Abstract
Multiple alignments of genome sequences are helpful guides to functional analysis, but predicting cis-regulatory modules (CRMs) accurately from such alignments remains an elusive goal. We predict CRMs for mammalian genes expressed in red blood cells by combining two properties gleaned from aligned, noncoding genome sequences: a positive regulatory potential (RP) score, which detects similarity to patterns in alignments distinctive for regulatory regions, and conservation of a binding site motif for the essential erythroid transcription factor GATA-1. Within eight target loci, we tested 75 noncoding segments by reporter gene assays in transiently transfected human K562 cells and/or after site-directed integration into murine erythroleukemia cells. Segments with a high RP score and a conserved exact match to the binding site consensus are validated at a good rate (50%–100%, with rates increasing at higher RP), whereas segments with lower RP scores or nonconsensus binding motifs tend to be inactive. Active DNA segments were shown to be occupied by GATA-1 protein by chromatin immunoprecipitation, whereas sites predicted to be inactive were not occupied. We verify four previously known erythroid CRMs and identify 28 novel ones. Thus, high RP in combination with another feature of a CRM, such as a conserved transcription factor binding site, is a good predictor of functional CRMs. Genome-wide predictions based on RP and a large set of well-defined transcription factor binding sites are available through servers at http://www.bx.psu.edu/.
Comprehensive discovery of functional DNA sequences in genomes requires both computational and experimental approaches (Collins et al. 2003). A particularly difficult challenge is identifying the cis-acting sequences, called cis-regulatory modules (CRMs), that are responsible for determining the amount, timing, and tissue specificity of gene expression. Unlike the situation for protein-coding genes, systematic rules for encoding CRMs in genomic DNA are not yet elucidated (Wasserman and Sandelin 2004), although various predictive methods are being explored. Methods that seek overrepresented motifs in co-expressed genes have limited but improving success (Tompa et al. 2005); however, most of these methods are not applicable to large genomic intervals. Consensus binding sites have been deduced for many transcription factors and are stored as positional weight matrices in databases such as TRANSFAC (Wingender et al. 2001) and JASPAR (Sandelin et al. 2004). Matches to the positional weight matrices in single DNA sequences far exceed the sites verified as being occupied by transcription factors (e.g., Grass et al. 2003). However, the number of predicted binding sites can be reduced substantially with increased specificity by requiring the matches to be conserved in multiple species (Berman et al. 2004; Gibbs et al. 2004).
DNA segments that appear to be under evolutionary constraint are good candidates for functional elements. This predictive method relies on the assumption that sequences carrying out similar functions in two related species are constrained to maintain a level of sequence similarity in excess of that seen for nonfunctional, or neutral, DNA (Pennacchio and Rubin 2001; Miller et al. 2004). Indeed, most DNA sequences known to be functional, such as exons and CRMs, align among human, mouse, and rat genomes (Waterston et al. 2002; Gibbs et al. 2004), but many CRMs fail to align between human and chicken (Hillier et al. 2004). Statistical methods that score multiple sequence alignments to find highly constrained elements are being developed (Stojanovic et al. 1999; Margulies et al. 2003; Cooper et al. 2005; Siepel et al. 2005). These discriminate very well between stringently constrained sequences and likely neutral DNA, but they are less effective for analyzing more diverse reference sets of CRMs (Hughes et al. 2005; King et al. 2005). Many studies demonstrate that constrained noncoding sequences can be used as guides to discovering functional binding sites and CRMs (e.g., Gumucio et al. 1996; Elnitski et al. 1997; Loots et al. 2000; Cliften et al. 2003; Kellis et al. 2003; Frazer et al. 2004), and, furthermore, function is strongly associated with evolutionary conservation in noncoding regulatory regions of Ciona (Johnson et al. 2004).
Another approach using aligned genomic sequences to predict CRMs is the computation of regulatory potential (RP), which captures context and pattern information in addition to conservation (Elnitski et al. 2003; Kolbe et al. 2004; Taylor et al. 2006). The statistical models used to compute RP scores are derived from a positive training set of alignments of known CRMs and a negative training set of alignments of ancestral repeats (a model for likely neutral DNA). A high RP score for an aligned block of sequences means that the patterns of alignment columns in it are more similar to the patterns observed in aligned CRMs than those seen in aligned ancestral repeats. The number of possible alignment columns is very large and increases exponentially with additional sequences. Patterns of alignment columns that are distinctive for different functional classes can be found only by grouping columns and training statistical models using these groups of alignment columns as a reduced representation of the alignments. Our current approach of incorporating phylogenetic information and utilizing performance to evaluate the many possible groupings insures that the reduced representation retains the information valuable for discrimination between functional classes (Taylor et al. 2006). The distinctive patterns that contribute to a high RP score are actually a series of groups of alignment columns. These capture multiple subtle contributions to discrimination rather than a single motif such as a nucleotide string needed to bind a single transcription factor. The RP scores perform better than constraint scores against a reference set of known regulatory elements from the HBB gene complex (King et al. 2005), and we have selected these as part of our strategy to predict CRMs.
The fraction of a mammalian genome whose conservation or RP score exceeds a predictive threshold (determined by equivalent sensitivity and specificity against a reference set) is larger than the lower-bound estimate of the fraction under purifying selection since the primate–rodent divergence (∼7% versus ∼5%) (Waterston et al. 2002; Chiaromonte et al. 2003; King et al. 2005). Thus, using RP or conservation alone for prediction should capture many CRMs, but either should also return many false positives. It is prudent to use an additional filter for predictions of CRMs (Berman et al. 2002, 2004).
We use conserved binding site motifs for the transcription factor GATA-1 as the additional filter, because most known erythroid CRMs have this binding site (Weiss and Orkin 1995), the binding specificity has been studied extensively (e.g., Ko and Engel 1993; Merika and Orkin 1993), and this protein is required for late erythroid maturation (Pevny et al. 1991). The mouse G1E cell line, derived from Gata1 knock-out embryonic stem cells, is blocked at the level of an immature committed erythroblast (Weiss et al. 1997) and undergoes terminal erythroid maturation when GATA-1 function is restored. Using this model system, we identified GATA-1-regulated erythroid genes by transcriptome analysis (Welch et al. 2004). In addition, we used similar approaches to identify patterns of altered gene expression in murine erythroleukemia cells induced to mature in vitro. We combined these studies to identify candidate genes that are likely to have GATA-1 and its binding site involved in regulation and applied our bioinformatics tools to predict CRMs. Many of the predicted CRMs had significant effects on the expression of reporter genes in transfected cells, showing the power of bioinformatic predictions based on RP scores plus conserved transcription factor binding sites.
Results
Cohorts of co-expressed genes from microarray expression analyses
Two somatic cell models of late erythroid maturation were used to find groups of genes whose expression levels increase or decrease during this process. Murine erythroleukemia (MEL) cells have properties of proerythroblasts, and are induced to mature into erythroblasts upon treatment with N,N′-hexamethylene-bis-acetamide (HMBA) (Reuben et al. 1976). Transcriptome analysis (Eisen et al. 1998) revealed a cohort of genes coexpressed with Hbb-b1 (encoding β-globin), which includes known markers of late erythroid differentiation, such as the heme biosynthetic gene Alas2 (May et al. 1995), the histone variant gene Hist1h1c (Brown et al. 1985; Cheng and Skoultchi 1989), and other genes not previously known to be in this cohort, such as Vav2, Btg2, and Hipk2. The Gata2 gene was down-regulated during maturation, as expected (Grass et al. 2003). A second cell culture model of erythroid maturation is the G1E line of murine immature erythroblasts that carry a knockout of the Gata1 gene (Weiss et al. 1997). The subline G1E-ER4 stably expresses an estrogen-activated form of GATA-1. Reactivation of GATA-1 function induces terminal erythroid maturation synchronously in all cells. From the results of a previous microarray analysis of gene expression after the restoration of GATA-1 in G1E-ER4 cells (Welch et al. 2004) we identified cohorts of up- and down-regulated genes. Many of these show similar patterns of expression in induced MEL cells.
Based on results from both cell lines, genes in the up-regulated cohort chosen for study were Alas2, Btg2, Vav2, Hist1h1c, Hipk2, and Hebp1. Gata2 was chosen for study as a down-regulated gene. Previous studies (Welch et al. 2004) also showed that Zfpm1 was an immediate target of GATA-1 in these cells, and thus this gene was also studied for predicted cis-regulatory modules. The patterns of expression for most genes were confirmed in an induced MEL cell line using RT-PCR analysis of RNA (Supplemental Fig. S1). At each target locus, we analyzed the gene of interest plus additional intergenic DNA extending to the flanking genes. A total of 1,012,000 bp (∼1 Mb) was included in the eight target loci.
Selection of conserved noncoding regionsto test as predicted CRMs
Mouse genomic DNA sequences whose alignment with four other mammals meet the following two criteria were predicted to be CRMs: (1) the RP score is greater than 0, and (2) the alignment contains a predicted match to a binding site for GATA-1. Only noncoding DNA sequences were used.
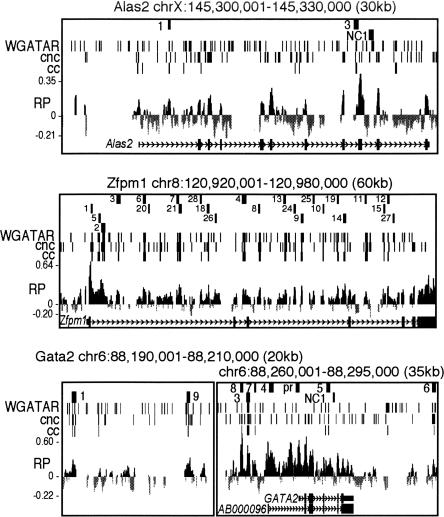
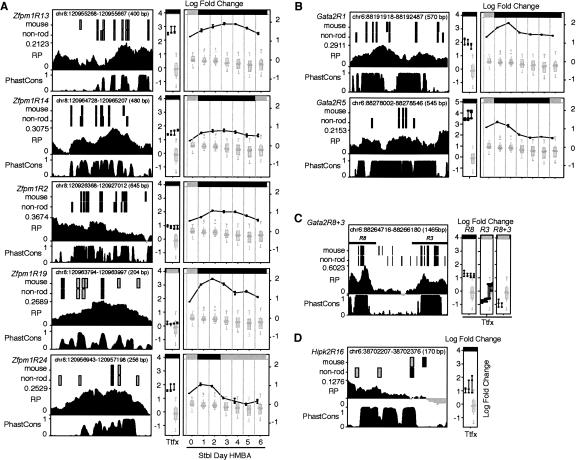
The RP scores were determined using the phylogeny-based method of Taylor et al. (2006) on TBA alignments (Blanchette et al. 2004) of the mouse DNA sequences with the orthologous sequences from rat, human, chimpanzee, and dog. Most loci have many genomic segments with positive RP scores (Fig. 1; Supplemental Fig. S2), which indicate patterns in the alignments similar to those that are distinctive for a training set of known regulatory regions. Individual loci differ in the distribution of RP scores, with a few loci, such as Zfpm1 (Fig. 1), enriched for positive scores and others, such as Hipk2, with primarily negative scores (Supplemental Fig. S2). (Custom tracks for interactive viewing of the RP scores and predicted binding sites along with other genome annotations are available at http://www.bx.psu.edu/~dcking/preCRMs/mm7/links.html). Overall, most DNA segments have negative RP scores, as expected from the genome-wide distributions (King et al. 2005; Taylor et al. 2006). Most loci also show additional strong peaks within introns or in flanking regions; these are candidates for enhancers or other distal CRMs.
Figure 1.
Maps of three target loci with preCRMs and genomic features. The tracks in each graph show chromosomal coordinates (mm7 assembly), positions and abbreviated names of preCRMs, positions of both ccGATA1BSs (matches to conserved consensus GATA-1 binding sites) and cncGATA1BSs (matches to position specific weight matrix of GATA-1 binding sites), a graph of the RP score based on five-species TBA alignments, and the gene exon–intron structure. Similar maps for Btg2, Hebp1, Hipk2, Hist1h1c, and Vav2 are in Supplemental Figure S2.
Matches to the binding site for GATA-1 fall into three categories. Of all the exact matches to the consensus motif (A/T)GATA(A/G) in the mouse sequence (Fig. 1), those also aligning with exact matches to the consensus in at least one non-rodent species (human, chimp, or dog) are called conserved consensus GATA-1 binding sites (ccGATA1BSs). Other matches to the binding site consensus are nonconserved consensus GATA-1 binding sites (nccGATA1BSs). The WGATAR motif was identified as a functional site in several erythroid regulatory elements as a site that is bound specifically by the GATA-1 protein in footprint assays (e.g., Plumb et al. 1989). Other experiments investigating the affinity of GATA-1 for DNA sequences in solution showed a broader specificity (Ko and Engel 1993; Merika and Orkin 1993). Thus, we also examined a third class, which are conserved sites that match the more general weight matrix description of a binding site but do not match exactly the consensus. The ccGATA1BSs were removed from that set, leaving conserved nonconsensus GATA-1 binding sites (cncGATA1BSs).
Genome-wide preCRMs generated by a similar method are provided at http://www.bx.psu.edu/~ross/dataset/DatasetHome.html. The file can be uploaded to genome browsers (Kent et al. 2002) to identify erythroid preCRMs anywhere in the mouse genome, or to databases (Giardine et al. 2003, 2005; Elnitski et al. 2005) and other resources for further analysis.
Transient expression assay for gene regulatory effects of preCRMs
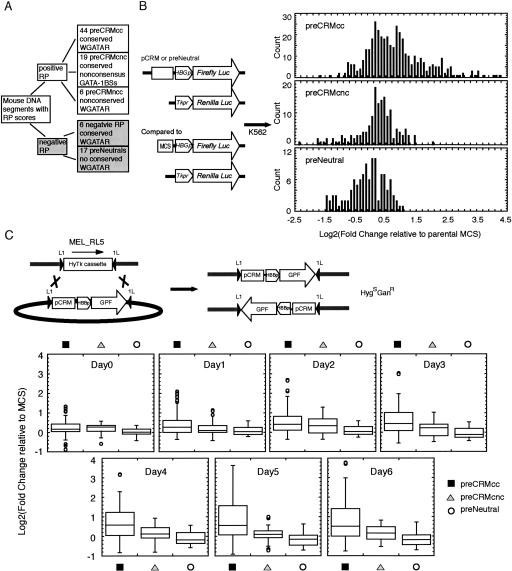
The RP scores and different classes of predicted GATA-1 binding sites were combined to identify distinctive groups of predicted cis-regulatory modules (preCRMs) for experimental tests (Fig. 2A). Within the eight target loci, we tested 44 noncoding DNA segments with a positive RP score and at least one ccGATA1BS (preCRMcc set); this included all noncoding segments with a mean RP score of at least 0.05 and a ccGATA1BS plus a sampling of those with a mean RP score between 0 and 0.05 and a ccGATA1BS. Other groups tested were 19 with a positive RP score and at least one cncGATA1BS (preCRMcnc set), six with positive RP and a nccGATA1BS, and six with negative RP but a ccGATA1BS. Another 17 DNA segments with negative RP and no ccGATA1BS served as predicted neutral fragments in the assays (preNeutral). The chromosomal coordinates and other properties of the tested DNA segments are listed in Suppplemental Table S1.
Figure 2.
Distribution of expression measurements after transient and site-directed stable transfection. (A) Partitioning DNA segments into classes of preCRMs or preNeutrals, based on RP score and predicted GATA-1 binding sites. (B) Transient transfection assay. (Left) Maps of the expression vectors show a firefly luciferase reporter gene expressed from the human A gamma globin gene promoter (HBG1pr) and a multiple cloning site (MCS) for inserting the preCRMs (pCRM). A co-transfection control plasmid has the Renilla luciferase gene expressed from the thymidine kinase promoter (TKpr). (Right) The distributions of luciferase activity measurements for three categories of tested DNA (preCRMcc, preCRMcnc and preNeutral) in transfected K562 cells. (C) Site-directed stable integration assay. The donor plasmid has an EGFP gene expressed from the human HBB promoter, preceded by a MCS into which preCRMs are inserted for test constructs. Cre-mediated exchange via loxP sites can replace the HyTK marker by the test expression cassette. The graphs show the distribution of EGFP fluorescence in pools of cells carrying expression constructs from the three major groups of tested DNAs, represented as log2 fold change, for each day during HMBA induction period. The distributions are depicted as box-plots, in which the box encompasses the values from the 25th to 75th percentiles, and the median is shown as a line across the box. The whiskers extend to the closer of two values: either the limit of the values in the distribution or 1.5 times the interquartile distance (the difference between the 75th and 25th percentiles). Values beyond 1.5 times the interquartile distance are plotted as circles.
The first assay tests for altered expression of a luciferase reporter after transient transfection of human K562 leukemia cells (Fig. 2B). After introduction into the cells, the reporter gene on an unintegrated plasmid is expressed for ∼2 d, at which time the cells are harvested. The recipient K562 cells have erythroid features and are readily transfectable (Benz Jr. et al. 1980). The luciferase reporter gene is driven by the promoter from the HBG1 gene, which is expressed in K562 cells.
Activity measurements from predicted neutral fragments (preNeutral) rarely exceed log2 of 0.7 (corresponding to a 1.6-fold increase, Fig. 2B), confirming that they have little if any biological effect. Very few activity measurements for the preCRMcnc constructs exceed those in the neutral distribution. In contrast, many preCRMcc constructs show a substantially increased activity (Fig. 2B). Using the Wilcoxon-Mann-Whitney test to compare the activity measurements for a preCRMs with those for the set of preNeutrals, a P-value threshold of ≤0.0001 was set for validation of activity for a preCRMs (see Methods).
Assay for effects of preCRMs after site-directed integration into MEL cells
One of the limitations of the transient expression assay is that the reporter plasmids do not assemble into a chromatin structure fully equivalent to that of a chromosome (Reeves et al. 1985). Thus, regulatory effects requiring a normal chromatin structure can be missed. We also tested the preCRMs in a reporter gene cassette after stable integration into a marked locus in MEL cells, using recombinase-mediated cassette exchange (Bouhassira et al. 1997). Targeting the expression cassettes to the same chromosomal location, using the Cre-loxP system (Fig. 2C), avoids large variations from position effects observed after random integration into mammalian cell lines (Bouhassira et al. 1997). We chose the RL5 locus of MEL cells (cell line MEL_RL5) because previous studies with components of the locus control region of the HBB gene complex showed that signals from enhancers were readily detected here and, in contrast to the RL4 and RL6 loci, orientation effects were minimal (Bouhassira et al. 1997; Molete et al. 2001). Additional studies in conjunction with these analyses showed that the RL5 locus is on mouse chromosome 4, between the Tal1 and Pdzk1ip1 (also known as Map17) genes (Supplemental material; Supplemental Fig. S3). Very similar effects were observed between pools of stably transfected cells and a large set of isolated clones carrying the same expression cassette (Supplemental material; Supplemental Fig. S4). Thus, most tests were done on triplicate pools.
At all stages of induction, the distribution of GFP fluorescence measurements for expression cassettes containing a preCRMcc is broader and shifted upward with respect to the distribution for cassettes with a predicted neutral segment (Fig. 2C), indicating that some of the preCRMcc constructs are functional. In contrast, the signals for the preCRMcnc set are more similar to those for the preNeutrals, indicating much less of an effect. As with the transient transfections, we use Wilcoxon-Mann-Whitney tests for validating preCRMs as functional by comparison with the preNeutrals (P ≤ 0.0065; see Methods). The site-directed integration assay extends our ability to validate enhancers by identifying segments that function after integration into a chromosome.
Validated preCRMs
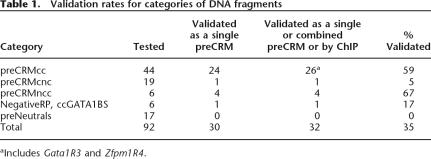
A total of 32 preCRMs was validated in the enhancer assays or by chromatin immunoprecipitation (Table 1). Four of these overlap previously described cis-regulatory elements, and all of these are validated in our assays (Supplemental Fig. S5; Supplemental Table S1). One is Alas2R3 (Fig. 1), an enhancer located in intron 8 of the gene Alas2 (Surinya et al. 1998). The other three, Gata2R7, Gata2R3, and Gata2R8 (Fig. 1), are in a hypersensitive site located ∼3 kb upstream from the erythroid promoter of Gata2. Previous work showed that GATA-2 and CBP are displaced from this region by GATA-1 during the down-regulation of Gata2 (Grass et al. 2003; Martowicz et al. 2005).
Table 1.
Validation rates for categories of DNA fragments
aIncludes Gata1R3 and Zfpm1R4.
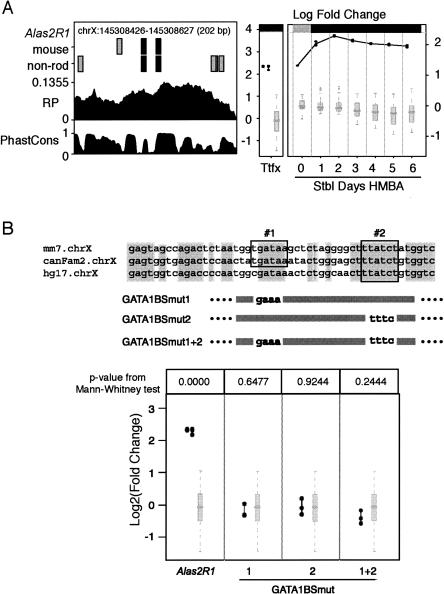
Several novel preCRMs have strong effects in both transient and site-directed stable integration assays. For example, Alas2R1, a predicted CRM in the first intron of the Alas2 gene, caused a fivefold increase in expression in transfected K562 cells in two separate experiments (Fig. 3A). This increase is highly significant when compared with the expression levels of constructs carrying predicted neutral fragments. Alas2R1 also caused an increase in expression from the cassette containing the HBB promoter and the EGFP gene when integrated at locus RL5 of MEL cells (Fig. 3A).
Figure 3.
Features, validation, and mutagenesis of a preCRM in the Alas2 locus. (A) Features and activities of preCRM Alas2R1. The graph on the left shows important features of the preCRM: the chromosomal position and size, segments that match the consensus GATA-1 binding site (black rectangles) and matches to the weight matrix for the GATA-1 binding site that are not matches to the consensus (gray rectangles) in mouse and in non-rodent (non-rod) sequences, regulatory potential scores (RP), and phastCons scores (PhastCons) across the genomic interval. The two graphs on the right summarize the activities of the preCRMs (connected dark filled circles), represented as log2 fold change and compared with that of the preNeutrals (light gray box-plot). The graph labeled “Ttfx” plots the values of the transient transfection experiments in K562 cells. The seven columns in the rightmost graph show the results for the induction time course (Stbl Days HMBA) of pools of cells after site-directed integration of the test expression cassette in MEL_RL5 cells. Significant differences of preCRM activity compared with the distribution of activity measurements for preNeutrals are denoted by a black bar along the top of the graphs; otherwise the bar is gray. (B) Mutagenesis and tests of GATA motifs in Alas2R1. The alignment of a subregion of mouse Alas2R1 with dog (canFam2) and human (hg17) shows two conserved consensus GATA-1 binding sites (outlined). Alas2R1 was mutated in individual (BSmut1 and BSmut2) and both (BSmut1 + 2) GATA-1 binding sites, with the block substitutions shown beneath the alignment. The activity levels of expression plasmids carrying the wild-type and mutated Alas2R1 preCRM are shown in the bottom panel, using the same conventions as in A, except that the numerical P-values for a difference of the activity of the test construct compared with the activities of preNeutrals is given at the top of each column.
The role of GATA-1 in this validated preCRM was tested by mutating the two matches to GATA-1 binding sites and measuring the effect of the altered preCRMs after transfection into K562 cells. Luciferase expression by the mutated expression plasmids decreased to the level of the parental plasmid (Fig. 3B), demonstrating an important role for these presumptive GATA-1 binding sites and supporting a role for GATA-1 in enhancement by this preCRM.
The gene Zfpm1 encodes a multiple zinc finger protein, FOG1, that cooperates with GATA-1 at some regulatory sites (Tsang et al. 1998; Crispino et al. 1999; Fox et al. 1999; Chang et al. 2002). This locus is an immediate target of GATA-1 in G1E-ER4 cells (Welch et al. 2004). Our bioinformatic approach predicts many preCRMs in Zfpm1 (Fig. 1), and five of them (R13, R14, R2, R10, and R12) are validated in both assays (Fig. 4A; Supplemental Fig. S5C). Others are validated only after site-directed integration (e.g., R19 and R7) or only by transient transfection (R24, R1, R18, R21, and R6). Validation by one assay but not the other could reflect the mechanism of the regulation conferred by the preCRMs; for example, those validated only after site-directed integration may act primarily through effects on chromatin structure. The different sets of transcription factors present in the recipient cells could also contribute to the differences, as well as the different promoters in the expression plasmids (Fig. 2). The general conclusion is that the results from the two types of assays are frequently independent, and thus it is important to perform both.
Figure 4.
Examples of validated preCRMs in three loci. Selected validated preCRMs from Zfpm1 (A), Gata2 single preCRMs (B), a combination of preCRMs from Gata2 (C), and Hipk2 (D) are shown; features and activities of other validated preCRMs are shown in the Supplemental figures. The conventions for displaying features and activities are the same as in Figure 3.
We chose Gata2 as an example of a gene whose expression is down-regulated in response to restoration of GATA-1 function in G1E-ER4 cells (Welch et al. 2004). The novel pCRMs Gata2R1 (∼50 kb upstream, Fig. 1) and Gata2R5 (in the fourth intron) are strongly validated in both assays (Fig. 4B). The activity of the preCRM Gata2R3 illustrates a context effect, requiring the presence of an activating sequence to show its role in repression in these assays. Previous studies have shown that Gata2R3 plays a negative role in regulation of Gata2 (Grass et al. 2003; Martowicz et al. 2005), but this preCRM alone has no significant effect in transient transfection assays (Fig. 4C). However, when it is present in the same fragment as Gata2R8, Gata2R3 counteracts the enhancement by Gata2R8.
All the individual preCRMs discussed so far contain at least one ccGATA1BS, and additional predicted CRMccs in three other loci, Vav2, Btg2, and Hebp1, were validated with strong effects (Supplemental Fig. S6A; Supplemental Table S1). Notably, four of the preCRMs for which the consensus GATA-1 binding site is present only in mouse or rodents (preCRMncc class) also were validated in transient transfection assays (Fig. 4D; Supplemental Fig. S6B), suggestive of lineage-specific regulation by GATA-1 (Valverde-Garduno et al. 2004). In the case of Hipk2R16, although the mouse binding site motif is not conserved, a nearby sequence is a conserved consensus GATA-1 binding site in several non-mouse species (Fig. 4D). This is consistent with turnover of the binding site in the mouse lineage, as has been documented in Drosophila (Ludwig et al. 2000) and mammals (Dermitzakis and Clark 2002).
Overall, the pCRMcc and pCRMncc classes had the highest validation rates, much higher than pCRMcnc or segments with negative RP (Table 1). This supports the use of a combination of high RP and a consensus binding site for a transcription factor in predicting CRMs. Also, deviation from the consensus binding site (pCRMcnc class) leads to less accurate predictions.
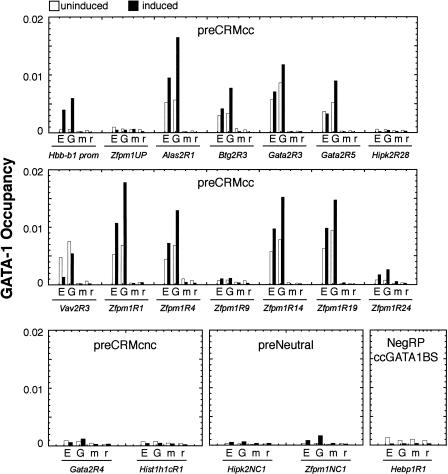
Site occupancy by GATA-1
A sampling of each class of preCRM was tested for occupancy by the protein GATA-1 in rescued G1E-ER4 cells using chromatin immunoprecipitation. Ten of 12 tested preCRMccs showed significant levels of GATA-1 protein bound (Fig. 5), and nine of those 10 have significant activity in the enhancer assays (including Gata2R3, which is active in combination with Gata2R8). Likewise, two preCRMccs that fail to be validated in enhancer assays, Hipk2R28 and Zfpm1R9, also show no significant binding by GATA-1. Thus, site occupancy by GATA-1 is positively associated with enhancer activity.
Figure 5.
Relative GATA-1 occupancy at a subset of preCRMs. Using antibodies directed against the estrogen-receptor moiety of the GATA-1-ER fusion protein (E) and the N terminus of GATA-1 (G), chromatin fragments were immunoprecipitated from untreated G1E-ER4 cells (uninduced) and G1E-ER4 cells treated with estradiol for 24 h (induced). Normal mouse (m) and rat (r) IgG controls were performed in parallel. Real-time polymerase chain reactions of each amplicon were performed in duplicate and quantified using SYBR green dye. Signals were referenced to a dilution series of the relevant input sample. Amplicons from the β-major globin promoter (Hbb-b1 prom) and an upstream region of the Zfpm promoter (Zfpm1UP) are shown as positive and negative controls.
An exception to this association is Zfpm1R4. This preCRMcc is bound by GATA-1, as previously reported by Welch et al. (2004), but it is not active in either enhancer assay. The site occupancy indicates that it is involved in some aspect of regulation, perhaps one displayed in the G1E-ER4 cells but not mimicked in the cell lines used for transfection.
Conservation of the consensus GATA-1 binding motif, WGATAR, is strongly associated with site occupancy. In contrast, neither of the preCRMs with a conserved nonconsensus site is occupied by GATA-1 (Fig. 5, preCRMcnc panel). Interestingly, a DNA fragment that contains a GATA-1 binding site motif but has a negative RP score, Hebp1R1, is not occupied by GATA-1 (Fig. 5). Thus, both conservation of the consensus motif and positive RP are associated with binding by GATA-1.
The preCRMccs Hipk2R28 and Zfpm1R9 appear to be false positives of our prediction pipeline. It is possible that they have a function for which we have not tested, but that function does not involve GATA-1 binding in G1E-ER4 cells.
Positive correlation of activity with RP and conserved consensus GATA-1 binding motif
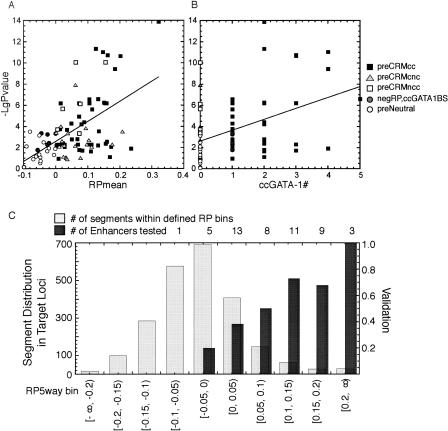
Both the activities in the transfection assays and the magnitude of the RP signals vary for the validated preCRMs, which raises the question of whether RP scores have a positive correlation with activity. Thus, the correlation between the negative log10 of the P-value for activity in the assays (transient or stable) and the mean RP score was examined for all DNA segments assayed. The segments with negative mean RP scores consistently have low activity (Fig. 6A). This includes some segments with conserved consensus GATA-1 binding sites, showing that they are not sufficient for activity. In contrast, many of the segments with high RP show strong activity and a tendency for a higher activity at higher RP scores. Although this is not uniformly true, with some segments of high RP showing no activity in these assays, the overall correlation is positive (R2 = 0.29). The enhancer activities from the tested fragments also show a positive correlation with the number of conserved consensus motifs (Fig. 6B), but with a lower R2 value (0.18) than that obtained for the correlation with RP.
Figure 6.
Correlation of enhancer activity with genomic features of preCRMs and distribution of segments by RP score and validation rates. (A) Correlation of enhancer activity with RP score. The negative log10 P-value for the difference in activity of each preCRM being different from that of preNeutrals is plotted against the mean RP score for the preCRM. The more significant activity from either the transient transfection or the site-directed stable integration assay was chosen for each preCRM. For stable transfection data, the smallest P-value during induction was used. (B) Correlation of enhancer activity with the number of ccGATA-1BSs in each preCRM. The values on the vertical axis are the same as in panel A, and the horizontal axis gives the number of matches to conserved consensus GATA-1 binding sites in each preCRM. (C) The distribution of DNA segments in the eight target loci after segmentation by RP score. Runs of at least 100 nucleotides of mouse DNA with mean RP scores within a designated range (bin in the histogram) were identified, and the number of segments in each bin of RP score is plotted. (Dark gray bars) Validation rate for DNA fragments containing at least one ccGATA1BS in each RP bin; the number of fragments tested in the enhancer assays is given on the top of each bin.
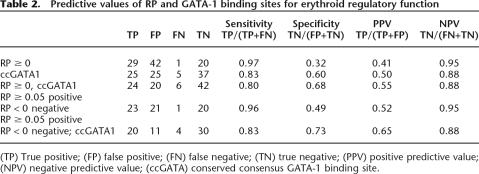
Another method for evaluating the relative contributions of RP score and conserved factor binding sites to the prediction of CRMs is to determine sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Using RP scores ≥0 or ccGATA1BSs separately to predict CRMs gives many false positives, as expected, and consequently the specificity and PPV are marginal (Table 2). The sensitivity and negative predictive values are quite good for these individual features. Combining the two features improves the specificity to 0.68 and the PPV to 0.55. Raising the RP threshold to at least 0.05 for a positive prediction while using RP < 0 for a negative prediction improves the performance. Combining the latter RP thresholds with a requirement for a ccGATA1BS gives the best mix of sensitivity and specificity (0.83 and 0.73) and of positive and negative predictive value (0.65 and 0.88). This analysis supports the contribution of both features to predictive value. However, only DNA segments with both high RP and ccGATA1BSs were examined comprehensively in our experiments, and segments in other categories were only sampled. Thus the actual values for these measures may be different in more comprehensive experiments.
Table 2.
Predictive values of RP and GATA-1 binding sites for erythroid regulatory function
(TP) True positive; (FP) false positive; (FN) false negative; (TN) true negative; (PPV) positive predictive value; (NPV) negative predictive value; (ccGATA) conserved consensus GATA-1 binding site.
Association of high RP segments with function
We also evaluated how well RP (in concert with ccGATA1BSs) works to enrich selections of noncoding DNA for function in regulation. All the noncoding mouse DNA in the eight target loci that aligned with other species was partitioned into bins by RP score (Fig. 6C). Most of these segments are in the bins with negative RP, as expected from the genome-wide distribution of RP scores (King et al. 2005; Taylor et al. 2006). Only a small fraction of conserved (aligned) DNA segments falls into high RP bins, but the segments that also have a ccGATA1BS are validated at a high rate (Fig. 6C). PreCRMccs with RP ≥ 0.05 are validated at rates from 50% to 100%. In contrast, many DNA segments have RP scores <0, and these are rarely validated. Thus, the bioinformatic predictions select a small subset of noncoding conserved DNA that has a high likelihood of functioning in gene regulation.
Discussion
We evaluated the effectiveness of using RP scores in combination with conserved binding sites for transcription factors to predict cis-regulatory modules for genes regulated during erythroid differentiation. Ninety-two DNA segments, covering a range of RP scores and varying in the presence or absence of predicted GATA-1 binding sites, were tested for enhancer activity. DNA segments with a high RP score (e.g., at least 0.05) and a conserved match to the consensus GATA-1 binding site were validated as enhancers at a high rate. The strength of the effects correlated positively both with RP score and with number of ccGATA1BSs. Thus, in a genome-wide application, RP > 0.05 and strict conservation of the GATA-1 binding site consensus should provide good specificity, >70% if the current rates are maintained (Table 2). However, some segments with RP scores between 0 and 0.05 are also validated, and thus greater sensitivity in studies of individual loci can be achieved by lowering the threshold for RP. For example, 18 out of 23 CRMs in the β-globin gene locus (King et al. 2005) pass an RP threshold of at least 0.05, computed using the method of Taylor et al. (2006). These include the CRMs with the strongest activity; the others have more subtle or lineage-specific effects. Thus, the thresholds for predictions should be set as appropriate to the scope and aims of the experiments.
Our experimental approaches may actually underestimate the number of preCRMs that play a role in regulation. Most of our current assays test only one preCRM at a time, requiring that an individual preCRM be sufficient to cause a phenotype in order to be validated. However, it is common for groups of CRMs to work together, as has been observed for the locus control region of the HBB complex (Bungert et al. 1995; Hardison et al. 1997; Li et al. 1999). Initial results reported here show that at least one of the preCRMs that is not validated in individual assays can act in concert with another CRM to modulate its effects. In addition, some preCRMs may be specific for a subset of erythroid promoters, and may not be active on the globin gene promoters employed in this study. The cell lines used in our study do not mimic all aspects of developmental regulation, and assays in whole animals will be required for full exploration of potential activities.
Positive and negative regulation are often exerted by the same CRMs, with changes in the trans-factors accomplishing the switch. This is the case for Gata2R3 (Grass et al. 2003; Martowicz et al. 2005). The inactivity of Gata2R3 alone in gene expression assays may result from complications of protein competition. This segment of DNA is located ∼2.8 kb upstream of an alternate, tissue-specific promoter in an erythroid DNase hypersensitive site, and it has been shown to enhance expression in G1E cells (Grass et al. 2003; Martowicz et al. 2005). It is bound initially by GATA-2 but is then replaced by GATA-1 at progressive stages of erythroid maturation, leading to an inhibition of expression of Gata2 (Grass et al. 2003; Martowicz et al. 2005). Both GATA-1 and GATA-2 are present in the two cell lines used in our transfection studies, and it is possible that the effects of these competing proteins offset each other, preventing an obvious effect on expression.
The 32 preCRMs validated in this study confirm four previously known erythroid CRMs and add 28 novel ones. Thus, this study increases substantially the set of confirmed erythroid CRMs, and they increase our knowledge about the regulation of individual genes. For example, our data reveal two more regulatory modules for Gata2, one located ∼70 kb upstream and another in intron 4. CRMs are distributed throughout the Zfpm1 gene, and a highly active cluster of them in intron 3 may be particularly important for regulation.
Our study demonstrates that the predictions based on RP scores and conserved transcription factor binding sites have good predictive value (Table 2), but improvements are still needed. Some strongly predicted regions show no effects in the assays used in this study. Further work with a wider range of assays is needed to determine whether these are actually false positives, or whether their function is not revealed by cell transfections.
Many stringently conserved noncoding regions, which align between human and fish sequences, have been shown to function as developmental enhancers (Aparicio et al. 1995; Plessy et al. 2005; Woolfe et al. 2005), with validation rates as high as 90% (Nobrega et al. 2003; Woolfe et al. 2005). However, most mammalian CRMs are not so highly constrained (Hillier et al. 2004); indeed, none of the eight loci investigated in our study shows substantial noncoding sequence matches between mouse and fish. Thus, we restricted our analysis to alignments of mammalian genome sequences, filtering these for positive RP scores and a conserved consensus GATA-1 binding site. Other recent studies have employed alignments along with an additional filter with good success. Donaldson et al. (2005) exploited detailed knowledge of critical binding site motifs and their spacing, along with human–rodent conservation and positive RP, to predict and validate novel enhancers for genes involved in mammalian hematopoiesis. Johnson et al. (2005) used a combination of clusters of predicted motifs and evolutionary conservation successfully to identify muscle CRMs in a genome-wide scan of Ciona. Thus, the approach of combining RP scores with another feature of CRMs, such as conserved motifs or clusters of motifs, should be broadly applicable to studies of gene regulation in complex genomes. Combining RP scores with new methods to identify clusters of conserved factor binding sites (Berman et al. 2004; Blanchette et al. 2006) may be particularly productive.
The phylogeny-based, multispecies RP scores (Taylor et al. 2006) improve prediction accuracy, compared with the initial implementations (Elnitski et al. 2003; Kolbe et al. 2004; Taylor et al. 2006). The current RP scores are systematically higher in the validated preCRMs. Three of the preCRMs that failed to validate had positive RP scores in the initial implementation but have negative scores by the current method. Thus, the improved methodology can reduce false positive predictions. In addition, requiring conservation of a stringent match to a GATA-1 binding site motif improves accuracy over use of conserved matches to GATA-1 binding site weight matrices. Another obvious limitation of our current prediction approach is that lineage-specific regulatory elements are invisible to techniques utilizing rodent–primate comparisons (Hughes et al. 2005; King et al. 2005). However, this limitation also could be overcome; lineage-specific CRMs may be identified utilizing a set of more closely related species, with techniques such as phylogenetic shadowing (Boffelli et al. 2003).
Methods
Conserved GATA-1 motifs
The motif-scorer scans aligned sequences with either the position-specific scoring matrix (PSSM, threshold = 0.85) for GATA-1 binding sites or pattern matching routines (searching for WGATAR), ignoring gap characters from the alignment rows in MAF format. The PSSM was generated by merging the PSSMs for GATA-1 binding sites in JASPAR (Sandelin et al. 2004) and TRANSFAC (Wingender et al. 2001). Sequence alignments with motif matches above the threshold in both mouse and at least one other non-rodent (human, chimp, or dog) comprised the conserved GATA1 binding site predictions.
Three different types of matches to the GATA-1 binding site were employed. The most stringent is an exact match to the consensus motif, WGATAR, in mouse and in the aligned positions in at least one non-rodent sequence (conserved consensus GATA-1 binding site, or ccGATA1BS). The second is a conserved non-consensus site (cncGATA1BS), which matches the merged PSSM for GATA-1 binding sites in mouse and at least one non-rodent sequence in the multiple alignment, but is not an exact match to WGATAR. The third is a nonconserved consensus site (nccGATA1BS), which has an exact match to WGATAR in mouse but not in an aligned non-rodent sequence.
Prediction of erythroid cis-regulatory modules and negative controls
Predicted cis-regulatory modules (preCRMs) in the intervals containing the target mouse genes have the following properties: They (1) align among mouse, rat, human, chimp, and dog; (2) do not contain exons; (3) have a five-way mouse–rat–human–chimp–dog RP score (Taylor et al. 2006) >0; and (4) contain at least one match to one category of GATA-1 binding site (see above). For the preCRMs with a ccGATA1BS, all those in the eight target loci with a mean RP score >0.05 and most of those with scores between 0 and 0.05 were tested.
Noncoding aligned DNA segments were initially predicted to be cis-regulatory modules (preCRMs) based on an empirically established threshold (Hardison et al. 2003) for mouse–human alignments (Elnitski et al. 2003; Schwartz et al. 2003) and their proximity to a predicted binding site for GATA-1 conserved among mouse, rat, and human (Schwartz et al. 2003). The advances in the availability of multiple sequences (Gibbs et al. 2004; Chimpanzee Sequencing and Analysis Consortium 2005; Dog Sequencing and Analysis Consortium 2005), greater sensitivity in multiple alignment (Blanchette et al. 2004), improved algorithms for Markov models (Kolbe et al. 2004; Taylor et al. 2006), and modified methods for determining conserved matches to transcription factor binding sites (see section above) allowed us to reevaluate previously predicted preCRMs in terms of RP score and conservation of GATA-1 binding sites. The mouse DNA sequence in the chosen eight loci was aligned with the orthologous sequences from rat, human, chimpanzee, and dog using TBA (Blanchette et al. 2004), and RP scores were recomputed using the phylogeny-based five-species implementation of Taylor et al. (2006). A modification to this method was implemented to address a missing data problem. Some of the genome assemblies are incomplete, and thus the alignments include some gaps corresponding to missing sequence (rather than real gaps in the alignment). Gaps likely to result from absence of sequence were replaced with a wildcard symbol. RP scores were then computed using the same models as the other five-way scores, but each column with a wildcard was assigned to the same reduced alphabet symbol as the nearest non-wildcard column based on distance between ancestral reconstructions (Taylor et al. 2006). The previously predicted CRMs were mapped onto the new five-way RP scores to obtain the mean RP score used in this report. The type of predicted GATA-1 binding site was also determined using the procedure described in the previous section.
Transient transfection and expression
The luciferase expression plasmid MCSγluc (Elnitski et al. 2001) containing the human A gamma globin (HBG1pr) gene promoter (from −260 to +35) fused to the firefly luciferase coding region of pGL3Basic (Promega) was modified to contain restriction endonuclease sites for MluI and NotI. Predicted CRMs and neutrals were amplified from MEL_RL5 genomic DNA and inserted into the MCS via these sites to make each test expression plasmid. Primer sequences for preCRMs and negative controls are in Supplemental Tables S2 and S3.
The plasmid DNAs were transiently transfected into K562 cells using the cationic lipid reagent Tfx50 (Promega) following the procedure as described in Elnitski and Hardison (1999) and Elnitski et al. (2001). Briefly, 0.8 μg of plasmid containing firefly luciferase reporter and 0.008 μg of cotransfection control plasmid expressing Renilla luciferase were transfected in triplicate into 4 × 105 cells at a 2:1 ratio (charge to mass) of Tfx50 to DNA. For a triplicate determination, a plasmid is prepared in three independent minipreps, each of which is transfected into the K562 cells. The entire triplicate experiment was done at least twice for each test plasmid.
Two days after the transfection, cell extracts were subject to a dual luciferase assay following the manufacturer’s protocol (Promega). For each of the triplicate samples, the firefly luciferase activity of the test plasmid (divided by the Renilla luciferase activity of the cotransfection control) was normalized by the firefly luciferase activity from the parental MCSγluc (divided by the Renilla luciferase activity of the cotransfection control) to obtain a fold change. The fold change is reported as its log (base 2).
Site-directed mutagenesis
Mutagenesis used the QuickChange Site-Directed Mutagenesis Kit (Stratagene), following manufacturer’s protocol. The mutagenic primers were designed as following:
QC1_Alas2R1_F: 5′-CCAGACTCTAATGGTGAAAAGCTC TAGGGGCTTTAT-3′
QC1_Alas2R1_R: 5′-ATAAAGCCCCTAGAGCTTTTCAC CATTAGAGTCTGG-3′
QC2_Alas2R1_F: 5′-TAAGCTCTAGGGGCTTTTTCTATGGTCT GCAGGCTC-3′
QC2_Alas2R1_R: 5′-GAGCCTGCAGACCATAGAAAAAGC CCCTAGAGCTTA-3′
The supercoiled double-stranded plasmid Alas2preCRM1γluc was used to amplify the mutated, nicked plasmid by pfuturbo DNA polymerase. The PCR product was treated with DpnI (McClelland and Nelson 1992) and then transformed into XL1-Blue supercompetent cells.
Recombinase-mediated cassette exchange and expressionafter stable transfection of MEL_RL5 cells
preCRMs were inserted into the plasmid L1-MCS2βEGFP-1L (Molete et al. 2001) containing MluI and NotI in the MCS, which expresses EGFP (Clontech) from the human β-globin gene (HBB) promoter (segment from −374 to +44 relative to the transcription start site). Thus, PCR-amplified DNA could be cloned into L1-MCS2βEGFP-1L as well as MCSγluc. The Cre expression plasmid pBS185 (CMV-CRE) was obtained from Clontech.
Expression cassettes containing the parental HBB promoter-EGFP construct with or without a preCRM or preNeutral were integrated at RL5 by site-specific recombination directed by CRE recombinase, following procedures as described in Bouhassira et al. (1997). Three pools of stably transfected cells carrying each expression cassette were isolated, and the median EGFP fluorescence was monitored by flow cytometry for several days to ascertain that the level was stable (Molete et al. 2001). The pools were then induced for erythroid maturation by incubating cultures of cells at a density of 2 × 105/mL in DMEM containing 4 mM N,N′-hexamethylene-bis-acetamide (HMBA) for 6 d at 37°C. The level of green fluorescence from EGFP was measured daily by flow cytometry. Each measurement on each pool of cells containing a preCRM (or preNeutral) was divided by the fluorescence measurement from cells carrying the parental cassette (MCS2βEGFP, which is also integrated independently for each experiment) to obtain a fold change. The log2 of the fold change is the expression value analyzed in this work.
Statistics for validation thresholds for enhancer activities
Validation of enhancer activity in either assay is based on comparison with the expression values from the set of predicted neutral fragments. Expression from a set of 17 preNeutrals (Supplemental Table S1) was measured in transiently transfected K562, with triplicate determinations and at least two experiments. This yielded a set of 38 measurements. Expression after stable, site-directed integration into MEL_RL5 cells, before and after induction, was measured for a set of 11 preNeutrals (a subset of those tested in transient transfections; Supplemental Table S1). This produced a set of at least 30 measurements for each day of the induction series. The sets of measurements (all as log2 fold change relative to the parental expression cassettes or plasmids) for the sets of preNeutrals are the comparison distributions for the expression from each preCRM in each assay.
A Wilcoxon-Mann-Whitney rank order test was applied for the set of expression measurements for each preCRM and preNeutral, comparing to the set of values from all preNeutrals for that assay. For the transient transfection assay, no preNeutral had a P-value <0.0001, so we consider a preCRM to be validated if its P-value in this test is ≤0.0001. For the site-directed integration assay, no preNeutral had a P-value <0.0065 at any day of induction, so we consider the activity from a preCRM to be significant if its P-value in this test is ≤0.0065. For the latter assay, we further require that a significant activity be observed for at least three consecutive days during the induction series to insure that a consistent effect is observed. The differences in P-value thresholds are influenced by the differences in numbers of measurements for both the preCRMs and the preNeutrals in the two assays, as well as differences in the dynamic range of values obtained.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (Welch et al. 2004). GATA-1 (N6) and ER (Ab-10) antibodies used for ChIP were obtained from Santa Cruz Biotechnology and Neomarkers, respectively.
Acknowledgments
This work was supported by NIH grants DK65806 (R.H.) and HG02238 (W.M.), Tobacco Settlement funds from the Commonwealth of Pennsylvania, and the Huck Institutes of Life Sciences of the Penn State University.
Footnotes
[Supplemental material is available online at www.genome.org. The expression profile data obtained during MEL cell differentiation have been submitted to GEO under accession no. GSE2217.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5353806
References
- Aparicio S., Morrison A., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Morrison A., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Chaudhuri C., Rigby P., Krumlauf R., Brenner S., Rigby P., Krumlauf R., Brenner S., Krumlauf R., Brenner S., Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl. Acad. Sci. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz E.J., Jr., Murnane M.J., Tonkonow B.L., Berman B.W., Mazur E.M., Cavallesco C., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Murnane M.J., Tonkonow B.L., Berman B.W., Mazur E.M., Cavallesco C., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Tonkonow B.L., Berman B.W., Mazur E.M., Cavallesco C., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Berman B.W., Mazur E.M., Cavallesco C., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Mazur E.M., Cavallesco C., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Cavallesco C., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Jenko T., Snyder E.L., Forget B.G., Hoffman R., Snyder E.L., Forget B.G., Hoffman R., Forget B.G., Hoffman R., Hoffman R. Embryonic-fetal erythroid characteristics of a human leukemic cell line. Proc. Natl. Acad. Sci. 1980;77:3509–3513. doi: 10.1073/pnas.77.6.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman B.P., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Nibu Y., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Pfeiffer B.D., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Tomancak P., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Celniker S.E., Levine M., Rubin G.M., Eisen M.B., Levine M., Rubin G.M., Eisen M.B., Rubin G.M., Eisen M.B., Eisen M.B. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman B.P., Pfeiffer B.D., Laverty T.R., Salzberg S.L., Rubin G.M., Eisen M.B., Celniker S.E., Pfeiffer B.D., Laverty T.R., Salzberg S.L., Rubin G.M., Eisen M.B., Celniker S.E., Laverty T.R., Salzberg S.L., Rubin G.M., Eisen M.B., Celniker S.E., Salzberg S.L., Rubin G.M., Eisen M.B., Celniker S.E., Rubin G.M., Eisen M.B., Celniker S.E., Eisen M.B., Celniker S.E., Celniker S.E. Computational identification of developmental enhancers: Conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Deblois G., Giguere V., Ferretti V., Bergeron D., Giguere V., Ferretti V., Bergeron D., Ferretti V., Bergeron D., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M., Ovcharenko I., Pachter L., Rubin E.M., Pachter L., Rubin E.M., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Bouhassira E.E., Westerman K., Leboulch P., Westerman K., Leboulch P., Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- Brown D.T., Wellman S.E., Sittman D.B., Wellman S.E., Sittman D.B., Sittman D.B. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol. Cell. Biol. 1985;5:2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungert J., Dave U., Lim K.-C., Kieuw K.H., Shavit J.A., Liu Q., Engel J.D., Dave U., Lim K.-C., Kieuw K.H., Shavit J.A., Liu Q., Engel J.D., Lim K.-C., Kieuw K.H., Shavit J.A., Liu Q., Engel J.D., Kieuw K.H., Shavit J.A., Liu Q., Engel J.D., Shavit J.A., Liu Q., Engel J.D., Liu Q., Engel J.D., Engel J.D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes & Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- Chang A.N., Cantor A.B., Fujiwara Y., Lodish M.B., Droho S., Crispino J.D., Orkin S.H., Cantor A.B., Fujiwara Y., Lodish M.B., Droho S., Crispino J.D., Orkin S.H., Fujiwara Y., Lodish M.B., Droho S., Crispino J.D., Orkin S.H., Lodish M.B., Droho S., Crispino J.D., Orkin S.H., Droho S., Crispino J.D., Orkin S.H., Crispino J.D., Orkin S.H., Orkin S.H. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl. Acad. Sci. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G.H., Skoultchi A.I., Skoultchi A.I. Rapid induction of polyadenylated H1 histone mRNAs in mouse erythroleukemia cells is regulated by c-myc. Mol. Cell. Biol. 1989;9:2332–2340. doi: 10.1128/mcb.9.6.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F., Weber R.J., Roskin K.M., Diekhans M., Kent W.J., Haussler D., Weber R.J., Roskin K.M., Diekhans M., Kent W.J., Haussler D., Roskin K.M., Diekhans M., Kent W.J., Haussler D., Diekhans M., Kent W.J., Haussler D., Kent W.J., Haussler D., Haussler D. The genome of Homo sapiens. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY.: 2003. The share of human genomic DNA under selection estimated from human–mouse genomic alignments; pp. 245–254. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Collins F.S., Green E.D., Guttmacher A.E., Guyer M.S., Green E.D., Guttmacher A.E., Guyer M.S., Guttmacher A.E., Guyer M.S., Guyer M.S. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Cooper G.M., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Stone E.A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Asimenos G., Green E.D., Batzoglou S., Sidow A., Green E.D., Batzoglou S., Sidow A., Batzoglou S., Sidow A., Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino J.D., Lodish M.B., MacKay J.P., Orkin S.H., Lodish M.B., MacKay J.P., Orkin S.H., MacKay J.P., Orkin S.H., Orkin S.H. Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: The GATA-1:FOG complex. Mol. Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E., Clark A., Clark A. Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Dog Sequencing Analysis Consortium Initial sequencing and analysis of the dog genome. Nature. 2005;438:803–819. [Google Scholar]
- Donaldson I.J., Chapman M., Kinston S., Landry J.R., Knezevic K., Piltz S., Buckley N., Green A.R., Göttgens B., Chapman M., Kinston S., Landry J.R., Knezevic K., Piltz S., Buckley N., Green A.R., Göttgens B., Kinston S., Landry J.R., Knezevic K., Piltz S., Buckley N., Green A.R., Göttgens B., Landry J.R., Knezevic K., Piltz S., Buckley N., Green A.R., Göttgens B., Knezevic K., Piltz S., Buckley N., Green A.R., Göttgens B., Piltz S., Buckley N., Green A.R., Göttgens B., Buckley N., Green A.R., Göttgens B., Green A.R., Göttgens B., Göttgens B. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum. Mol. Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D., Spellman P.T., Brown P.O., Botstein D., Brown P.O., Botstein D., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L., Hardison R., Hardison R. Efficient and reliable transfection of mouse erythroleukemia cells using cationic lipids. Blood Cells Mol. Dis. 1999;25:299–304. doi: 10.1006/bcmd.1999.0257. [DOI] [PubMed] [Google Scholar]
- Elnitski L., Miller W., Hardison R., Miller W., Hardison R., Hardison R. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the β-globin locus control region. Role of basic helix-loop-helix proteins. J. Biol. Chem. 1997;272:369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- Elnitski L., Li J., Noguchi C.T., Miller W., Hardison R., Li J., Noguchi C.T., Miller W., Hardison R., Noguchi C.T., Miller W., Hardison R., Miller W., Hardison R., Hardison R. A negative cis-element regulates the level of enhancement by hypersensitive site 2 of the β-globin locus control region. J. Biol. Chem. 2001;276:6289–6298. doi: 10.1074/jbc.M009624200. [DOI] [PubMed] [Google Scholar]
- Elnitski L., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Schwartz S., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L., Giardine B., Shah P., Zhang Y., Riemer C., Weirauch M., Burhans R., Miller W., Hardison R.C., Giardine B., Shah P., Zhang Y., Riemer C., Weirauch M., Burhans R., Miller W., Hardison R.C., Shah P., Zhang Y., Riemer C., Weirauch M., Burhans R., Miller W., Hardison R.C., Zhang Y., Riemer C., Weirauch M., Burhans R., Miller W., Hardison R.C., Riemer C., Weirauch M., Burhans R., Miller W., Hardison R.C., Weirauch M., Burhans R., Miller W., Hardison R.C., Burhans R., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Improvements to GALA and dbERGE II: Databases featuring genomic sequence alignment, annotation and experimental results. Nucleic Acids Res. 2005;33:D466–D470. doi: 10.1093/nar/gki045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A.H., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M., Holmes M., Kowalski K., Mackay J., Crossley M., Kowalski K., Mackay J., Crossley M., Mackay J., Crossley M., Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Tao H., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R., Tao H., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R., de Jong P.J., Chen X., Doherty M.F., Cox D.R., Chen X., Doherty M.F., Cox D.R., Doherty M.F., Cox D.R., Cox D.R. Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res. 2004;14:367–372. doi: 10.1101/gr.1961204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Schwartz S., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. GALA, a database for genomic sequence alignments and annotations. Genome Res. 2003;13:732–741. doi: 10.1101/gr.603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Miller W., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Miller W., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Miller W., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Miller W., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Miller W., Shah P., Zhang Y., Blankenberg D., Albert I., Miller W., Zhang Y., Blankenberg D., Albert I., Miller W., Blankenberg D., Albert I., Miller W., Albert I., Miller W., Miller W., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., Scott G., Steffen D., Worley K.C., Burch P.E., Steffen D., Worley K.C., Burch P.E., Worley K.C., Burch P.E., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Grass J.A., Boyer M.E., Pal S., Wu J., Weiss M.J., Bresnick E.H., Boyer M.E., Pal S., Wu J., Weiss M.J., Bresnick E.H., Pal S., Wu J., Weiss M.J., Bresnick E.H., Wu J., Weiss M.J., Bresnick E.H., Weiss M.J., Bresnick E.H., Bresnick E.H. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio D., Shelton D., Zhu W., Millinoff D., Gray T., Bock J., Slightom J., Goodman M., Shelton D., Zhu W., Millinoff D., Gray T., Bock J., Slightom J., Goodman M., Zhu W., Millinoff D., Gray T., Bock J., Slightom J., Goodman M., Millinoff D., Gray T., Bock J., Slightom J., Goodman M., Gray T., Bock J., Slightom J., Goodman M., Bock J., Slightom J., Goodman M., Slightom J., Goodman M., Goodman M. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the β-like globin genes. Mol. Phylogenet. Evol. 1996;5:18–32. doi: 10.1006/mpev.1996.0004. [DOI] [PubMed] [Google Scholar]
- Hardison R., Oeltjen J., Miller W., Oeltjen J., Miller W., Miller W. Long human–mouse sequence alignments reveal novel regulatory elements: A reason to sequence the mouse genome. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- Hardison R.C., Chiaromonte F., Kolbe D., Wang H., Petrykowska H., Elnitski L., Yang S., Giardine B., Zhang Y., Riemer C., Chiaromonte F., Kolbe D., Wang H., Petrykowska H., Elnitski L., Yang S., Giardine B., Zhang Y., Riemer C., Kolbe D., Wang H., Petrykowska H., Elnitski L., Yang S., Giardine B., Zhang Y., Riemer C., Wang H., Petrykowska H., Elnitski L., Yang S., Giardine B., Zhang Y., Riemer C., Petrykowska H., Elnitski L., Yang S., Giardine B., Zhang Y., Riemer C., Elnitski L., Yang S., Giardine B., Zhang Y., Riemer C., Yang S., Giardine B., Zhang Y., Riemer C., Giardine B., Zhang Y., Riemer C., Zhang Y., Riemer C., Riemer C., et al. Global predictions and tests of erythroid regulatory regions. Cold Spring Harb. Symp. Quant. Biol. 2003;68:335–344. doi: 10.1101/sqb.2003.68.335. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Bork P., Burt D.W., Groenen M.A., Delany M.E., Burt D.W., Groenen M.A., Delany M.E., Groenen M.A., Delany M.E., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hughes J.R., Cheng J.F., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Cheng J.F., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D.R., De Gobbi M., de Jong P., Rubin E., Higgs D.R., de Jong P., Rubin E., Higgs D.R., Rubin E., Higgs D.R., Higgs D.R. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc. Natl. Acad. Sci. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., Davidson B., Brown C.D., Smith W.C., Sidow A., Davidson B., Brown C.D., Smith W.C., Sidow A., Brown C.D., Smith W.C., Sidow A., Smith W.C., Sidow A., Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., Zhou Q., Yagi K., Satoh N., Wong W., Sidow A., Zhou Q., Yagi K., Satoh N., Wong W., Sidow A., Yagi K., Satoh N., Wong W., Sidow A., Satoh N., Wong W., Sidow A., Wong W., Sidow A., Sidow A. De novo discovery of a tissue-specific gene regulatory module in a chordate. Genome Res. 2005;15:1315–1324. doi: 10.1101/gr.4062605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Chiaromonte F., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L.J., Engel J.D., Engel J.D. DNA-binding specifities of the GATA transcription factor family. Mol. Cell. Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe D., Taylor J., Elnitski L., Eswara P., Li J., Miller W., Hardison R., Chiaromonte F., Taylor J., Elnitski L., Eswara P., Li J., Miller W., Hardison R., Chiaromonte F., Elnitski L., Eswara P., Li J., Miller W., Hardison R., Chiaromonte F., Eswara P., Li J., Miller W., Hardison R., Chiaromonte F., Li J., Miller W., Hardison R., Chiaromonte F., Miller W., Hardison R., Chiaromonte F., Hardison R., Chiaromonte F., Chiaromonte F. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 2004;14:700–707. doi: 10.1101/gr.1976004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Harju S., Peterson K.R., Harju S., Peterson K.R., Peterson K.R. Locus control regions: Coming of age at a decade plus. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- Loots G.G., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Locksley R.M., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Blankespoor C.M., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Wang Z.E., Miller W., Rubin E.M., Frazer K.A., Miller W., Rubin E.M., Frazer K.A., Rubin E.M., Frazer K.A., Frazer K.A. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M., Bergman C., Patel N.H., Kreitman M., Patel N.H., Kreitman M., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Blanchette M., Haussler D., Green E.D., Blanchette M., Haussler D., Green E.D., NISC Comparative Sequencing Program. Haussler D., Green E.D., Green E.D. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martowicz M.L., Grass J.A., Boyer M.E., Guend H., Bresnick E.H., Grass J.A., Boyer M.E., Guend H., Bresnick E.H., Boyer M.E., Guend H., Bresnick E.H., Guend H., Bresnick E.H., Bresnick E.H. Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J. Biol. Chem. 2005;280:1724–1732. doi: 10.1074/jbc.M406038200. [DOI] [PubMed] [Google Scholar]
- May B.K., Dogra S.C., Sadlon T.J., Bhasker C.R., Cox T.C., Bottomley S.S., Dogra S.C., Sadlon T.J., Bhasker C.R., Cox T.C., Bottomley S.S., Sadlon T.J., Bhasker C.R., Cox T.C., Bottomley S.S., Bhasker C.R., Cox T.C., Bottomley S.S., Cox T.C., Bottomley S.S., Bottomley S.S. Molecular regulation of heme biosynthesis in higher vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 1995;51:1–51. doi: 10.1016/s0079-6603(08)60875-2. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M., Nelson M. Effect of site-specific methylation on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1992;20:2145–2157. doi: 10.1093/nar/20.suppl.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M., Orkin S.H., Orkin S.H. DNA-binding specificy of GATA family transcription factors. Mol. Cell. Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W., Makova K.D., Nekrutenko A., Hardison R.C., Makova K.D., Nekrutenko A., Hardison R.C., Nekrutenko A., Hardison R.C., Hardison R.C. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- Molete J.M., Petrykowska H., Bouhassira E.E., Feng Y.Q., Miller W., Hardison R.C., Petrykowska H., Bouhassira E.E., Feng Y.Q., Miller W., Hardison R.C., Bouhassira E.E., Feng Y.Q., Miller W., Hardison R.C., Feng Y.Q., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Sequences flanking hypersensitive sites of the β-globin locus control region are required for synergistic enhancement. Mol. Cell. Biol. 2001;21:2969–2980. doi: 10.1128/MCB.21.9.2969-2980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M., Ovcharenko I., Afzal V., Rubin E.M., Afzal V., Rubin E.M., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Rubin E.M., Rubin E.M. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.-F., D’Agati V., Orkin S.H., Costantini F., Simon M.C., Robertson E., Klein W.H., Tsai S.-F., D’Agati V., Orkin S.H., Costantini F., Robertson E., Klein W.H., Tsai S.-F., D’Agati V., Orkin S.H., Costantini F., Klein W.H., Tsai S.-F., D’Agati V., Orkin S.H., Costantini F., Tsai S.-F., D’Agati V., Orkin S.H., Costantini F., D’Agati V., Orkin S.H., Costantini F., Orkin S.H., Costantini F., Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Plessy C., Dickmeis T., Chalmel F., Strahle U., Dickmeis T., Chalmel F., Strahle U., Chalmel F., Strahle U., Strahle U. Enhancer sequence conservation between vertebrates is favoured in developmental regulator genes. Trends Genet. 2005;4:207–210. doi: 10.1016/j.tig.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Plumb M., Frampton J., Wainwright H., Walker M., Macleod K., Goodwin G., Harrison P., Frampton J., Wainwright H., Walker M., Macleod K., Goodwin G., Harrison P., Wainwright H., Walker M., Macleod K., Goodwin G., Harrison P., Walker M., Macleod K., Goodwin G., Harrison P., Macleod K., Goodwin G., Harrison P., Goodwin G., Harrison P., Harrison P. GATAAG; a cis-control region binding an erythroid-specific nuclear factor with a role in globin and non-globin gene expression. Nucleic Acids Res. 1989;17:73–92. doi: 10.1093/nar/17.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Gorman C.M., Howard B., Gorman C.M., Howard B., Howard B. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985;13:3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben R.C., Wife R.L., Breslow R., Rifkind R.A., Marks P.A., Wife R.L., Breslow R., Rifkind R.A., Marks P.A., Breslow R., Rifkind R.A., Marks P.A., Rifkind R.A., Marks P.A., Marks P.A. A new group of potent inducers of differentiation in murine erythroleukemia cells. Proc. Natl. Acad. Sci. 1976;73:862–866. doi: 10.1073/pnas.73.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Engstrom P., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Kent W.J., Miller W., Haussler D., Bejerano G., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Kent W.J., Miller W., Haussler D., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Kent W.J., Miller W., Haussler D., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Kent W.J., Miller W., Haussler D., Hou M., Rosenbloom K., Clawson H., Kent W.J., Miller W., Haussler D., Rosenbloom K., Clawson H., Kent W.J., Miller W., Haussler D., Clawson H., Kent W.J., Miller W., Haussler D., Kent W.J., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Evolutionarily conserved elements in vertebrate, fly, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovic N., Florea L., Riemer C., Gumucio D., Slightom J., Goodman M., Miller W., Hardison R., Florea L., Riemer C., Gumucio D., Slightom J., Goodman M., Miller W., Hardison R., Riemer C., Gumucio D., Slightom J., Goodman M., Miller W., Hardison R., Gumucio D., Slightom J., Goodman M., Miller W., Hardison R., Slightom J., Goodman M., Miller W., Hardison R., Goodman M., Miller W., Hardison R., Miller W., Hardison R., Hardison R. Comparison of five methods for finding conserved sequences in multiple alignments of gene regulatory regions. Nucleic Acids Res. 1999;27:3899–3910. doi: 10.1093/nar/27.19.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surinya K.H., Cox T.C., May B.K., Cox T.C., May B.K., May B.K. Identification and characterization of a conserved erythroid-specific enhancer located in intron 8 of the human 5-aminolevulinate synthase 2 gene. J. Biol. Chem. 1998;273:16798–16809. doi: 10.1074/jbc.273.27.16798. [DOI] [PubMed] [Google Scholar]
- Taylor J., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F., King D.C., Hardison R.C., Miller W., Chiaromonte F., Hardison R.C., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify fuctional elements. Genome Res. 2006 doi: 10.1101/gr.4537706. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Favorov A.V., Frith M.C., Fu Y., Kent W.J., Frith M.C., Fu Y., Kent W.J., Fu Y., Kent W.J., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Fujiwara Y., Hom D.B., Orkin S.H., Fujiwara Y., Hom D.B., Orkin S.H., Hom D.B., Orkin S.H., Orkin S.H. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes & Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Garduno V., Guyot B., Anguita E., Hamlett I., Porcher C., Vyas P., Guyot B., Anguita E., Hamlett I., Porcher C., Vyas P., Anguita E., Hamlett I., Porcher C., Vyas P., Hamlett I., Porcher C., Vyas P., Porcher C., Vyas P., Vyas P. Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: Implications for cis-element identification. Blood. 2004;104:3106–3116. doi: 10.1182/blood-2004-04-1333. [DOI] [PubMed] [Google Scholar]
- Wasserman W.W., Sandelin A., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weiss M.J., Orkin S.H., Orkin S.H. GATA transcription factors: Key regulators of hematopoiesis. Exp. Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- Weiss M.J., Yu C., Orkin S.H., Yu C., Orkin S.H., Orkin S.H. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.J., Watts J.A., Vakoc C.R., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J., Watts J.A., Vakoc C.R., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J., Vakoc C.R., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J., Blobel G.A., Chodosh L.A., Weiss M.J., Chodosh L.A., Weiss M.J., Weiss M.J. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., Krull M., Matys V., Michael H., Ohnhauser R., Matys V., Michael H., Ohnhauser R., Michael H., Ohnhauser R., Ohnhauser R., et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]