL of the reaction mix to 900 μL of 2% (w/v) Na2CO3. The amount of fluorescent product 4-methylumbelliferyl produced in the reaction was determined using a fluorometer. The GUS activity was expressed as pmoles of 4-methylumbelliferyl formed mg total protein−1 min−1.
Histochemical Localization of GUS Expression
Histochemical staining for GUS activity was done according to the protocol described by Raghothama et al. (1997), with slight modifications. Whole seedlings or parts of plants to be stained were incubated in GUS reaction mix (25 mg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 50 mL of 100 mm sodium Pi buffer with 0.1% [v/v] Triton X-100) for 6 to 17h. The stained seedlings were transferred to 70% (v/v) alcohol to remove chlorophyll. Microscopic analysis and photographing of GUS expression in seedlings and organs such as flowers and fruits was done with a dissecting scope. Representative samples of GUS stained tissues were taken for histochemical analysis of the reporter gene expression. The tissue fixation and subsequent processing for the sectioning were essentially the same as that described by Takechi et al. (1999). Stained tissues were vacuum infiltrated with 4% (v/v) formaldehyde in 50 mm potassium Pi buffer for 20 min, followed by an overnight incubation at 4°C in the same medium with gentle shaking. The fixed tissues were thoroughly washed with Pi buffer and dehydrated in alcohol series before embedding in Tecnovit resin (Energy Beam Science, Inc., Agawam, MA). Tissue sections (8 μ) were photographed under a microscope utilizing the differential interference contrast.
Analysis of GFP Expression
Expression of GFP was visualized with the Sterio Fluorescent system (Lecia, McHenry, IL) using the standard fluorescein isothiocyanate filter. The optimal exposure time for capturing the images was 30 s. The light source was provided by an HBO-50W high-pressure mercury lamp. The maximum excitation and emission wavelengths for the GFP2 and GFP3 filter set were 510 nm (range of 480–440 nm) and 525/550 nm (range of 470–440 nm), respectively. Images were automatically transferred to a computer and analyzed using the SPOT version 3.0.4 software (Apple Event 3.0, Sterling Heights, MI).
ACKNOWLEDGMENTS
We sincerely appreciate the technical advice and help of Dr. Mary Alice Webb in preparing microscopy pictures. We thank Drs. Peter B. Goldsbrough, Angus Murphy, and Cary Mitchell for critical review of the manuscript.
Footnotes
This work was supported by the U.S. Department of Agriculture (grant no. 00–35100–9370 to K.G.R.). This is journal paper no. 16,860 of the Purdue University Agricultural Research Program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.020007.
LITERATURE CITED
- Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19:529–538. [Google Scholar]
- Becker D, Kemper E, Shell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Borch K, Bouma TJ, Lynch JP, Brown KM. Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 1999;22:425–431. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Scattergood CB. Growth and phosphate-transport in barley and tomato plants during the development of, and recovery from, phosphate-stress. J Exp Bot. 1982;33:865–875. [Google Scholar]
- Clarkson DT, Serson J, Scattergood CB. Influence of phosphate-stress on phosphate absorption and translocation by various parts of root-system of Hordeum vulgare L (barley) Planta. 1978;139:47–53. doi: 10.1007/BF00390809. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Daram P, Brunner S, Persson BL, Amrhein N, Bucher M. Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta. 1998;206:225–233. doi: 10.1007/s004250050394. [DOI] [PubMed] [Google Scholar]
- Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M. Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell. 1999;11:2153–2166. doi: 10.1105/tpc.11.11.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L. The role of ethylene in root hair growth in Arabidopsis. J Plant Nutr Soil Sci. 2001;164:141–145. [Google Scholar]
- Drew MC, Saker LR. Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: evidence of non-allosteric regulation. Planta. 1984;160:500–507. doi: 10.1007/BF00411137. [DOI] [PubMed] [Google Scholar]
- Furihata T, Suzuki M, Sakurai H. Kinetic characterization of 2 phosphate-uptake systems with different affinities in suspension-cultured Catharanthus roseus protoplasts. Plant Cell Physiol. 1992;33:1151–1157. [Google Scholar]
- Gahoonia TS, Neilsen NE. Direct evidence on participation of root hairs in phosphorus (32P) uptake from soil. Plant Soil. 1998;198:147–152. [Google Scholar]
- Gilbert GA, Knight JD, Vance CP, Allan DL. Proteoid root development of phosphorus deficient lupin is mimicked by auxin and phosphonate. Ann Bot. 2000;85:921–928. [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong LM, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. Gus fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RJ, Wilson KJ. The GUS gene fusion system. In: Gelvin SB, Schilperoort RA, Verma DPS, editors. Plant Molecular Biology Manual. Boston: Kluwer Academic Publishers; 1991. pp. 1–33. [Google Scholar]
- Johnson JF, Vance CP, Allan DL. Phosphorus deficiency in Lupinus albus. Altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase. Plant Physiol. 1996;112:31–41. doi: 10.1104/pp.112.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupier D, Schuit J, Kupier PJC. Effect of internal and external cytokinin concentrations on root growth and shoot to root ratio of Plantago major spp pleiosperma at different nutrient condition. Plant Soil. 1988;111:231–236. [Google Scholar]
- Leggewie G, Willmitzer L, Riesmeier JW. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998a;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Trieu AT, Blaylock LA, Harrison MJ. Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant-Microbe Interact. 1998b;11:14–22. doi: 10.1094/MPMI.1998.11.1.14. [DOI] [PubMed] [Google Scholar]
- Lynch JP. Root architecture and phosphorus acquisition efficiency in common bean. In: Flores HE, Lynch JP, Eissenstat D, editors. Radical Biology: Advances and Perspectives on the Function of Plant Roots. Rockville, MD: American Society of Plant Physiology; 1997. pp. 81–92. [Google Scholar]
- Lynch J, Brown KM. Ethylene and plant responses to nutritional stress. Physiol Plant. 1997;100:613–619. [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ. 2001;24:459–467. [Google Scholar]
- Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Pena A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- McPharlin IR, Bieleski RL. Phosphate-uptake by Spirodela and Lemna during early phosphorus deficiency. Aust J Plant Physiol. 1987;14:561–572. [Google Scholar]
- Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D. Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc Natl Acad Sci USA. 1997;94:7098–7102. doi: 10.1073/pnas.94.13.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Raghothama KG. Transcriptional regulation of plant phosphate transporters. Proc Natl Acad Sci USA. 1999;96:5868–5872. doi: 10.1073/pnas.96.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukatira UT, Liu C, Varadarajan DK, Raghothama KG. Negative regulation of phosphate starvation-induced genes. Plant Physiol. 2001;127:1854–1862. [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Neumann G, Massonneau A, Langlade N, Dinkelaker C, Romheld V, Martinoia E. Physiological aspects of cluster root function and development in phosphorus-deficient white lupin (Lupinus albus L.) Ann Bot. 2000;85:909–919. [Google Scholar]
- Ow DW, Wood KV, Deluca M, Dewet JR, Helinski DR, Howell SH. Transient and stable expression of the firefly luciferase gene in plant-cells and transgenic plants. Science. 1986;234:856–859. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Kunze R, De Vries S, Bisseling T. Isolation of total, poly (A) and polysomal RNA from plant tissues. In: Gelvin SB, Shilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–13. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate transport and signaling. Curr Opin Plant Biol. 2000a;3:182–187. [PubMed] [Google Scholar]
- Raghothama KG. Phosphorus acquisition: plant in the driver's seat! Trends Plant Sci. 2000b;5:412–413. doi: 10.1016/s1360-1385(00)01746-5. [DOI] [PubMed] [Google Scholar]
- Raghothama KG, Maggio A, Narasimhan ML, Kononowicz AK, Wang GL, Durzo MP, Hasegawa PM, Bressan RA. Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol Biol. 1997;34:393–402. doi: 10.1023/a:1005812217945. [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W. From faith to fate: ethylene signaling in morphogenic responses to P and Fe deficiency. J Plant Nutr Soil Sci. 2001;164:147–154. [Google Scholar]
- Shimogawara K, Usuda H. Uptake of inorganic-phosphate by suspension-cultured tobacco cells-kinetics and regulation by Pi starvation. Plant Cell Physiol. 1995;36:341–351. [Google Scholar]
- Smith FW, Ealing PM, Dong B, Delhaize E. The cloning of two Arabidopsis genes belonging to a phosphate transporter family. Plant J. 1997;11:83–92. doi: 10.1046/j.1365-313x.1997.11010083.x. [DOI] [PubMed] [Google Scholar]
- Tadano T, Sakai H. Secretion of acid-phosphatase by the roots of several crop species under phosphorus-deficient conditions. Soil Sci Plant Nutr. 1991;37:129–140. [Google Scholar]
- Takechi K, Sakamoto W, Katsuhara M, Murata M, Motoyoshi F. In situ RNA hybridization using Technovit resin in Arabidopsis thaliana. Plant Mol Biol Rep. 1999;17:43–51. [Google Scholar]