Abstract
Heterochromatin is important for gene regulation and chromosome structure, but the genes that are occupied by heterochromatin proteins in the mammalian genome are largely unknown. We have adapted the DamID method to systematically identify target genes of the heterochromatin proteins HP1 and SUV39H1 in human and mouse cells. Unexpectedly, we found that CBX1 (formerly HP1β) and SUV39H1 bind to genes encoding KRAB domain containing zinc finger (KRAB-ZNF) transcriptional repressors. These genes constitute one of the largest gene families and are organized in clusters in the human genome. Preference of CBX1 for this gene family was observed in both human and mouse cells. High-resolution mapping on human chromosome 19 revealed that CBX1 coats large domains 0.1–4 Mb in size, which coincide with the position of KRAB-ZNF gene clusters. These domains show an intricate CBX1 binding pattern: While CBX1 is globally elevated throughout the domains, it is absent from the promoters and binds more strongly to the 3′ ends of KRAB-ZNF genes. KRAB-ZNF domains contain large numbers of LINE elements, which may contribute to CBX1 recruitment. These results uncover a surprising link between heterochromatin and a large family of regulatory genes in mammals. We suggest a role for heterochromatin in the evolution of the KRAB-ZNF gene family.
Heterochromatin plays key roles in chromosome structure and gene regulation and is marked by a set of specialized nonhistone proteins and specific histone modifications (Li et al. 2002; Dimitri et al. 2005). In the past decades, several protein components of heterochromatin have been identified and characterized. Heterochromatin Protein 1 (HP1) was first discovered in Drosophila as a protein that is associated with pericentric heterochromatin (James et al. 1989). Humans have three homologs of HP1: CBX1 (HP1β), CBX3 (HP1γ), and CBX5 (HP1α), which are known as Cbx1 (HP1β or M31), Cbx3 (HP1γ or M32), and Cbx5 (HP1α) in mice. Mammalian HP1 proteins are also enriched in pericentric heterochromatin (Maison and Almouzni 2004).
HP1 proteins contain two conserved protein domains, the N-terminal chromodomain (CD) and the C-terminal chromoshadow-domain (CSD). Both domains have been shown to contribute to stable association of HP1 proteins with heterochromatin in vivo (Platero et al. 1995; Thiru et al. 2004). The CD binds di- or trimethylated lysine 9 of histone H3 (H3K9) (Maison and Almouzni 2004). The CSD mediates homo- and heterodimerization of HP1 proteins (Nielsen et al. 2001a) and interacts with various other proteins (Li et al. 2002), including SUV39H1 (Aagaard et al. 1999). SUV39H1 and SUV39H2 are histone methyl transferases essential for H3K9 methylation in heterochromatin (O’Carroll et al. 2000; Rea et al. 2000). The interactions between HP1 proteins, SUV39H1, and methylated H3K9 might stabilize the protein complexes at heterochromatic locations.
Although in most species a large fraction of heterochromatin is concentrated in regions surrounding the centromeres, evidence is accumulating that heterochromatin proteins also play a role in the regulation of genes throughout the genome (Nielsen et al. 2001b; Liu et al. 2005). The role of heterochromatin in gene regulation appears to be context dependent: Some genes are repressed, while others may be activated by heterochromatin (Dimitri et al. 2005). The silencing effect of heterochromatin has been well described. The classical example is the transcriptional silencing of genes that are artificially placed close to heterochromatic regions, a phenomenon known as position effect variegation (Weiler and Wakimoto 1995). In addition, there is evidence indicating that heterochromatin is involved in gene activation (Piacentini et al. 2003; Cryderman et al. 2005). For example, Drosophila has active genes embedded in heterochromatin that are immune to silencing and, in some cases, need the heterochromatic localization for their correct expression (Dimitri et al. 2005).
A striking feature of heterochromatin is its ability to coat large genomic regions. Detailed molecular mapping in Schizosaccharomyces pombe revealed telomeric and centromeric heterochromatic domains of up to ∼20 kb (Noma et al. 2001; Cam et al. 2005). In Arabidopsis thaliana and Drosophila heterochromatin domains of more than 0.5 Mb were found, although these domains can be interrupted by “islands” with euchromatic characteristics (Greil et al. 2003; Lippman et al. 2004). These heterochromatic domains contain a variety of repeat elements and genes. Results of microscopy experiments are suggestive of pericentric heterochromatin domains on mammalian chromosomes (Minc et al. 1999; Hayakawa et al. 2003). Molecular mapping of heterochromatin proteins in mammalian cells has been limited to individual genes (Nielsen et al. 2001b; Ayyanathan et al. 2003; Vakoc et al. 2005), and so far it remains unknown if heterochromatin proteins also form domains outside pericentric regions in mammalian chromosomes.
Here, we report the detailed mapping of the binding sites of heterochromatin proteins CBX1 and SUV39H1 in the human and mouse genome. The results demonstrate that CBX1 forms large heterochromatin domains of up to 4 Mb in size. These domains specifically harbor genes that encode KRAB-ZNF proteins, which form one of the largest families of transcriptional regulators. Based on these results, we suggest a role for heterochromatin in the evolution of the KRAB-ZNF gene family.
Results
DamID in mammalian cells
For the identification of target loci of human heterochromatin proteins we used the DamID method, which has successfully been applied in Drosophila to map the genomic binding sites of heterochromatin proteins and various other regulatory proteins (van Steensel and Henikoff 2000; van Steensel et al. 2001, 2003; Greil et al. 2003; Orian et al. 2003; Sun et al. 2003; Bianchi-Frias et al. 2004; de Wit et al. 2005; for review, see Orian 2006). In short, DamID involves the low-level expression of a fusion protein consisting of DNA adenine methyltransferase (Dam) and a chromatin protein of interest. This fusion protein is targeted to the native binding sites of the chromatin protein, where Dam methylates adenines in the surrounding DNA. Subsequently, the methylated DNA fragments are isolated and amplified by selective PCR, labeled with a fluorescent dye, and hybridized to microarrays. To correct for unspecific Dam binding and local differences in chromatin accessibility, methylated DNA from cells that were transfected with Dam alone is amplified, labeled with a different dye, and cohybridized. The ratio of the fluorescent signals on the array indicates the amount of targeted Dam methylation and denotes the level of binding of the Dam-fusion protein to the probed sequences (van Steensel et al. 2001). Supplemental Figure S1 gives an outline of the DamID experimental design. Recently it was reported that DamID and chromatin immunoprecipitation yield very similar results in the case of Drosophila GAGA factor (Moorman et al. 2006; Negre et al. 2006) and PcG proteins (Negre et al. 2006; Tolhuis et al. 2006).
We adapted the DamID technique for use in human and mouse cells (detailed protocols can be found at http://www.nki.nl/nkidep/vansteensel). For DamID the expression level of Dam-fusion proteins in cells should be kept very low to avoid mistargeting of the fusion protein and to suppress nontargeted background methylation (van Steensel and Henikoff 2000; van Steensel et al. 2001). For this purpose, we chose the pIND expression-vector system (Invitrogen) that exhibits a very low basal expression and can be induced by addition of a synthetic steroid hormone (No et al. 1996). We found that leaky expression from the uninduced vectors is sufficient for DamID experiments, while high expression after induction allows for the detection of Dam proteins by Western blotting or immunofluorescence microscopy. Low-level expression of Dam protein had no effect on cell growth in MCF7 cells (Supplemental Fig. S2).
CBX1 binding map in human MCF7 cells
For the mapping of CBX1 binding sites we made a CBX1-Dam construct (Fig. 1A). By immunofluorescence microscopy we confirmed that this fusion protein is correctly targeted to heterochromatic regions in mouse cells (Supplemental Fig. S3), suggesting that the CBX1 is still functional when fused to Dam. The correct size of Dam fusion proteins was confirmed by Western blotting (data not shown).
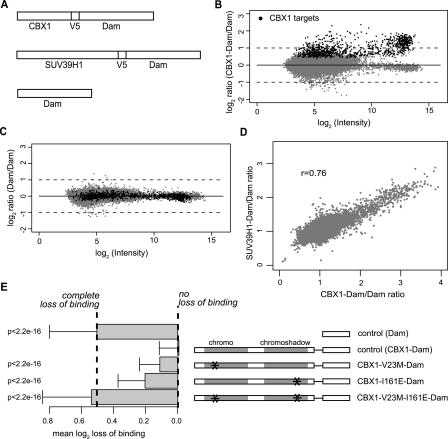
Figure 1.
CBX1 and SUV39H1 binding profiles. (A) Schematic drawing of CBX1-Dam-fusion protein, SUV39H1-Dam-fusion protein, and the Dam protein constructs that are used to generate the binding profiles. To facilitate detection of the Dam-fusion proteins a V5 epitope tag was added. Both CBX1 and SUV39H1 as well as control DamID profiles consist of the combined data of four independent experiments hybridized on four oligonucleotide arrays. (B) CBX1 DamID binding profile. For each probed sequence, the average log2 CBX1-Dam/Dam ratio is plotted against the average fluorescence intensity (log2 √(Cy5 × Cy3). Statistically significant CBX1 target loci (n = 933) are indicated by black dots. Gray dots represent nontarget loci. (C) Control DamID profile. Black dots represent CBX1 target loci projected on control data. Gray dots represent nontarget loci. (D) Bivariate scatter plot of CBX1 and SUV39H1 binding data. (r) Pearson correlation coefficient. (E) Molecular mechanism of CBX1 targeting. Overview of CBX1 mutants (right side) and bar plot showing their average loss of binding to CBX1 target loci (left side). Error bars indicate standard deviations. P-values compared to control (CBX1-wt) according to Wilcoxon rank sum test. The values representing “complete loss of binding” and “no loss of binding” were inferred from the Dam and CBX1-Dam controls, respectively. DamID profiles of CBX1-mutants consist, for each protein, of data of two independent experiments. For details on the mutants and controls, see text.
For the detection of targeted Dam methylation, we first used oligoarrays containing 28,756 oligonucleotides corresponding to 16,260 unique genes. The oligonucleotides match the 3′ end of coding regions and do not provide information about intergenic regions. However, in Drosophila HP1 often binds to the transcribed region of genes (Sun et al. 2003; de Wit et al. 2005) and HP1 binding could readily be detected by DamID using cDNA arrays (Greil et al. 2003).
We mapped the genomic distribution of CBX1 in MCF7 human breast carcinoma cells. The CBX1 binding profile was generated by combining data from four independent experiments. The CBX1 binding ratios are presented in Figure 1B as a function of the microarray spot intensities. The normalized CBX1 binding ratios are somewhat skewed toward positive values, due to the presence of loci that are preferentially bound by CBX1. A linear error model was used to determine the statistical significance of CBX1 binding (see Methods). At a significance cutoff of P < 0.05 (corrected for multiple testing), we were able to identify 933 oligonucleotides that correspond to targets of CBX1. The identity of the target loci will be discussed below. We also constructed a control profile (Fig. 1C) where we hybridized amplified methylated DNA from cells expressing Dam alone. As expected, the CBX1 targets show no binding in the control profile.
To confirm the specificity of the CBX1 binding profile, we used previously described CBX1 point mutants that do not localize to chromatin correctly. HP1 proteins localize to chromatin via CD and CSD interactions and both protein domains are important for correct localization. Mutation V23M in the CD is known to abolish interaction with methylated H3K9 (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001), while mutation I161E blocks dimerization and interactions of the CSD with non-HP1 proteins (Brasher et al. 2000; Thiru et al. 2004). To test the effects of these mutations, we monitored the genome-wide changes in binding of each mutant Dam-fusion by direct comparison to wild-type CBX1-Dam (Fig. 1E). In this set of experiments we also included unfused Dam, which in this context can be regarded as a “null mutation” of CBX1, allowing for an estimate of the change in signals that can be expected in case of complete loss of CBX1 binding activity. We also included a “self–self” comparing wild-type CBX1-Dam to wild-type CBX1-Dam, allowing for an estimate of the random biological and technical noise. The results (Fig. 1E) show that both single point mutants displayed significant but partial loss of binding to target genes. In other words, these mutants can still bind to the target genes, although not as efficiently as wild-type CBX1 proteins. We note that these experiments were performed in cells that express endogenous wild-type HP1 proteins. Therefore, the residual targeting of CBX1V23M-Dam could be accounted for by dimerization with endogenous protein. In contrast, the double point mutant had completely lost its ability to bind to target genes. These results demonstrate that the observed binding profile with wild-type CBX1 is not the result of nonspecific protein–DNA interactions.
To further confirm the specificity of the CBX1 binding profile, we tested if the CBX1 binding profile was dependent on the specific Dam enzyme that was used. To do this we generated CBX1 binding maps using Dam enzymes from Escherichia coli and phage T4 (which have an overall protein sequence identity of ∼23%) and found that these overlap to a great extent (Supplemental Fig. S4A). In addition, we found that C and N terminal Dam-fusion proteins give comparable CBX1 binding maps (data not shown). Taken together, these results indicate that we have identified specific target loci of CBX1 using DamID in human cells.
Human HP1 homologs have overlapping target specificities
The three human HP1 homologs CBX1, CBX3, and CBX5 have been described to have at least partially overlapping microscopic subnuclear distributions (Nielsen et al. 1999; Minc et al. 2000; Maison et al. 2002; Festenstein et al. 2003; Gilbert et al. 2003). We confirmed these observations in human MCF7 cells as well as in mouse NIH3T3 cells (Supplemental Fig. S5). In addition, the three HP1 homologs have been shown to form heterodimers (Nielsen et al. 2001a). To investigate whether CBX1, CBX3, and CBX5 bind to the same loci in MCF7 cells we compared their binding maps. Scatter plots of CBX1, CBX3, and CBX5 show significant correlations between binding profiles of all three proteins (Supplemental Fig. S4B–D), indicating they have similar target gene specificities. However, given the inevitable experimental noise present in these data sets, we cannot rule out that there are quantitative differences in the binding patterns of the three proteins.
Strong overlap of SUV39H1 and CBX1 binding patterns
Based on the reported colocalization and functional interactions between HP1 proteins and SUV39H1, we expected to find a strong overlap of their target specificities. To test this we used DamID to generate a profile of SUV39H1 binding and compared it to the CBX1 binding profile (Fig. 1D). Indeed, the binding patterns of CBX1 and SUV39H1 strongly correlate with each other (r = 0.76), indicating that most genes are bound by both proteins. Thus, within the probed gene set, CBX1 and SUV39H1 bind predominantly to the same genes.
CBX1 and SUV39H1 localize to LINEs but not to SINEs
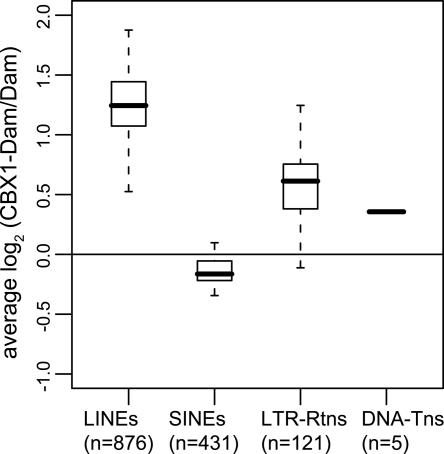
Besides oligonucleotides that match unique genes, the array also contains 1434 oligonucleotides consisting of sequences homologous to transposable elements (TEs). The “TE-loci” displayed high spot intensities on the microarray, probably because they match sequences that occur multiple times in the genome. We noticed that many of these TE-loci showed significant binding by CBX1, and can be seen as a separate “cloud” in the CBX1 binding profile (Fig. 1B). We categorized the TE-derived sequences according to the four major classes of TEs (Lander et al. 2001) and compared the average binding of CBX1 per TE class (Fig. 2). This revealed that CBX1 associates with LINE elements but not with SINE elements and only weakly with LTR retrotransposons. Similar results were obtained for SUV39H1 (data not shown). We note that this preference for LINEs is not a DamID artifact caused by the high copy number of LINEs, because SINEs are even more abundant in the human genome, yet do not show elevated binding. (Copy numbers are estimated to be ∼1.5 × 106 for SINEs and 0.85 × 106 for LINEs; Weiner 2002). Because of the homology between LINE sequences in the human genome and the expected cross-hybridization to probes on the array, we cannot determine the genomic location of LINE elements that are bound by CBX1.
Figure 2.
CBX1 binds to LINEs but not SINEs. Box-and-whisker plot of CBX1 binding to various repeat classes. Average log2 binding of CBX1 to 1,434 repeat containing oligonucleotides. The number of oligonucleotides per repeat class is indicated between parentheses. (LINEs) long interspersed nucleotide elements. (SINEs) short interspersed nucleotide elements. (LTR-Rtns) long terminal repeat retrotransposons. (DNA-Tns) DNA transposons. Horizontal lines represent the smallest value (outliers excepted), the 25th, 50th (median), and 75th percentiles and the largest value (outliers excepted).
We analyzed the ability of the CBX1 mutants (Fig. 1E) to locate to LINE elements. We find that the point mutations affect the binding to single-copy loci and to LINEs to an equal extent (data not shown). This suggests that the molecular targeting mechanism may be similar for most target loci.
CBX1 and SUV39H1 preferentially bind genes encoding KRAB-domain containing zinc finger proteins
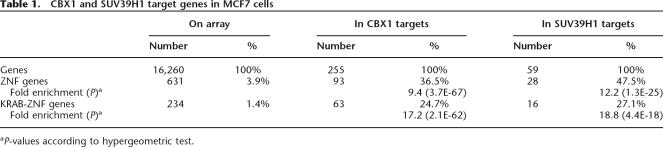
We were able to detect 255 genes (∼1.6% of all probed genes) that were significantly bound by CBX1 and 59 genes that were bound by SUV39H1 (Table 1). The absolute binding ratios of CBX1 were somewhat higher than SUV39H1 binding ratios (Fig. 1D), which may be the reason that fewer SUV39H1 targets than CBX1 targets could be identified at the same significance level. Surprisingly, inspection of the identity of the CBX1 and SUV39H1 target genes revealed that 37% of the CBX1 target genes and 48% of the SUV39H1 target genes encode C2H2 Zinc Finger (ZNF) proteins (Table 1). ZNF genes form a large gene superfamily encoding putative transcription factors, with ∼800 members in humans (Knight and Shimeld 2001; Grimwood et al. 2004).
Table 1.
CBX1 and SUV39H1 target genes in MCF7 cells
aP-values according to hypergeometric test.
About one-third of the ZNF proteins in the human genome have an N-terminal KRAB domain (KRAB-ZNF proteins). The KRAB domain is only found in tetrapod vertebrate genomes and is a potent transcriptional repressor domain (Urrutia 2003). Closer inspection of the target ZNF genes revealed that the majority are KRAB-ZNF genes (Table 1). In total, the CBX1 targets are ∼17-fold enriched for KRAB-ZNF genes. Not only ZNF genes with a KRAB-domain but also some ZNF genes without KRAB-domain are bound by CBX1 (Table 1; data not shown). No common features were found in the other target genes. Thus, CBX1 and SUV39H1 have a striking preference for KRAB-ZNF genes. We decided to analyze the binding to the KRAB-ZNF genes in detail.
CBX1 forms large domains on chromosome 19
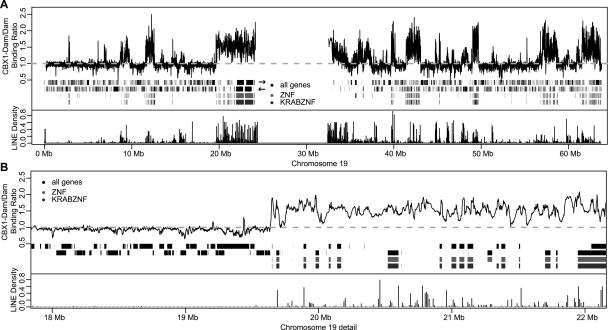
About 56% of all human KRAB-ZNF genes are located in several clusters on chromosome 19 (Chr19). To study the targeting of heterochromatin proteins to these genes in detail, we generated a high-resolution CBX1-binding map of Chr19. For this purpose, we designed a tiling path array that covers all nonrepetitive parts of the entire chromosome, with 60-mer oligonucleotide probes positioned on average every 200 bp. We then used this tiling array to detect CBX1 binding (Fig. 3A). Strikingly, CBX1 binding forms ∼20 large domains, ranging in size from ∼0.1–4 Mb. These domains are 2–3 orders of magnitude larger than the estimated resolution of DamID (van Steensel and Henikoff 2000; Sun et al. 2003). The majority of these domains coincide with KRAB-ZNF gene clusters. Detailed inspection (Fig. 3B) indicates that within these domains, CBX1 can be detected both in genes and in intergenic regions. Although local variation occurs, almost all parts of the domains show increased levels of CBX1 association compared to regions outside the domains. This suggests that CBX1 may associate with most of the chromatin fiber within these domains.
Figure 3.
CBX1 binding forms large domains on chromosome 19. (A) Chromosomal map of CBX1 binding along Chr19. (Top panel) The black line represents the running mean (window size = 20 probes) of CBX1 binding to Chr19. From top to bottom: Black bars represent Ensembl annotated genes on the + and − strands. Light gray bars are zinc finger genes and dark gray bars are KRAB-ZNF genes. (Bottom panel) Bars show the LINE density per 10 kb along Chr19. (B) Same as in A, enlargement of 4 Mb of the chromosomal map. Data are the average of two experiments.
According to the tiling array data, virtually all KRAB-ZNF genes on Chr19 show elevated binding by CBX1 (Fig. 4A). This is in contrast with the data obtained with the coding region oligonucleotide arrays, where we could identify only 23% (44 out of 195) of the KRAB-ZNF genes on Chr19 as high-confidence CBX1 targets. Thus, the tiling array data point out that the majority of KRAB-ZNF genes are bound by CBX1. This result suggests that DamID is particularly sensitive when used in combination with genomic tiling arrays. The enrichment of CBX1 binding was much less prominent for ZNF genes without a KRAB domain, of which roughly 30% showed CBX1 association (Fig. 4A). Chr19 contains four nonclustered KRAB-ZNF genes, which show relatively weak CBX1 binding (black arrowheads, Fig. 4A)
Figure 4.
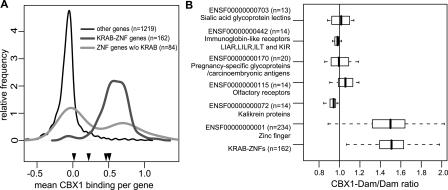
CBX1 binds to the KRAB-ZNF gene family. (A) Most KRAB-ZNF genes on Chr19 are bound by CBX1. Frequency distribution of CBX1 binding to KRAB-ZNF genes (dark gray line), ZNF genes without KRAB domain (light gray line), and non-ZNF genes (thin black line) on Chr19. Black arrowheads indicate average CBX1 binding to four nonclustered KRAB-ZNF genes, i.e., ZNF114, ZNF333, ZNF175, and ENSG00000142528 from left to right. (B) Box-and-whisker plot of CBX1 binding to gene families on Chr19. All gene families that have >10 members on the Chr19 genomic tiling array are shown. To define gene families we used Ensembl protein family IDs (e.g., ENSF00000000001). For comparison we included the KRAB-ZNFs family (identified on the basis of their InterPro IDs; see Methods). The number of genes per family on Chr19 is given in parentheses. Vertical lines represent the smallest value (outliers excepted), the 25th, 50th (median), and 75th percentiles and the largest value (outliers excepted).
Besides the KRAB-ZNF genes, Chr19 contains several other clusters of related genes, such as clusters of olfactory receptor genes, a cluster of immunoglobin-like receptor genes, and a cluster of sialic acid glycoprotein lectin genes (Grimwood et al. 2004). To exclude that CBX1 simply binds to clusters of related genes, we systematically analyzed binding to all known gene families with 10 or more members on Chr19 (Fig. 4B). We found that KRAB-ZNF genes represent the only gene family that is almost completely associated with CBX1 (Fig. 4B). Thus, the binding of CBX1 to KRAB-ZNF gene clusters is highly specific.
LINE elements are abundant in CBX1 domains
Because CBX1 associates with LINE elements, we wondered whether LINEs might be particularly abundant in the CBX1 domains on Chr19. Indeed, comparison of the chromosomal distributions shows that LINE density is strongly elevated in the CBX1 domains (Fig. 3A). However, at high resolution there seems to be no obvious relationship between LINE positioning and CBX1 levels (Fig. 3B). Note that our Chr19 tiling array only contains nonrepetitive sequences and therefore does not directly provide information about the binding of CBX1 to individual LINE elements on this chromosome. Taken together, these data demonstrate that CBX1 domains not only harbor KRAB-ZNF gene clusters, but are also marked by a high density of LINEs.
We investigated whether the high LINE density is unique to KRAB-ZNF clusters. We calculated the average LINE density of genes (and 300 kb of flanking sequence) belonging to the different families indicated in Figure 4B. No correlation was observed between LINE density and CBX1 binding for these families (data not shown). For example, olfactory receptor genes have LINE densities similar to those of KRAB-ZNF genes (data not shown), but negligible levels of CBX1 binding (Fig. 4B). This indicates that the enrichment of LINEs in KRAB-ZNF gene domains is not sufficient to explain the CBX1 binding.
Detailed analyses of CBX1 binding along KRAB-ZNF genes and LINEs
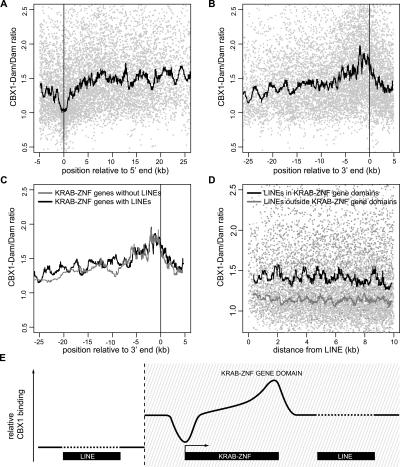
Visual inspection of binding along KRAB-ZNF genes suggested that CBX1 binding is not uniform. To investigate whether CBX1 binding follows a specific pattern, we aligned the transcriptional start or end sites of KRAB-ZNF genes on Chr19 and plotted CBX1 binding along these genes. A sliding window median curve was added to highlight the consensus profile (Fig. 5A,B). This revealed two striking features of the distribution of CBX1. First, CBX1 is generally absent from the 5′ ends of KRAB-ZNF genes (Fig. 5A). This region of depletion is ∼1–2 kb in size and centered around the transcription start site. Second, CBX1 is enriched toward the 3′ ends of KRAB-ZNF genes, with prominent binding in the final ∼2 kb (Fig. 5B). The 3′ end enrichment is independent of gene length (data not shown). The peak of enrichment is located within the genes rather than downstream, indicating that the 3′ ends of KRAB-ZNF transcription units contain a signal that recruits CBX1.
Figure 5.
Detailed analysis of CBX1 binding to KRAB-ZNF genes and LINEs. (A,B) CBX1 is depleted from promoter regions and enriched at 3′ ends of genes. All KRAB-ZNF genes on Chr19 were aligned by their 5′ (A) or 3′ (B) ends, and combined CBX1 binding data were plotted. Each light gray dot represents one tiling array probe and only tiling array probes that are located <5 kb upstream or within genes (A) or <5 kb downstream or within genes (B) are plotted. Black lines depict the sliding window median value (window size = 101 probes). (C) Enrichment at 3′ ends of KRAB-ZNF genes is independent of LINE sequences. Sliding window medians are shown for 95 KRAB-ZNF genes that do not contain any LINE-related sequences (gray) and 78 KRAB-ZNF genes that contain at least one LINE-related sequence (black). (D) LINEs in KRAB-ZNF gene domains are embedded in CBX1-bound sequences. Each dot represents one tiling array probe. Lines depict sliding window median CBX1 binding values of probes surrounding LINE-sequences (defined as having at least 1000 bp LINE homology) that are located <200 kb (black line, dark gray dots) or >200 kb (gray line, light gray dots) from a KRAB-ZNF gene. (E) Scheme of CBX1 binding to KRAB-ZNF genes and LINEs within and outside KRAB-ZNF gene domains.
Because mammalian genes often contain (remnants of) LINEs in their intronic sequences (Li et al. 2001), we wondered whether intragenic LINEs might be responsible for CBX1 recruitment to the KRAB-ZNF genes. We therefore compared the CBX1 binding profiles of LINE-containing KRAB-ZNF genes and LINE-free KRAB-ZNF genes (Fig. 5C). No clear differences could be observed, indicating that KRAB-ZNF genes recruit CBX1 independently of the presence of LINEs (Fig. 5C).
Even though the previous result did not support a direct role for LINEs in the recruitment of CBX1, we performed an additional analysis to address this issue. The Chr19 tiling array does not directly provide information about the binding of CBX1 to LINE elements. However, if LINEs are important recruitment sites for CBX1, then it may be expected that sequences immediately flanking LINEs show elevated binding ratios, because targeted Dam methylation spreads ∼1–2 kb from a protein binding site (van Steensel and Henikoff 2000; Sun et al. 2003). To test this, we aligned all LINE elements on Chr19 and plotted CBX1 binding to neighboring sequences (Fig. 5D). We calculated a sliding window curve of binding next to LINEs that are located within KRAB-ZNF gene domains and a second curve for LINEs outside these domains. As expected, within KRAB-ZNF gene domains the sequences neighboring LINEs showed elevated CBX1 binding, while this was not the case on the remainder of Chr19. No gradient of binding was observed in the first 2 kb from the LINE elements, suggesting that binding of CBX1 to LINEs is not strongly enriched relative to the surrounding sequences.
Figure 5E summarizes these results schematically. Our data show that within KRAB-ZNF gene domains, binding of CBX1 is generally elevated. Only at transcription start sites is CBX1 binding reduced, and at 3′ ends of KRAB-ZNF genes the binding is enriched. Binding to LINEs may only occur in KRAB-ZNF gene domains, although direct measurements at individual LINE copies will be required to confirm this.
CBX1 binding to KRAB-ZNF genes is conserved in mouse
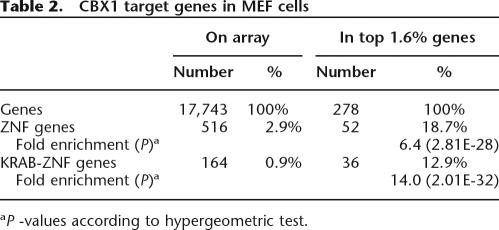
KRAB-ZNF gene clusters undergo rapid evolution. Homologous human and mouse KRAB-ZNF gene clusters contain different numbers of genes and show evidence of gene loss and gain since the divergence of the primate and rodent lineages (Dehal et al. 2001). To test whether CBX1 also binds to KRAB-ZNF genes in mouse, we mapped CBX1 binding in Mouse Embryonic Fibroblasts (MEFs). For detection of CBX1-targeted methylation, we used microarrays with 26,962 oligonucleotides corresponding to coding regions of the mouse genome. In MCF7 cells we found that 1.6% of probed genes were targets of CBX1 (Table 1). To allow direct comparison between the mouse and human data, we selected the top 1.6% of genes bound by Cbx1 in MEFs. This gene list is also significantly enriched for KRAB-ZNF genes (Table 2). This result indicates that the preferential binding of CBX1 to KRAB-ZNF genes is conserved between mouse and human cells. In addition, because MEFs are a different cell type than human MCF7 cells, this result shows that binding of CBX1 to KRAB-ZNF genes is not limited to a single cell type.
Table 2.
CBX1 target genes in MEF cells
aP -values according to hypergeometric test.
KRAB-ZNF genes are partially coexpressed in human tissues
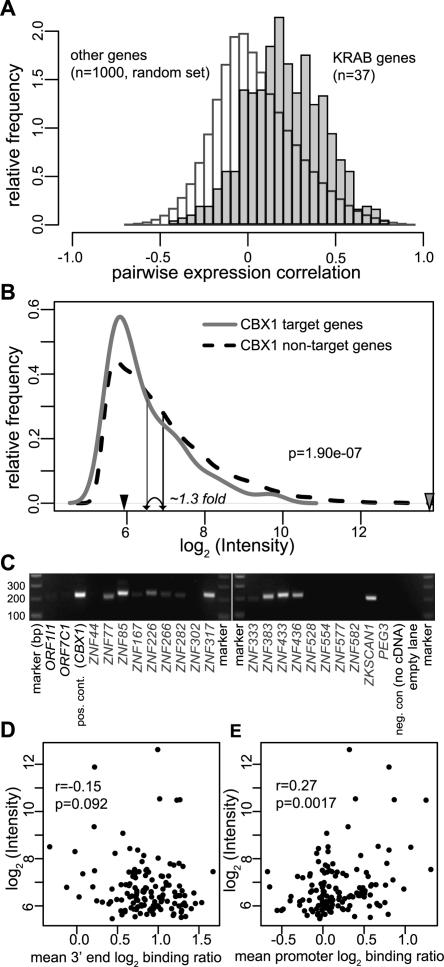
Our observation that most KRAB-ZNF genes are located in heterochromatin domains raised the possibility that the expression of these genes is coordinately regulated, because they share a similar chromatin environment. We therefore asked whether KRAB-ZNF genes show a certain degree of coexpression. We compared the expression profiles of KRAB-ZNF genes and other genes in a large microarray expression data set from 79 different human tissues (Su et al. 2004). We restricted our analysis to a hand-curated set of microarray probes to KRAB-ZNF genes with minimal predicted cross-hybridization. We reasoned that if KRAB-ZNF genes are coordinately regulated, their expression patterns across these tissues should be correlated. We therefore calculated all pairwise correlation coefficients from the expression profiles of all KRAB-ZNF genes and of a control set of 1000 randomly picked non-KRAB-ZNF genes. The correlation coefficient for any pair of genes can range from −1 (mutually exclusive expression pattern) to +1 (identical expression pattern). The distributions of these pairwise correlation coefficients (Fig. 6A) show that KRAB-ZNF genes have mostly positively correlated expression patterns in human tissues. Although the average correlation between KRAB-ZNF genes is modest (mean r = 0.20), it is clearly higher than a set of randomly selected control genes. The correlation in expression of KRAB-ZNF genes that reside in different domains is almost as strong as the correlation between genes within the same domain (mean r = 0.20 and r = 0.22, respectively), indicating that the coexpression of KRAB-ZNFs is not limited to genes within the same cluster. These results show that the family of KRAB-ZNF genes has a modest degree of overall coexpression in the human body, possibly because the entire family is fine-regulated by heterochromatin.
Figure 6.
Expression of KRAB-ZNF genes and CBX1 target genes. (A) KRAB-ZNF genes are coexpressed in 79 human tissues. Microarray expression data for 37 KRAB-ZNF genes or 1000 randomly picked non-KRAB-ZNF genes were obtained from Su et al. (2004). For each gene pair, the pairwise Pearson correlation in expression throughout 79 different tissues was calculated. Histograms present pairwise correlation coefficients between KRAB-ZNF genes (gray) or non-KRAB-ZNF genes (white). (B) CBX1 target genes have slightly lower expression levels than CBX1 nontarget genes in MCF7 cells. MCF7 expression profiles were made on coding region oligonucleotide arrays and the density distribution of spot intensities (log2 √(Cy5 × Cy3)) of CBX1 target genes (gray line) and CBX1 nontarget genes (dashed black line) is shown. The average spot intensity per group is indicated by vertical arrows. P-value is according to the Wilcoxon rank sum test. The expression levels of an inactive gene (Testis-Specific Serine Kinase Substrate) and a highly active gene (beta-actin) are indicated by black and gray arrowheads, respectively. (C) The majority of KRAB-ZNF genes is expressed in MCF7 cells. RT-PCR of 19 KRAB-ZNF genes. ZNF436, ZNF167, ZKSCAN1, and ZNF282 are located on chromosomes 1, 3, 7, and 7, respectively. The other KRAB-ZNF genes are located on Chr19. Controls include the ORF1I1 and ORF7C1 olfactory receptor genes that are expected to be repressed in MCF7 cells, CBX1 that is expressed in MCF7 cells, and a negative control with CBX1 primers without cDNA. (D,E) Correlation between expression level and CBX1 binding to 3′ end (D) or promoters (E) of KRAB-ZNF genes. Average CBX1 binding to the last 3′ 2 kb or to the 2 kb surrounding the transcriptional start site of 139 KRAB-ZNF genes was linked to the expression data from B. P-values are according to Spearman’s ρ statistic.
Expression levels of CBX1 target genes
The previous analysis does not address whether heterochromatin might repress or activate the KRAB-ZNF genes. To investigate the transcriptional status of the CBX1 target genes, we generated expression profiles by hybridizing labeled mRNA from MCF7 cells to our coding region oligonucleotide array, and used the spot signal intensities as an estimate of the transcript levels in the cells. Figure 6B shows the distribution of expression levels of CBX1 target genes. On average CBX1 target genes had 1.3-fold lower expression levels than nontarget genes. While this difference is statistically significant (P < 1.90 × 10−7, Wilcoxon rank test), it indicates that overall the CBX1 target genes have only slightly lower expression levels than nontarget genes. This was observed for KRAB-ZNF target genes as well as for non-KRAB-ZNF target genes (data not shown). We also investigated the expression status of KRAB-ZNF genes directly by RT-PCR in MCF7 cells. In accordance with the wide distribution of expression levels that was detected on the microarray (Fig. 6B), we could detect transcripts for 12 out of 19 KRAB-ZNFs tested (Fig. 6C).
Because CBX1 binding was most prominent at the 3′ ends of KRAB-ZNF genes (Fig. 5B), we tested whether the binding of CBX1 on the final 2 kb of the Chr19 KRAB-ZNF genes correlated with their expression levels. Only a weak and nonsignificant negative correlation was observed (Fig. 6D). This result is in agreement with Figure 6B, which shows slightly reduced expression of CBX1 targets throughout the genome, which were identified using oligonucleotide probes mostly located at 3′ ends of genes.
Although on average the CBX1 binding to promoters of KRAB-ZNF genes was low, some promoters displayed higher CBX1 levels than others (Fig. 5A). Surprisingly, we found a moderate but significant positive correlation between promoter binding of CBX1 and KRAB-ZNF gene expression levels (Fig. 6E). The opposite correlations between gene expression levels and promoter binding versus 3′ end binding suggests a multifaceted role of CBX1 in gene regulation.
Removal of CBX1 or CBX5 by RNA interference did not detectably affect the expression of the target genes (data not shown), possibly due to redundancy of CBX1 with CBX3 and CBX5. Attempts to circumvent this redundancy by simultaneous RNA interference of all three HP1 proteins have been unsuccessful so far (data not shown). Overexpression of GFP-tagged CBX1 or CBX5 did not detectably affect the expression of target genes either (data not shown). Nevertheless, our expression profiling and RT-PCR results indicate that the majority of CBX1 target genes are expressed, at somewhat lower levels than nontarget genes.
Discussion
Here, we show by genome-wide mapping that the heterochromatin proteins CBX1 and SUV39H1 have a striking preference for KRAB-ZNF genes, which constitute one of the largest gene families in the human genome. Preference for this gene family was observed in human and mouse cells. High resolution mapping of human chromosome 19 revealed that CBX1 forms large domains that coincide with clusters of KRAB-ZNF genes.
Regulatory role of heterochromatin proteins
Heterochromatin can mediate gene silencing as well as gene activation. In a variety of species, insertion of euchromatic reporter genes into heterochromatin leads to silencing or variegation of expression. This has led to the notion that heterochromatin constitutes a repressive environment. In contrast, several observations link heterochromatin to active transcription. For example, some pericentric genes require a heterochromatic environment for their correct expression (Wakimoto and Hearn 1990), and HP1 proteins are found on active genes (Greil et al. 2003; Piacentini et al. 2003; Vakoc et al. 2005). In S. pombe heterochromatic loci are transcribed (Hall et al. 2002; Volpe et al. 2002) and RNA Polymerase II is required for siRNA-mediated heterochromatin formation (Kato et al. 2005). The molecular basis for these opposing roles of heterochromatin in gene regulation is not known. Our data suggest that HP1 proteins may perhaps have different regulatory roles in promoters and 3′ regions. Because the overall correlations between HP1 binding and expression are rather weak, we suggest that the heterochromatic environment may be involved in fine-tuning of the expression of these genes. In addition, heterochromatin may play a role in the evolution of the KRAB-ZNF gene family (see below).
Cooperative mechanism of heterochromatin protein targeting?
Evidence from a variety of organisms indicates that repetitive sequences play a key role in the recruitment of heterochromatin proteins (Henikoff 1998; Martienssen 2003). In agreement with the association between heterochromatin and repetitive sequences, we identified LINE elements as targets of CBX1. We deduced that LINE elements in KRAB-ZNF gene domains are bound by CBX1, whereas LINE elements outside these domains are not. This finding resembles the pattern of HP1 binding to TEs in Drosophila, where individual TE copies located in repeat-poor genomic areas are not bound by HP1 whereas TE copies in repeat-dense regions are. This has led to the proposal that cooperative action of multiple neighboring repeats is necessary for the establishment of a heterochromatin domain (de Wit et al. 2005). Analogous to the situation in Drosophila, LINE elements located in repeat-poor regions outside KRAB-ZNF gene domains may not be able to recruit CBX1. Similarly, the nonclustered KRAB-ZNF genes on Chr19 show relatively weak CBX1 binding. We suggest that cooperative recruitment of heterochromatin proteins may occur in KRAB-ZNF domains, with contributions by the KRAB-ZNF genes themselves, the LINEs, and possibly other repetitive sequences that have been found in these domains (Eichler et al. 1998; Grimwood et al. 2004).
CBX1 binding along KRAB-ZNF genes
Interestingly, we found that CBX1 binding is excluded from the transcription start sites and is especially enriched at the 3′ ends of KRAB-ZNF genes. The absence of CBX1 from transcription start sites may be linked to the previously reported enrichment of “active” histone modifications, such as H3K4 methylation and H3K9/K14 acetylation, at these positions (Bernstein et al. 2005). In particular, H3K9 acetylation is incompatible with CBX1 binding to methylated H3K9, because acetylation and methylation cannot occur on the same lysine residue. Alternatively, the 5′ end of genes may be devoid of nucleosomes (Chen et al. 2005). The enrichment of CBX1 at 3′ ends of KRAB-ZNF genes may be due to a specific recruitment signal in the DNA sequence or be linked to the final stage of transcription elongation or to termination. Future studies may identify the signal that underlies the specific recruitment of heterochromatin proteins to KRAB-ZNF genes.
Heterochromatin and the evolution of KRAB-ZNF genes
A striking feature of the KRAB-ZNF gene family is its rapid expansion in recent evolution. Since its emergence in early tetrapod vertebrates, this gene family has been subject to constant reshaping by gene losses and gene duplications and has expanded to a family of more than 400 genes in the human genome (Dehal et al. 2001; Shannon et al. 2003). For example, a large cluster of KRAB-ZNF genes located in the pericentromeric region of human Chr19 appears to be primate specific (Bellefroid et al. 1995). Sequence comparisons of KRAB-ZNF genes in different clusters indicate that in situ tandem duplications play a prominent role in the creation of new genes (Dehal et al. 2001).
We propose that heterochromatin proteins might function to stabilize KRAB-ZNF gene clusters. In Saccharomyces cerevisiae, heterochromatin protein Sir2 is needed for the genomic maintenance of repeats of ribosomal genes (rDNA). Without Sir2 the rDNA repeats are lost by recombination (Gottlieb and Esposito 1989). Similar to the stabilizing function of Sir2 on rDNA repeats in yeast, heterochromatin in mammals may prevent recombination-mediated deletion of recently duplicated KRAB-ZNF genes and thereby have facilitated the expansion of this gene family.
Several observations are in agreement with this model. Analysis of three Drosophila species has indicated that the size of the histone gene cluster is correlated with heterochromatic localization (Fitch et al. 1990), and indeed the histone and rDNA gene arrays in D. melanogaster are bound by HP1 (van Steensel and Henikoff 2000). Comparative studies of genome organization have shown that pericentric and telomeric heterochromatin are hotbeds of genomic rearrangements in a variety of species (Eichler and Sankoff 2003). More specifically, it has been argued that heterochromatic β-satellite repeat sequences have played an important role in the rapid expansion of one primate specific cluster of KRAB-ZNF genes (Eichler et al. 1998). A unique aspect of this cluster is the occurrence of blocks of classical centromeric β-satellite sequences that are interspersed between the ZNF genes, indicating that these structures were coordinately duplicated (Eichler et al. 1998).
We propose a model in which an early KRAB-ZNF gene acquired CBX1 binding, most likely through a recruitment signal located in the 3′ end of the gene. This gene could successfully duplicate because the tandemly repeated DNA was stabilized by the heterochromatic environment. The heterochromatin-induced stability of KRAB-ZNF genes in successive rounds of duplication may have facilitated the spectacular expansion of this gene family. Our observation that virtually all KRAB-ZNF genes on Chr19 are bound by CBX1 is consistent with this model. While the mechanism by which heterochromatin stabilizes tandem repeats is still unclear, it is interesting to note that heterochromatin proteins inhibit incorrect recombination in the mating type locus and rDNA repeats of S. pombe (Klar and Bonaduce 1991; Cam et al. 2005) and meiotic recombination in Drosophila (Westphal and Reuter 2002). These effects may be related to the hypothesized stabilization of gene duplications by heterochromatin.
Taken together, we suggest a self-reinforcing cycle of events that facilitated expansion of KRAB-ZNF gene clusters during the evolution of tetrapods. In this cycle, heterochromatin may stabilize KRAB-ZNF gene repeats by preventing recombination, and in turn these repeats strengthen heterochromatin binding by cooperative recruitment. Genome-wide maps of heterochromatin will provide a basis for further studies of the roles of heterochromatin in genome evolution and gene regulation.
Methods
Vectors
Maps and sequences of vectors that are used to express N- or C-terminal Dam-fusion proteins can be found at http://www.nki.nl/nkidep/vansteensel. Dam fusion protein expressing vectors were made by cloning phage T4 DAM (kindly provided by Dr. Stanley Hattman, Univ. of Rochester, NY) or E. coli Dam into pIND/V5-HisA (Invitrogen). Subsequently the mouse Cbx1, human CBX3 and CBX5, and SUV39H1 ORFs were added upstream of Dam. Note that mouse Cbx1 and human CBX1 are identical at protein sequence level. Lentiviral Dam-fusion protein transfer constructs (pL) were created by adaptation of HIV-CS-CG (Miyoshi et al. 1998). In addition, we made versions of the pIND and pL vectors that are compatible with the Gateway recombinational cloning system (Rual et al. 2004). This allows fast and easy cloning of open reading frames from several entry-clone collections into our vectors. The pIND and pL vectors contain five hybrid Ecdysone/glucocorticoid Response Elements (E/GREs) and the minimal heat shock promoter (PΔHSP) for inducible expression of the fusion protein. To facilitate detection of the Dam-fusion proteins, a V5 epitope tag was added. The CBX1 point mutations were created with the QuikChange Site-Directed Mutagenesis kit (Stratagene). All constructs were verified by sequencing. GFP-HP1 fusion protein expressing vectors were a kind gift of Dr. R. Dirks (Leiden Univ. Medical Center, The Netherlands).
Tissue culture and immunofluorescence microscopy
Human MCF7 and 293T cells and mouse INK4A−/− embryonal fibroblasts (MEFs), NIH3T3, and B78 cells were maintained in DMEM medium supplemented with 10% FCS and antibiotics. Lentiviral infections were performed as previously described (Dirac and Bernards 2003).
For immunofluorescence microscopy, B78 and NIH3T3 cells were transfected with Dam expressing vectors together with regulator vector pVgRXR (Invitrogen) and induced for 24 h with 2 μM Ponasterone A (Invitrogen). Cells were fixed with 2% formaldehyde and fusion proteins were stained with mouse anti-V5 antibody (Invitrogen) and FITC-conjugated anti-mouse antibody. For confocal microscopy MCF7 and NIH3T3 cells were transfected with GFP-HP1 expressing vectors. Endogenous HP1 proteins were stained with mouse anti-HP1 antibodies (anti-CBX1, 1MOD-1A9; anti-CBX3, 2MOD-1G6 and anti-CBX5, 2HP-1H5; Euromedex), and TexasRed-conjugated anti-mouse antibody. DNA was counterstained with DAPI.
DamID, microarrays, expression profiling, RT-PCR
Supplemental Figure S1 gives an overview of a DamID experiment. Detailed protocols of the DamID procedure are available at http://www.nki.nl/nkidep/vansteensel.
We have used three different microarray platforms in this study: (1) 70-mer oligonucleotide arrays containing the Operon v3 human 35K or mouse 32K libraries (Operon Biotechnologies); (2) an 18k human cDNA array containing PCR-amplified cDNAs from the Research Genetics Human Sequence Verified library (Laoukili et al. 2005); these arrays were printed by the NKI microarray facility; (3) genomic tiling arrays that cover the whole Chr19 excluding repetitive regions, the ENCODE regions (hg16 based on NCBI build 34) that were obtained from http://genome.ucsc.edu/ENCODE/, a few selected other regions, and promoters with flanking sequences of 22,612 genes (NCBI build 34, v31) with on average 4.6 probes per promoter. This array was designed by us and printed and hybridized by NimbleGen. The average 60-mer probe spacing is 200 bp. Probe sequences do not contain GATC, the recognition sequence of Dam. Expression profiles of MCF7 cells were made as described (Laoukili et al. 2005) using the human 35k oligonucleotide array. Fluorescence intensities were quantified using ImaGene software (BioDiscovery Inc.), background corrected, and normalized using a LOWESS fit per subarray, as described earlier (Yang et al. 2002). Detailed protocols for RNA isolation, amplification, labeling, and hybridization can be found at http://microarrays.nki.nl/download/protocols.html. The RT-PCR was performed on MCF7 cDNA that was prepared from DNAse-treated total RNA from MCF7 cells. Primer pairs were designed to produce ∼210 bp products and span one or more introns, except for ORF1I1 and ORF7C1, which are intronless genes.
DamID normalization and error model
All statistical analyses were performed in the R language (http://www.r-project.org/).
The CBX1 and SUV39H1 DamID data were normalized using R-packages limma (Smyth 2004) 2.0.8 and vsn (Huber et al. 2002) 1.6.3 (available at http://www.r-project.org/). Raw data files were loaded in R and the weight of control spots and poor quality spots were set to zero, to exclude them from the results. We did not apply a background correction to the data. DamID data were normalized between different arrays (method = “vsn”). We only considered features with average positive log2 binding ratios to be targets. Limma’s linear modeling approach was used to determine significance levels of fusion protein binding for each probed sequence. P-values were corrected for multiple testing in limma using the Benjamini and Hochberg method (adjust = “fdr”). DamID data from cDNA arrays was normalized as described above, except that within-array normalization (method = “loess”) was used. Genomic tiling array data were normalized as described above, except that raw data files were loaded in R and within-array normalization (method = “loess”) and between array normalization (method = “Aquantile”) were used.
Annotations
The coding region oligonucleotide arrays were designed using previous releases of the human and mouse genome sequences. Therefore, we reannotated these arrays using the most recent sequence releases (NCBI build 35 for human, NCBI build 34 for mouse). Oligonucleotide sequences were matched to the human or mouse genomes using MEGABLAST (using default settings). Ensembl gene annotations were used (v31 for human, v33 for mouse genes). Human oligonucleotides with multiple MEGABLAST hits in the genome were subsequently matched by MEGABLAST to RepBase Update (humrep.ref and humsub.ref, volume 10, issue 11) (Jurka et al. 2005) and manually classified into LINEs, SINEs, LTR retrotransposons, and DNA transposons. ZNF genes were defined as genes with InterPro annotation IPR007087. KRAB-ZNF genes were defined as genes with both IPR007087 and IPR001909. To define other gene families we used Ensembl (v36) Gene family IDs.
Coexpression analysis
For analysis of coexpression of genes we used a previously published compendium of expression profiles from 79 different human tissues (Su et al. 2004). Of these profiles, we used only data obtained with the Affymetrix HG-U133A array. To remove probe sets with poorly reproducible signals, we calculated Pearson correlation coefficients (r) between the first and second tissue duplicates accross the 79 tissues, and discarded probe sets with r < 0.3. Ensembl gene identifiers corresponding to the HG-U133A probe sets, as well as their annotations, were downloaded from Ensembl BioMart (v31). Probe sets matching multiple Ensembl genes were discarded, while data from multiple probe sets matching the same gene were averaged. Finally, tissue duplicates were averaged to obtain 79 tissue expression values for each gene. This cleaned-up data set contained expression data for 10,722 unique Ensembl genes. Analysis of KRAB-ZNF gene expression was restricted to a list of hand-curated probe sets with minimal predicted cross-hybridization (http:/znf.llnl.gov).
Acknowledgments
We thank S. Hattman, P. Verschuure, R. Dirks, and I. Verma for plasmids; A. Dirac for help with lentivirus production; members of the NKI Microarray Facility for arrays and technical support; D. Sie for help with GEO submission of microarray data; M. Heimerikx for microarray expression profiling; W. Meuleman for statistical advice; L. Oomen for technical support with microscopy experiments; J. Romans for help with Adobe software; and L. Stubbs, F. van Leeuwen, M. Fornerod, M. van Lohuizen, and members of our laboratory for stimulating discussions. This work was supported by the Dutch Cancer Society and the Netherlands Organization for Scientific Research.
Footnotes
[Supplemental material is available at www.genome.org. The microarray data from this study have been submitted to GEO under accession no. GSE5445. Detailed protocols of the DamID procedure are available at http://www.nki.nl/ nkidep/vansteensel.]
Article published online before print. Article and publication data are at http://www.genome.org/cgi/doi/10.1101/gr.5391806
References
- Aagaard L., Laible G., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Laible G., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Selenko P., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Schmid M., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Dorn R., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Schotta G., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Kuhfittig S., Wolf A., Lebersorger A., Singh P.B., Wolf A., Lebersorger A., Singh P.B., Lebersorger A., Singh P.B., Singh P.B., et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanathan K., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Tanaka K., Torigoe K., Rauscher III F.J., Torigoe K., Rauscher III F.J., Rauscher III F.J. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Thomas J.O., Allshire R.C., Kouzarides T., Allshire R.C., Kouzarides T., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bellefroid E.J., Marine J.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Marine J.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Martial J.A., Ihle J.N., Ward D.C., Ihle J.N., Ward D.C., Ward D.C. Emergence of the ZNF91 Kruppel-associated box-containing zinc finger gene family in the last common ancestor of anthropoidea. Proc. Natl. Acad. Sci. 1995;92:10757–10761. doi: 10.1073/pnas.92.23.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Karlsson E.K., Kulbokas III E.J., Gingeras T.R., Kulbokas III E.J., Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D., Orian A., Delrow J.J., Vazquez J., Rosales-Nieves A.E., Parkhurst S.M., Orian A., Delrow J.J., Vazquez J., Rosales-Nieves A.E., Parkhurst S.M., Delrow J.J., Vazquez J., Rosales-Nieves A.E., Parkhurst S.M., Vazquez J., Rosales-Nieves A.E., Parkhurst S.M., Rosales-Nieves A.E., Parkhurst S.M., Parkhurst S.M. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:e178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasher S.V., Smith B.O., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Smith B.O., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Fogh R.H., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Nietlispach D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Thiru A., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Nielsen P.R., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Broadhurst R.W., Ball L.J., Murzina N.V., Laue E.D., Ball L.J., Murzina N.V., Laue E.D., Murzina N.V., Laue E.D., Laue E.D. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I., Chen X., FitzGerald P.C., Grewal S.I., FitzGerald P.C., Grewal S.I., Grewal S.I. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Chen X., Wang J., Woltring D., Gerondakis S., Shannon M.F., Wang J., Woltring D., Gerondakis S., Shannon M.F., Woltring D., Gerondakis S., Shannon M.F., Gerondakis S., Shannon M.F., Shannon M.F. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell. Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman D.E., Grade S.K., Li Y., Fanti L., Pimpinelli S., Wallrath L.L., Grade S.K., Li Y., Fanti L., Pimpinelli S., Wallrath L.L., Li Y., Fanti L., Pimpinelli S., Wallrath L.L., Fanti L., Pimpinelli S., Wallrath L.L., Pimpinelli S., Wallrath L.L., Wallrath L.L. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- Dehal P., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Land M., Terry A., Ecale Zhou C.L., Rash S., Terry A., Ecale Zhou C.L., Rash S., Ecale Zhou C.L., Rash S., Rash S., et al. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- de Wit E., Greil F., van Steensel B., Greil F., van Steensel B., van Steensel B. Genome-wide HP1 binding in Drosophila: Developmental plasticity and genomic targeting signals. Genome Res. 2005;15:1265–1273. doi: 10.1101/gr.3198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P., Corradini N., Rossi F., Verni F., Corradini N., Rossi F., Verni F., Rossi F., Verni F., Verni F. The paradox of functional heterochromatin. Bioessays. 2005;27:29–41. doi: 10.1002/bies.20158. [DOI] [PubMed] [Google Scholar]
- Dirac A.M., Bernards R., Bernards R. Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J. Biol. Chem. 2003;278:11731–11734. doi: 10.1074/jbc.C300023200. [DOI] [PubMed] [Google Scholar]
- Eichler E.E., Sankoff D., Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Eichler E.E., Hoffman S.M., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Hoffman S.M., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., McCready P., Lamerdin J.E., Mohrenweiser H.W., Lamerdin J.E., Mohrenweiser H.W., Mohrenweiser H.W. Complex β-satellite repeat structures and the expansion of the zinc finger gene cluster in 19p12. Genome Res. 1998;8:791–808. doi: 10.1101/gr.8.8.791. [DOI] [PubMed] [Google Scholar]
- Festenstein R., Pagakis S.N., Hiragami K., Lyon D., Verreault A., Sekkali B., Kioussis D., Pagakis S.N., Hiragami K., Lyon D., Verreault A., Sekkali B., Kioussis D., Hiragami K., Lyon D., Verreault A., Sekkali B., Kioussis D., Lyon D., Verreault A., Sekkali B., Kioussis D., Verreault A., Sekkali B., Kioussis D., Sekkali B., Kioussis D., Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- Fitch D.H., Strausbaugh L.D., Barrett V., Strausbaugh L.D., Barrett V., Barrett V. On the origins of tandemly repeated genes: Does histone gene copy number in Drosophila reflect chromosomal location? Chromosoma. 1990;99:118–124. doi: 10.1007/BF01735327. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Boyle S., Sutherland H., de Las Heras J., Allan J., Jenuwein T., Bickmore W.A., Boyle S., Sutherland H., de Las Heras J., Allan J., Jenuwein T., Bickmore W.A., Sutherland H., de Las Heras J., Allan J., Jenuwein T., Bickmore W.A., de Las Heras J., Allan J., Jenuwein T., Bickmore W.A., Allan J., Jenuwein T., Bickmore W.A., Jenuwein T., Bickmore W.A., Bickmore W.A. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 2003;22:5540–5550. doi: 10.1093/emboj/cdg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R.E., Esposito R.E. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Greil F., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B., van Driel R., Henikoff S., van Steensel B., Henikoff S., van Steensel B., van Steensel B. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes & Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwood J., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Gordon L.A., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Olsen A., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Terry A., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Schmutz J., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Lamerdin J., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Hellsten U., Goodstein D., Couronne O., Tran-Gyamfi M., Goodstein D., Couronne O., Tran-Gyamfi M., Couronne O., Tran-Gyamfi M., Tran-Gyamfi M., et al. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Hall I.M., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I., Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I., Noma K., Ayoub N., Cohen A., Grewal S.I., Ayoub N., Cohen A., Grewal S.I., Cohen A., Grewal S.I., Grewal S.I. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Haraguchi T., Masumoto H., Hiraoka Y., Haraguchi T., Masumoto H., Hiraoka Y., Masumoto H., Hiraoka Y., Hiraoka Y. Cell cycle behavior of human HP1 subtypes: Distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 2003;116:3327–3338. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Huber W., von Heydebreck A., Sultmann H., Poustka A., Vingron M., von Heydebreck A., Sultmann H., Poustka A., Vingron M., Sultmann H., Poustka A., Vingron M., Poustka A., Vingron M., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. (Suppl. 1) [DOI] [PubMed] [Google Scholar]
- Jacobs S.A., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Taverna S.D., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Zhang Y., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Briggs S.D., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Li J., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Eissenberg J.C., Allis C.D., Khorasanizadeh S., Allis C.D., Khorasanizadeh S., Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T.C., Eissenberg J.C., Craig C., Dietrich V., Hobson A., Elgin S.C., Eissenberg J.C., Craig C., Dietrich V., Hobson A., Elgin S.C., Craig C., Dietrich V., Hobson A., Elgin S.C., Dietrich V., Hobson A., Elgin S.C., Hobson A., Elgin S.C., Elgin S.C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kato H., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y., Goto D.B., Martienssen R.A., Urano T., Furukawa K., Murakami Y., Martienssen R.A., Urano T., Furukawa K., Murakami Y., Urano T., Furukawa K., Murakami Y., Furukawa K., Murakami Y., Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Klar A.J., Bonaduce M.J., Bonaduce M.J. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2–mat3 “cold spot” of fission yeast. Genetics. 1991;129:1033–1042. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R.D., Shimeld S.M., Shimeld S.M. Identification of conserved C2H2 zinc-finger gene families in the Bilateria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-research0016. research0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T., O’Carroll D., Rea S., Mechtler K., Jenuwein T., Rea S., Mechtler K., Jenuwein T., Mechtler K., Jenuwein T., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laoukili J., Kooistra M.R., Bras A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H., Kooistra M.R., Bras A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H., Bras A., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H., Kauw J., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H., Kerkhoven R.M., Morrison A., Clevers H., Medema R.H., Morrison A., Clevers H., Medema R.H., Clevers H., Medema R.H., Medema R.H. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Li W.H., Gu Z., Wang H., Nekrutenko A., Gu Z., Wang H., Nekrutenko A., Wang H., Nekrutenko A., Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- Li Y., Kirschmann D.A., Wallrath L.L., Kirschmann D.A., Wallrath L.L., Wallrath L.L. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. 2002;99:16462–16469. doi: 10.1073/pnas.162371699. (Suppl. 4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Lavine K., Mittal V., May B., Kasschau K.D., Mittal V., May B., Kasschau K.D., May B., Kasschau K.D., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Liu L.P., Ni J.Q., Shi Y.D., Oakeley E.J., Sun F.L., Ni J.Q., Shi Y.D., Oakeley E.J., Sun F.L., Shi Y.D., Oakeley E.J., Sun F.L., Oakeley E.J., Sun F.L., Sun F.L. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat. Genet. 2005;37:1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- Maison C., Almouzni G., Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G., Taddei A., Lachner M., Jenuwein T., Almouzni G., Lachner M., Jenuwein T., Almouzni G., Jenuwein T., Almouzni G., Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Martienssen R.A. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- Minc E., Allory Y., Worman H.J., Courvalin J.C., Buendia B., Allory Y., Worman H.J., Courvalin J.C., Buendia B., Worman H.J., Courvalin J.C., Buendia B., Courvalin J.C., Buendia B., Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- Minc E., Courvalin J.C., Buendia B., Courvalin J.C., Buendia B., Buendia B. HP1γ associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Blomer U., Takahashi M., Gage F.H., Verma I.M., Blomer U., Takahashi M., Gage F.H., Verma I.M., Takahashi M., Gage F.H., Verma I.M., Gage F.H., Verma I.M., Verma I.M. Development of a self-inactivating lentivirus vector. J. Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C., Sun L.V., Wang J., de Wit E., Talhout W., Ward L.D., Greil F., Lu X., White K.P., Bussemaker H.J., Sun L.V., Wang J., de Wit E., Talhout W., Ward L.D., Greil F., Lu X., White K.P., Bussemaker H.J., Wang J., de Wit E., Talhout W., Ward L.D., Greil F., Lu X., White K.P., Bussemaker H.J., de Wit E., Talhout W., Ward L.D., Greil F., Lu X., White K.P., Bussemaker H.J., Talhout W., Ward L.D., Greil F., Lu X., White K.P., Bussemaker H.J., Ward L.D., Greil F., Lu X., White K.P., Bussemaker H.J., Greil F., Lu X., White K.P., Bussemaker H.J., Lu X., White K.P., Bussemaker H.J., White K.P., Bussemaker H.J., Bussemaker H.J., et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc. Natl. Acad. Sci. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Hennetin J., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G., Hennetin J., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G., Sun L.V., Lavrov S., Bellis M., White K.P., Cavalli G., Lavrov S., Bellis M., White K.P., Cavalli G., Bellis M., White K.P., Cavalli G., White K.P., Cavalli G., Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., Ortiz J.A., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., You J., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., Oulad-Abdelghani M., Khechumian R., Gansmuller A., Chambon P., Losson R., Khechumian R., Gansmuller A., Chambon P., Losson R., Gansmuller A., Chambon P., Losson R., Chambon P., Losson R., Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R., Ortiz J.A., Remboutsika E., Chambon P., Losson R., Remboutsika E., Chambon P., Losson R., Chambon P., Losson R., Losson R. Heterochromatin formation in mammalian cells: Interaction between histones and HP1 proteins. Mol. Cell. 2001a;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- Nielsen S.J., Schneider R., Bauer U.M., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., Schneider R., Bauer U.M., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., Bauer U.M., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., Firestein R., Cleary M., Jenuwein T., Herrera R.E., Cleary M., Jenuwein T., Herrera R.E., Jenuwein T., Herrera R.E., Herrera R.E., et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001b;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- No D., Yao T.P., Evans R.M., Yao T.P., Evans R.M., Evans R.M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Allis C.D., Grewal S.I., Allis C.D., Grewal S.I., Grewal S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- O’Carroll D., Scherthan H., Peters A.H., Opravil S., Haynes A.R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Scherthan H., Peters A.H., Opravil S., Haynes A.R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Peters A.H., Opravil S., Haynes A.R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Opravil S., Haynes A.R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Haynes A.R., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Laible G., Rea S., Schmid M., Lebersorger A., Jerratsch M., Rea S., Schmid M., Lebersorger A., Jerratsch M., Schmid M., Lebersorger A., Jerratsch M., Lebersorger A., Jerratsch M., Jerratsch M., et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A. Chromatin profiling, DamID and the emerging landscape of gene expression. Curr. Opin. Genet. Dev. 2006;16:157–164. doi: 10.1016/j.gde.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., Li L., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., Sawado T., Williams E., Loo L.W., Cowley S.M., Yost C., Williams E., Loo L.W., Cowley S.M., Yost C., Loo L.W., Cowley S.M., Yost C., Cowley S.M., Yost C., Yost C., et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L., Fanti L., Berloco M., Perrini B., Pimpinelli S., Fanti L., Berloco M., Perrini B., Pimpinelli S., Berloco M., Perrini B., Pimpinelli S., Perrini B., Pimpinelli S., Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J.S., Hartnett T., Eissenberg J.C., Hartnett T., Eissenberg J.C., Eissenberg J.C. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Mechtler K., Ponting C.P., Allis C.D., Ponting C.P., Allis C.D., Allis C.D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Hirozane-Kishikawa T., Hao T., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Hirozane-Kishikawa T., Hao T., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Hao T., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Rosenberg J., Lamesch P., Vidalain P.O., Lamesch P., Vidalain P.O., Vidalain P.O., et al. Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M., Hamilton A.T., Gordon L., Branscomb E., Stubbs L., Hamilton A.T., Gordon L., Branscomb E., Stubbs L., Gordon L., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.V., Chen L., Greil F., Negre N., Li T.R., Cavalli G., Zhao H., Van Steensel B., White K.P., Chen L., Greil F., Negre N., Li T.R., Cavalli G., Zhao H., Van Steensel B., White K.P., Greil F., Negre N., Li T.R., Cavalli G., Zhao H., Van Steensel B., White K.P., Negre N., Li T.R., Cavalli G., Zhao H., Van Steensel B., White K.P., Li T.R., Cavalli G., Zhao H., Van Steensel B., White K.P., Cavalli G., Zhao H., Van Steensel B., White K.P., Zhao H., Van Steensel B., White K.P., Van Steensel B., White K.P., White K.P. Protein–DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. 2003;100:9428–9433. doi: 10.1073/pnas.1533393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiru A., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Nietlispach D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Mott H.R., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Okuwaki M., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Lyon D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Nielsen P.R., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Hirshberg M., Verreault A., Murzina N.V., Laue E.D., Verreault A., Murzina N.V., Laue E.D., Murzina N.V., Laue E.D., Laue E.D. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., Muijrers I., de Wit E., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., Muijrers I., de Wit E., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., de Wit E., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., Talhout W., van Steensel B., van Lohuizen M., van Steensel B., van Lohuizen M., van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A., Mandat S.A., Olenchock B.A., Blobel G.A., Olenchock B.A., Blobel G.A., Blobel G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Henikoff S., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Henikoff S., Delrow J., Henikoff S., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Bussemaker H.J., Delrow J., Bussemaker H.J., Bussemaker H.J. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. 2003;100:2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Hall I.M., Teng G., Grewal S.I., Martienssen R.A., Teng G., Grewal S.I., Martienssen R.A., Grewal S.I., Martienssen R.A., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wakimoto B.T., Hearn M.G., Hearn M.G. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler K.S., Wakimoto B.T., Wakimoto B.T. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. SINEs and LINEs: The art of biting the hand that feeds you. Curr. Opin. Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- Westphal T., Reuter G., Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P., Lin D.M., Peng V., Ngai J., Speed T.P., Peng V., Ngai J., Speed T.P., Ngai J., Speed T.P., Speed T.P. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]