Abstract
We have characterized the tomato (Lycopersicon esculentum Mill.) MADS box gene TM29 that shared a high amino acid sequence homology to the Arabidopsis SEP1, 2, and 3 (SEPALLATA1, 2, and 3) genes. TM29 showed similar expression profiles to SEP1, with accumulation of mRNA in the primordia of all four whorls of floral organs. In addition, TM29 mRNA was detected in inflorescence and vegetative meristems. To understand TM29 function, we produced transgenic tomato plants in which TM29 expression was down-regulated by either cosuppression or antisense techniques. These transgenic plants produced aberrant flowers with morphogenetic alterations in the organs of the inner three whorls. Petals and stamens were green rather than yellow, suggesting a partial conversion to a sepalloid identity. Stamens and ovaries were infertile, with the later developing into parthenocarpic fruit. Ectopic shoots with partially developed leaves and secondary flowers emerged from the fruit. These shoots resembled the primary transgenic flowers and continued to produce parthenocarpic fruit and additional ectopic shoots. Based on the temporal and spatial expression pattern and transgenic phenotypes, we propose that TM29 functions in floral organ development, fruit development, and maintenance of floral meristem identity in tomato.
Flower development has been the subject of intensive studies over the last decade, particularly in the model plants Arabidopsis and snapdragon (Antirrhinum majus). These studies led to the formulation of the ABC model of floral organ identity, which explained the activities of three classes of genes in specifying the identity of floral organs (Weigel and Meyerowitz, 1994). This model has been supported by genetic and molecular data in a wide range of angiosperm species.
According to the ABC model, expression of a class A gene specifies the formation of sepals (the first whorl organ); in combination with the class B genes expression, specifies petal formation. Expression of class B genes and a class C gene specifies stamen identity, whereas expression of C alone determines a carpel identity (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). Most of the ABC genes belong to the MADS box family (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994).
Although the ectopic expressions of the ABC genes are sufficient to determine various floral organ identities within the floral meristem, they are insufficient to convert vegetative leaves to floral organs. This suggested that other regulators, in addition to the ABC genes, are required for floral organ specification. Recently, a group of three related MADS box genes SEP 1, 2, and 3 (SEPALLATA 1, 2, and 3) were shown to be necessary for the activity B and C class genes in the control of floral organ formation. First, the SEP1, SEP2, and SEP3 (formerly AGL2, AGL4, and AGL9) redundantly control the activities of the B and C organ identity genes in Arabidopsis because the triple mutant sep1sep2sep3 flower consists entirely of sepals. The sep1/2/3 triple mutant phenotype strikingly resembles bc (ap3 ag and pi ag) double mutants, indicating the B and C organ identity genes are inactive in the triple mutant (Pelaz et al., 2000). Second, it has been shown that ectopic expressions of SEP3 together with B and C genes can convert leaves into floral organs (Honma and Goto, 2001; Pelaz et al., 2001b).
An additional phenotype of the triple sep1/2/3 mutant is that a secondary sepallata flower replaces the fourth whorl of the primary flower. The formation of the secondary flower indicates that the sep1/2/3 triple mutant loses floral meristem determinacy, a phenotype that is described for the ag mutant (Yanofsky et al., 1990). Under noninductive photoperiod (short day), the ag mutant produces ectopic shoots after the formation of several whorls of floral organs, indicating that the floral meristem is reprogrammed and undergoes floral meristem reversion (Okamuro et al., 1996). However, floral meristem reversion has not yet reported for the triple sep1/2/3 mutant.
Although the three SEP genes clearly have overlapping functions, they may have separate functions independent of each other (Pelaz et al., 2001a). Similarly, there are significant functional differences observed among the SEP orthologues in different species. The down-regulation of GRCD1 (Gerbera Regulator of Capitulum Development 1), an SEP1 homolog, caused homeotic changes in the sterile staminoides of female florets to petals (Kotilainen et al., 2000). Three SEP-like MADS box genes (OsMADS5, OsMADS7, and OsMADS8) in rice (Oryza sativa) are found to control flowering time (Kang and An, 1997; Kang et al., 1997). The down-regulation of tomato (Lycopersicon esculentum Mill.) MADS box 5 (TM5), a SEP3 homolog resulted in additional whorls per flower, the wrong number of organs in each whorl and alterations in the inner three whorls of transgenic flowers with green petals, dialytic stamens, and sterile carpels (Pneuli et al., 1994a). Also, the cosuppression of FBP2 (Petunia Floral Binding Protein 2), a SEP3 homolog, resulted in green petals and homeotic replacement of stamens by green petaloid structures as well as the development of ectopic inflorescences in the fourth whorl (Angenent et al., 1994).
To analyze SEP functions in tomato and gain insight into conservation of SEP function among different species, we characterized a SEP homolog from tomato, TM29 (Tomato MADS-box 29) and examined its function using cosuppression and antisense techniques. TM29 is expressed in over a wide range of developmental stages in the tomato flower. Down-regulation of TM29 results in aberrant phenotypes in the inner whorls that resembled those obtained with down-regulation of FBP2 and TM5, two other SEP-like genes. Ectopic shoots that form in the fourth whorl produced flowers and leaf structures, suggesting that TM29 may not only function in floral organ development but also in maintenance of a determinate floral identity.
RESULTS
TM29 Groups to the SEP Subfamily
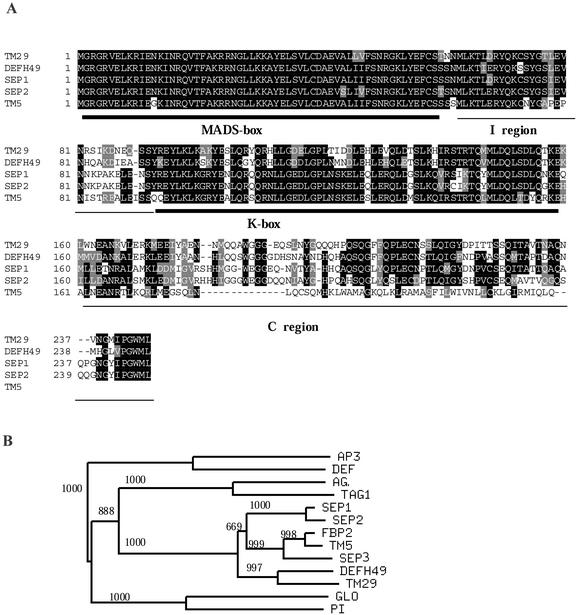
TM29 was isolated from a young tomato fruit cDNA library as part of experiments to identify MADS box genes regulating fruit development. The longest cDNA clone of TM29 identified was 1,231 bp long and potentially encodes 246 amino acids (EMBL database accession no. AJ302015), which comprise the four regions of typical plant MADS box genes: the MADS box, intergenic region, the K box, and the C-terminal region (Fig. 1A). Southern-blot hybridization analysis suggests that there is only one copy of TM29 in the tomato genome (data not shown).
Figure 1.
Sequence analysis of TM29. A, An alignment of predicted protein sequences from related MADS box genes using ClustalW analysis. Gaps were introduced to maximize alignment. The MADS box, K box, I region, and C region are identified. The five amino residues at the 3′ terminal were conserved among TM29, DEFH49, AGL2, and AGL4. B, Phylogenetic analysis of selected MADS box proteins using sequences from the MADS box, the I region, and the K box regions. Bootstrap values are shown on branches. Branches with less than 50% of bootstrap support are collapsed. Accession numbers: AG, P17839; FBP2, JQ1690; SEP1, P29382; SEP2, P29384; SEP3, O22456; AP3, P35632; DEF, P23706; DEFH49, S78015; GLO, Q03378; PI, P48007; TAG1, Q40168; and TM5, Q42464.
TM29 predicted protein has 78% identity to DEFH49 (Davies et al., 1996) of snapdragon and 68%, 63%, and 58% identity to SEP1 (AGL2), SEP2 (AGL4), and SEP3 (AGL9), respectively (Ma et al., 1991; Mandel and Yanofsky, 1998) of Arabidopsis. Among the known MADS box genes in tomato, TM5 (Pneuli et al., 1994a) was the closest in identity (72% over the M, I, and K regions) to TM29. Phylogenetic analysis using amino acid sequences from the MADS box, the I box, and the K box regions assigned TM29 to the SEP subfamily (Fig. 1B). Within the SEP subfamily, conserved residues at the tail end of the C-terminal region were revealed (Fig. 1A).
Spatial and Temporal Expression of TM29 in Tomato
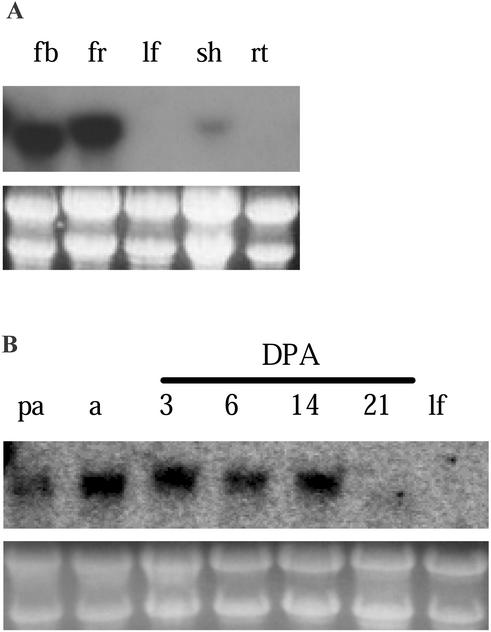
TM29 transcripts were detected in various plant organs using northern-blot hybridization. The transcripts accumulated to a high level in flower buds (0.1–3-mm diameter) and young fruits (1–7 d old; Fig. 2A). However, the level of TM29 mRNA was low in shoot tips and undetectable in leaves and roots. In a further experiment with fruit tissues, TM29 transcripts were detected in ovaries (the initial fruit tissues) pre-anthesis and at anthesis, and in 3-, 6-, and 14-d-old fruits, but not in 21-d-old fruits or young leaves (Fig. 2B). Together, these northern results show TM29 expression occurs from the early stages of flower development to the early stages of fruit development.
Figure 2.
Northern analysis of TM29 expression. Northern blots were probed with a TM29 cDNA fragment. Loading levels of RNA samples are shown by the gel photograph of stained rRNA bands. A, Total RNA extracted from tomato flower buds (fb), 1- to 7-DPA fruits (fr), young leaves (lf), shoot tips (sh), and roots (rt). B, RNA extracted from ovary at pre-anthesis (pa) and anthesis (a); fruit at 3, 6, 14, and 21 DPA; and young leaves (lf).
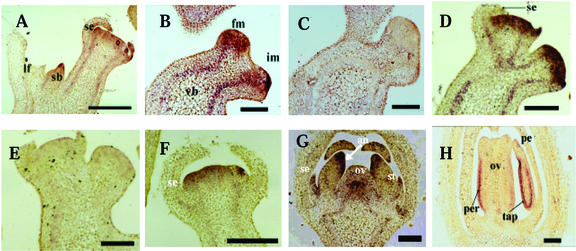
In situ hybridization was used to further examine the temporal and spatial expression pattern of TM29 in meristems and developing floral organs. The vegetative meristems in tomato are responsible for primary and sympodial shoot growth until termination by their conversion to inflorescence meristems (Schmitz and Theres, 1999). The inflorescence meristem then divides to give a floral meristem and an inflorescence meristem (Allen and Sussex, 1996). TM29 transcripts were detected in vegetative meristems at the tip of the sympodial meristem (Fig. 3A) and also in inflorescence and floral meristems before the emergence of floral organ primordia (Fig. 3B). In contrast, SEP1 transcripts are floral specific and absent from inflorescence and vegetative meristems (Flanagan and Ma, 1994). The presence of TM29 transcripts in the vegetative and inflorescence meristems suggests that TM29 may have additional functions compared with SEP1.
Figure 3.
In situ analysis of TM29 expression in different tissues in wild-type (WT) tomato. A, Sympodial bud in the axil of a leaf showing TM29 expression at the tip. B, Bifurcating structure with a floral (fm) and an inflorescence (im) meristem. TM29 is expressed uniformly in the floral meristem and strongly at the tip of the inflorescence meristem. Transcripts are also seen in the vascular bundles (vb). C, Tissue section as in B probed with sense RNA as negative control. D, Section showing floral meristems with emerging sepal primordia at different stages. E, Tissue section as in D probed with sense RNA as negative control. F, A floral meristem with elongated sepals. TM29 expression in the sepals is markedly reduced at this stage. G, Flower bud with all four floral organ primordia. TM29 expression is reduced in the petals and is localized to the tip of the stamens where the anthers (an) are formed. Expression continues to be seen in the center of the ovary (ov). H, Flower bud at about 4 d pre-anthesis. TM29 expression is localized to the pericarp (per) region of the ovary and in the tapetum (tap) regions of the stamens. an, Anther region; fm, floral meristem; im, inflorescence meristem; lf, leaf; ov, ovary primordium; per, pericarp region; sb, sympodial bud; se, sepal primordium; st, stamen; tap, tapetal region; vb, vascular bundle. Scale bar = 1.5 mm.
TM29 expression was observed in the primordia of all four types of floral organs. TM29 mRNA was detected in the sepal primordia when they emerged on the flanks of the floral meristem (Fig. 3D) but not in older sepal primordia or mature sepals (Figs. 3, D and F). TM29 expression during petal development was similar to that of sepal, i.e. transcripts were detected in emerging petal primordia but not in mature petals (Fig. 3, F–H). In a similar fashion, TM29 expression was detected in emerging stamen primordia (not shown). The expression was observed in young anthers (Fig. 3G) and in the tapetal region of developed anthers (Fig. 3H). TM29 was also expressed in the region occupied by the fourth whorl throughout flower development (Figs. 3, D, F, and G). There was uniform expression in the ovary primordium at early stages, but the expression was mainly in the peripheral region of the well-differentiated ovary (Fig. 3H). Tissues that were probed with sense RNA as negative controls did not show any signals above background level (Fig. 3, C and E). Together, these results show that TM29 mRNA level is high in primordia of all floral organs and diminishes as each organ develops and mature. This expression pattern is similar to that of SEP1 in Arabidopsis.
TM29 Antisense and Cosuppression Transgenic Plants Produced Sepallata-Like Flowers, Parthenocarpic Fruit, and Ectopic Shoots
To examine the function of TM29, tomato transgenic plants were generated to express the sense and antisense RNA of TM29 under control of the constitutive 35S promoter. Plants that were regenerated and rooted on kanamycin-containing medium were established in a greenhouse and confirmed to be transgenic by PCR analysis. Of the 22 sense transgenic plants (S/01–S/22), one (S/05) showed aberrant phenotype. A higher proportion of transgenic plants with the antisense gene, six often, showed similar alterations as the S/05 plant (Table I). There were no significant changes to vegetative organs of these transgenic plants. The alterations in morphogenetic features were evident in flowers and fruit of the transgenic lines.
Table I.
Characteristics of transgenic lines showing altered phenotypes
| Plant Labela | Severity of Flower Phenotypeb | Fruits Showing Ectopic Inflorescence or Swollen Carpelsc | Seeds in Fruit |
|---|---|---|---|
| % | |||
| S/05 | +++ | 31.5 (19) | No |
| AS/01 | ++ | 22.2 (11) | No |
| AS/38 | +++ | 9.7 (31) | No |
| AS/45 | +++ | 37.7 (27) | No |
| AS/69 | ++ | 8.7 (23) | No |
| AS/70 | + | 6.7 (15) | No |
| AS/83 | ++ | 5.7 (35) | No |
| WT control | – | 0 (33) | Yes |
S, Sense; AS, antisense.
Severity of flower phenotype was measured as follows: –, normal flower phenotype, yellow petals and stamens; +, petals and stamens are yellow with green streaks; ++, petals have yellowish margins but green midrib; stamen has yellowish green color; and +++, both petals and stamens are completely green.
Nos. in parentheses indicate total nos. of fruit observed on each plant.
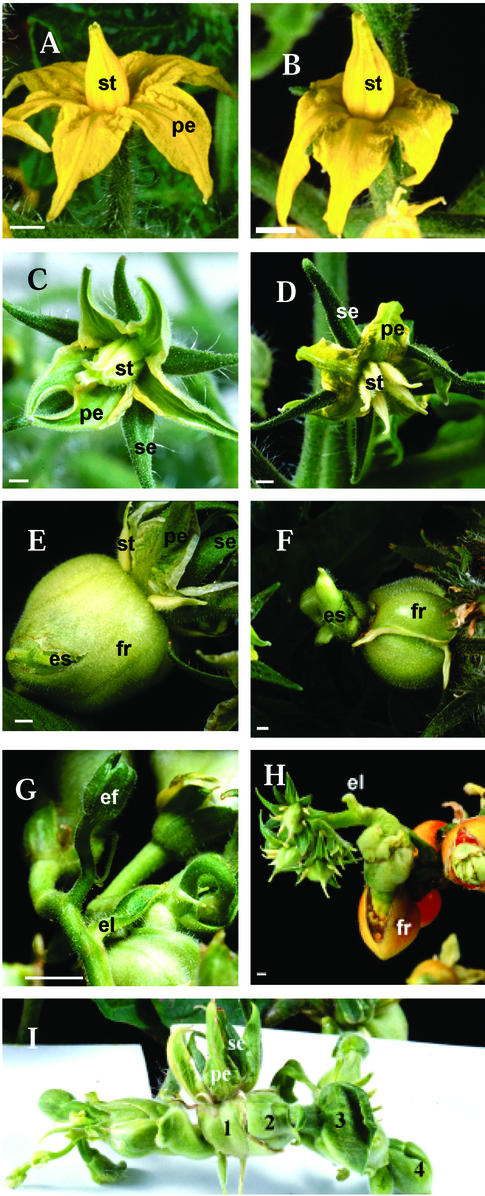
The WT tomato flower consists of four whorls of floral organs with five to six green sepals in the outer whorl, five yellow petals in the second whorl, and five yellow stamens in the third whorl that form a cone surrounding the pistil (Fig. 4A). At 3 DPA, the stamens still formed a cone (Fig. 4B). The fourth whorl is occupied by a carpel with multilocular ovary, a protruded style, and a stigma (Lozano et al., 1998). Tomato flowers normally require pollination and fertilization to set fruit.
Figure 4.
Phenotypes of transgenic tomato plants. A, WT tomato flower at anthesis showing yellow petals and yellow stamens. B, WT flower at 3 DPA with fused stamens. C, Antisense transgenic flower at anthesis showing bigger sepals, green petals, and green stamens forming a loose cone. D, Transgenic flower displaying dialytic stamens. E, Ectopic shoot emerging from the fruit. F, Poorly developed ectopic shoot. G, Ectopic shoot produced flowers and leaf-like struc- tures. H, Ectopic flowers reiterated the development of the primary flower. I, Ectopic shoot showing successive generations (1–4) of ectopic flowers. el, Ectopic leaf; es, ectopic shoot; fr, fruit;. p, petals, se, sepals, st, stamens. Bars = 2 mm.
The sepals in the transgenic flowers largely resembled the WT. Scanning electron micrographs revealed no significant change in the epidermal cells. However, the transgenic sepals were significantly larger (7.99 ± 0.3 mm long and 1.81 ± 0.03 mm wide) than the WT sepals (6.01 ± 0.22 long and 1.44 ± 0.10 mm wide; Table II). The size of the cells in transgenic sepals, inferred from electron micrographs, was not significantly bigger than the WT cells; therefore, this increase in sepal size was attributed to an increase in cell number.
Table II.
Effect of TM29 down-regulation on floral organ sizea
| Sepal
|
Petal
|
Stamen
|
Ovary
|
|||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Length | Width | Length | Width | Length | Width | Length | Width |
| mm | ||||||||
| Control | 6.01 ± 0.22 | 1.44 ± 0.10 | 6.01 ± 0.18 | 1.88 ± 0.11 | 4.68 ± 0.10 | 1.18 ± 0.12 | 5.06 ± 0.10 | 0.89 ± 0.09 |
| S/05 | 8.33 ± 0.27 | 1.75 ± 0.19 | 7.58 ± 0.1 | 1.91 ± 0.08 | 4.70 ± 0.21 | 1.08 ± 0.12 | 5.49 ± 0.20 | 1.58 ± 0.13 |
| AS/01 | 7.66 ± 0.04 | 1.83 ± 0.10 | 7.83 ± 0.30 | 2.49 ± 0.15 | 4.74 ± 0.15 | 0.91 ± 0.11 | 5.41 ± 0.14 | 1.49 ± 0.05 |
| AS/38 | 7.83 ± 0.16 | 1.81 ± 0.11 | 6.91 ± 0.19 | 2.33 ± 0.21 | 4.58 ± 0.16 | 0.99 ± 0.08 | 5.52 ± 0.22 | 1.61 ± 0.10 |
| AS/45 | 8.16 ± 0.20 | 1.83 ± 0.13 | 7.16 ± 0.22 | 2.44 ± 0.17 | 4.99 ± 0.05 | 1.08 ± 0.12 | 5.61 ± 0.13 | 1.59 ± 0.10 |
Values are expressed as mean ± sd; sample size (no. of flowers) = 5.
The transgenic petals were green with a thick cauline texture and tapered sharply toward the apex (Fig. 4C). Likewise, the transgenic stamens were green, formed a loose cone, and did not produce pollen as in the WT (Fig. 4C). At 2 to 3 DPA, the transgenic stamens became dialytic and separated from each other while still attached to the flower (Fig. 4D). The petals and stamens remained on the flower and did not senesce until 25 DPA. Senescence was only observed after this point in the form of yellowing and wilting. However, these organs did not abscise from the flower. In the non-transgenic tomato flower, petals and stamens senesced 4 to 5 DPA and abscised after 7 to 8 DPA.
Electron micrographs revealed the presence of stomata on the abaxial surface of the transgenic petal, unlike in the WT petal, where they were rare (Fig. 5, A and B). The transgenic petals were bigger in size than the non-transgenic petal (Table II). The average length and width at anthesis were 7.37 ± 0.41 and 2.29 ± 0.34 mm, respectively, for the transgenic petal and 6.01 ± 0.18 and 1.88 ± 0.11 mm for the non-transgenic petal.
Figure 5.
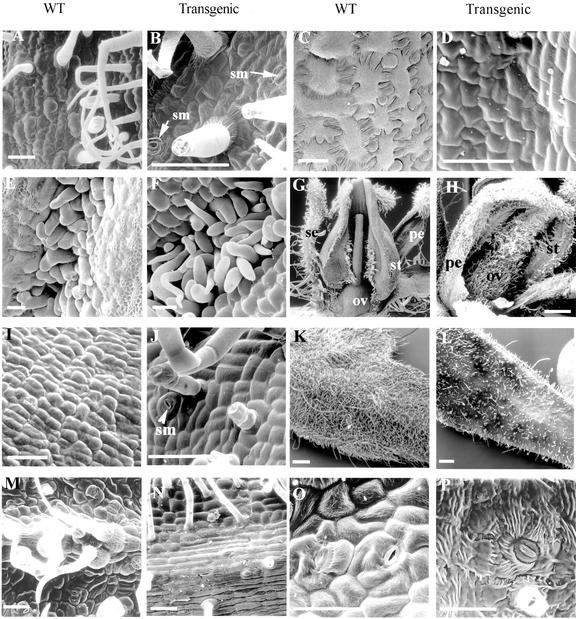
Scanning electron microscopy (SEM) photographs of floral and leaf tissues of WT and transgenic plants. A, Abaxial surface of WT petal. B, Abaxial surface of transgenic petal showing stomata (sm). C, Abaxial surface of WT stamen. D, Abaxial surface of transgenic stamen. E, Lateral hairs on WT stamens are tightly interweaved. F, Lateral hairs on transgenic stamens showing poor interweaving. G, WT flower showing hairless ovary. H, Transgenic ovary covered with hairs. I, Epidermal surface of WT ovary. J, Epidermal surface of transgenic ovary showing stomata, glandular, and non-glandular hairs. K, SEM of WT leaflet. L, SEM of ectopic leaf of a transgenic plant. M, Abaxial surface of WT leaflet. N, Abaxial surface of ectopic leaf. O, Adaxial surface of WT leaflet. P, Adaxial surface of ectopic leaf. ov, ovary; pe, petal; se, sepal; st, stamen, sm, stoma(ta). Bars in A through F, I, J, and M through P = 50 μm; in K and L, bars = 0.5 mm.
Epidermal cells in the transgenic stamens were arranged differently from the interlocking arrangement of the WT cells (Figs. 5, C and D). In WT stamens, rows of lateral and adaxial hairs present on adjacent stamens interweave to form the staminal cone (Sekhar and Sawhney, 1984; Fig. 5E). Similar hairs were present on the transgenic stamens; however, these did not interweave strongly between stamens and could account for the loose fusion between these aberrant stamens (Fig. 5F). No pollen grains were observed from the transgenic stamens either under light or electron microscope (data not shown).
The morphology of the transgenic ovary displayed some different features from the WT ovary (Fig. 5, G and H). The surface of the ovary and style was covered by glandular and non-glandular trichomes, unlike the WT ovary (Fig. 5, G and H). Electron micrographs revealed the presence of stomata on the epidermal surface of the transgenic style and ovary but not on the WT ovary (Fig. 5, I and J). However, there was no significant change in cell shape to suggest homeotic change. The average size of the transgenic carpel measured by pistil length and ovary width at the stage equivalent to anthesis was bigger than the non-transgenic carpel (Table II).
The transgenic ovary developed into parthenocarpic fruit without pollination. Repeated attempts to cross-pollinate with WT pollen failed to produce seed, an indication of ovary sterility. The size of the transgenic fruit at 5 DPA (2.97- ± 0.26-mm diameter) was bigger than the WT at the same stage (1.55- ± 0.03-mm diameter). Transgenic fruits showed a delayed ripening process and remained green for 4 to 6 weeks after reaching final fruit size. Overall, the average diameter of mature transgenic fruits measured at the breaker stage (2.8 ± 0.42 cm) was much greater than the non-transgenic Microtom tomato fruit (1.5 ± 0.30 cm).
The transgenic fruits became swollen and misshapen due to growth of internal tissues. The pericarp was broken and ectopic shoot emerged from these fruits (Fig. 4, E and F). In the lines with severe phenotype, the ectopic shoot produced flowers and leaf-like structures (Fig. 4, G and H). The ectopic flowers displayed the same phenotype as the aberrant primary flowers described previously. The ovary of the ectopic flower also swelled, reiterating the ectopic characteristics of the primary flowers (Fig. 4H). This cycle was often repeated to give three to four generations of ectopic shoots (Fig. 4I). The ectopic leaf-like organs were small in size and were present as simple leaf structures with short petioles arising directly from the ectopic shoot below the ectopic flowers (Figs. 4G and 5L). These ectopic leaves were morphologically different from the unipinnate compound leaf of the WT (Janssen et al., 1998). However, they possessed features common to the WT leaf, i.e. stomata on the abaxial and adaxial surfaces, midrib cells, glandular and non-glandular trichomes, and similar epidermal cell morphology as the WT leaflet (Fig. 5, K–P).
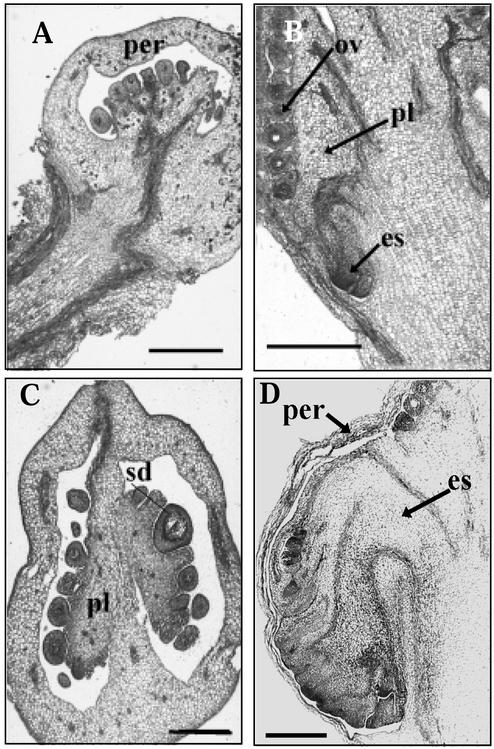
To observe the early developmental stages of the ectopic shoot, thin ovary sections from the AS/45 transformant were stained with toluidine blue. In normal tomato fruit development, the placenta and ovules occupy the entire locular cavity (Fig. 6, A and C). Inside the transgenic ovary at 2 DPA, the ectopic shoot is seen developing within the ovary (Fig. 6B). The ectopic shoot then develops to displace the internal organs within the ovary at 6 DPA (Fig. 6D).
Figure 6.
Early development of ectopic shoot. A, Longitudinal section of WT tomato fruit at 4 DPA. B, Transgenic fruit (line AS/45) at 3 DPA showing ectopic shoot development. C, WT fruit at 10 DPA. D, Transgenic fruit at 6 DPA. The ectopic shoot displaces the placenta and ovules within the fruit. es, Ectopic inflorescence; ov, ovule; per, pericarp; pl, placenta; sd, seed. Bars = 500 μm.
Down-Regulation of TM29 Expression Accounts for the Transgenic Phenotypes
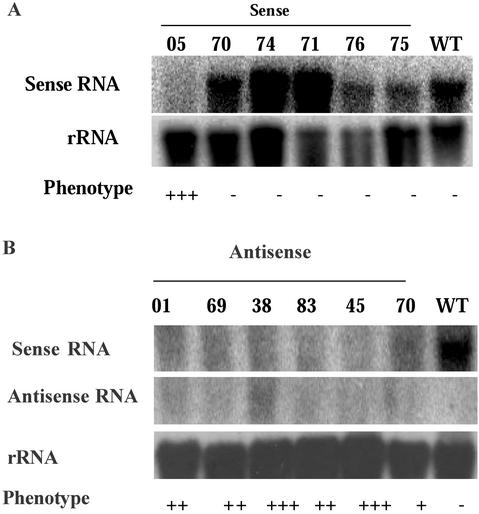
To determine that the transgenic phenotypes are related to an altered TM29 expression, we assessed the levels of TM29 transcripts present in selected sense and antisense transgenic lines using single-strand RNA probes. For the sense-transformed plants, six lines were examined together with the WT non-transgenic tomato using an antisense probe, which can detect both endogenous and transgenic copies of the TM29 transcripts. Two lines showed a higher level of mRNA than the control, two lines showed a lower level than the control, and one line showed a similar level to the control. This indicates that at least two lines overexpressed TM29 mRNA. All five lines showed normal phenotype, indicating that overexpression of TM29 did not give the transgenic phenotype. In contrast, TM29 transcripts were virtually absent in the S/05 line that showed an altered phenotype (Fig. 7A). This suggested that cosuppression of the TM29 transcript had occurred in the S/05 line and could account for the transgenic phenotype.
Figure 7.
The expressions of sense and antisense TM29 transcripts in transgenic lines. A, Total RNA extracted from six sense transgenic plants and the WT plant was probed with antisense probe. Lane 1 contained RNA from the line S/05 showing altered phenotype. Lanes 2 through 6 contained RNA samples from five plants showing no altered phenotypes. B, Total RNA extracted from six antisense transgenic plants and the WT plant was hybridized with antisense and sense probes separately to detect endogenous sense and transgene-expressed antisense transcripts, respectively. The loading levels are shown by hybridization with rRNA gene probe. For symbols of the phenotype, refer to Table I.
For the antisense-transformed plants, six lines that showed the range of phenotypes were examined together with the WT control plant using both antisense and sense probes. The antisense probe detected a high level of TM29 sense transcripts in the control plants, but very low level in all the transgenic lines. The sense probe did not detect any RNA from the WT control plant as expected and detected very low levels of antisense transcripts in the transgenic lines (Fig. 7B). This result suggests both sense and antisense RNA are down-regulated in the antisense transgenic lines examined. The expression analysis results for both sense and antisense transgenic lines are strong evidence that down-regulation of TM29 expression contributes to the transgenic phenotype.
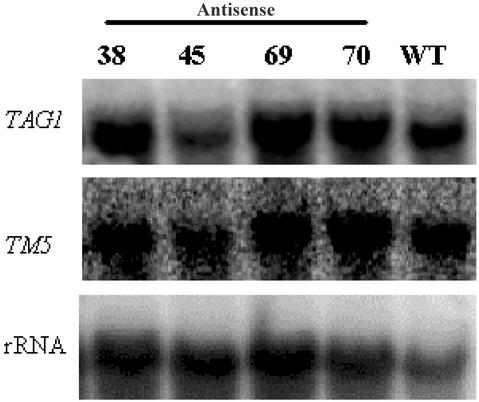
To examine whether other MADS box genes have also been down-regulated in the transgenic lines, we assessed the expression of two other tomato MADS box genes, TM5 and TAG1, in the antisense lines. These two genes were selected because TM5 has the highest sequence identity to TM29 among the known tomato MADS box genes, and, second, the antisense phenotypes of TM5 and TAG1 have already been described (Pneuli et al., 1994a, 1994b). Transcripts of these two genes were detected in three of the four antisense lines tested at a comparable level with the WT control, and in one line at a slightly reduced level (Fig. 8). This result indicates that other related MADS box genes were not generally down-regulated in the antisense transgenic lines and further supports that down-regulation of TM29 expression accounts for the transgenic phenotypes.
Figure 8.
The expressions of TM5 and TAG1 in antisense transgenic plants. Total RNA extracted from flowers of four antisense transgenic plants (AS38, 45, 69, and 70) and the WT plant was sequentially probed with TM5, TAG1, and 18S rRNA probes.
DISCUSSION
In this study, we have examined the function of a new tomato MADS box gene using both cosuppression and antisense techniques. Both techniques are commonly used to reduce the expression of endogenous genes such that the resultant phenotype mimics that of a knockout mutant. They have been successfully used in studying the functions of MADS box genes. These include FPBP7 (FLORAL BINDING PROTEIN 7) and FBP11 MADS box genes of petunia (Petunia hybrida), PETUNIA FLOWERING GENE and Gerbera GRCD1 (Mizukami and Ma, 1992, 1995; Pneuli et al., 1994a, 1994b; Angenent et al., 1995; Immink et al., 1999; Kotilainen et al., 2000). In our study, we have used a similar transgenic approach to demonstrate that TM29 is required for normal development of the inner three whorls of floral organs as well as maintenance of floral meristem identity. In seven independent transgenic lines in which TM29 was suppressed, both development of the three inner floral whorls and the maintenance of the floral state were compromised. In contrast, no consistent changes in the expression of two closely related MADS box genes, TM5 and TAG1, were observed, suggesting that changes can be attributed specifically to TM29.
TM29 expression in the floral meristem occurs before the emergence of any of the floral organs and the onset of ABC floral organ identity gene expression. The pattern of TM29 expression in the floral organs resembles that of SEP1 (Flanagan and Ma, 1994) and suggests that it may be required in the early stages of floral development to mediate the activities of the ABC organ identity genes. The morphological aberrations caused by down-regulating TM29 are consistent with its spatio-temporal expression in the WT flower. The reduction in TM29 RNA increased the size of all floral organs. Petals were thick textured, green in color with stomata, delayed in senescence, and failed to abscise. The stamens exhibited a similar green color, were dialytic with poor interweaving of lateral hairs, and did not abscise. The carpels were covered with glandular and non-glandular trichomes with stomata. The green petals, green stamens, and the glandular trichomes all suggested the homeotic conversion of these organs to sepal identity (Smyth, 2000). Upon closer examination, we identified some changes to epidermal cell morphology of transgenic floral organs, which support such a change of identity. This included the presence of stomata on transgenic petals, style, and ovary, as well as the unusual hairiness of the transgenic ovary, which were rare on the WT organs. However, these observed features, by themselves, are not strong evidence of homeotic conversion of the inner floral organs to sepals because stomata can be found on petals of some tomato cultivars (Sekhar and Sawhney, 1984).
The alterations in the inner three whorls of TM29 transgenic flowers are similar to what was observed in antisense TM5 transgenic flowers (Pneuli et al., 1994a). Similarly, in petunia, the cosuppression of FBP2 (SEP3 orthologue) also caused aberrations in the inner three whorls. Petals were reduced in size and the color changed from white to green, stamens were transformed into green petaloid structures, and the pistil lacked ovules and placental tissue (Angenent et al., 1994). The phenotypes of the TM29, TM5, and FBP2 transgenic flowers resemble the sepallata flower (Pelaz et al., 2000). The three SEPALLATA genes (SEP1, SEP2, and SEP3) redundantly control the B and C floral organ identity functions in Arabidopsis. The triple mutant of these three genes is a phenocopy of the double mutant of pistillata and agamous, characterized by homeotic conversions of petals and stamens to sepals (Bowman et al., 1991; Pelaz et al., 2000). The sequence, expression, and transgenic phenotypes of TM29, TM5, and FBP2 suggest they are orthologues of the SEP genes.
The SEP orthologues in other angiosperm species may operate differently from the Arabidopsis SEP genes. In Arabidopsis, the single SEP mutants do not display any significant change in phenotype (Pelaz et al., 2000); similarly, transgenic Arabidopsis plants expressing antisense SEP1 or SEP2 did not show any alterations in flower phenotype (C. Ampomah-Dwamena, B. Veit, and J.-L. Yao, unpublished data). TM5 is the designated orthologue of SEP3 and judging by sequence and expression analyses, TM29 is a likely orthologue of SEP genes. The similarity between the TM29 and TM5 transgenic flower phenotypes suggests that both genes are required for normal tomato flower development. To confirm this, northern analysis of TM5 transcripts in our TM29 transgenic plants showed that TM5 expression was not affected. Therefore, the functions of TM29 and TM5 may overlap but may not be completely redundant.
Reproductive Defects in TM29 Transgenic Flowers
The expression of TM29 in mature floral buds was confined to the reproductive organs (i.e. the stamens and ovary and absent from the perianth organs). Consistent with this pattern, reproductive defects were observed in transgenic stamens and ovary. The transgenic stamens did not produce pollen. Because TM29 transcript accumulation in the WT stamens localized to the endothecial and tapetal cells, its down-regulation may have affected the pollen mother cells and disrupted pollen formation.
The WT stamens are joined by interwoven rows of lateral and adaxial hairs to form a cone around the pistil (Sekhar and Sawhney, 1987). Separation of stamens is not seen in WT flowers. In contrast, tomato mutants such as dl (dialytic; Llop-Tous et al., 1999) and pat (parthenocarpic fruit; Mazzucato et al., 1998) display stamens that are not united. These mutants are characterized by suppressed hair growth on the stamens. The transgenic stamens in our study formed a loose cone and separated from each other later in development. They remained on the flower and did not abscise. The separate nature of transgenic stamens might be attributed one or more of the following three factors: (a) absence or poor growth of lateral and adaxial hairs on these stamens, (b) lack of interweaving between the hairs of adjacent stamens, or (c) pressure exerted by growth of the ovary combined with failure to abscise. SEM was used to discriminate between these possibilities. Lateral and adaxial hairs were present on the transgenic stamens; however, these hairs did not interweave strongly among themselves and might be responsible for the loose fusion of the transgenic stamens. Because the stamens delayed in senescence and failed to abscise from the flower, the pressure that the developing ovary exerted on these organs might also have caused stamen separation. On the whole, it appears that transgenic stamens are held weakly together by the poorly interwoven adaxial and lateral hairs. The delay in stamen senescence and abscission contributed to this dialytic phenotype.
The WT tomato ovary normally forms a fruit after self-pollination. Lack of pollination often leads to the termination of ovary growth and subsequent fruit abortion. The parthenocarpic fruit development in transgenic flowers suggests that TM29 may function as a negative regulator of parthenocarpic fruit development. Consistent with such a repressor activity is a mutation in the Arabidopsis fwf (fruit without fertilization) gene, which causes development of seedless fruit in the absence of pollination. Parthenocarpy is recessive in fwf plants and suggests that the activity of FWF is involved in processes that repress silique development in the absence of fertilization (Vivian-Smith et al., 2001). However, the presence of other floral organs inhibited parthenocarpy in fwf, suggesting a role for interorgan communication to control this trait (Vivian-Smith et al., 2001). This is in contrast to the transgenic TM29 tomato flowers in which parthenocarpic fruit were still able to develop in the presence of other floral organs. Similar to the TM29 transgenic flowers, parthenocarpic fruit development occurred frequently in tomato flowers expressing antisense TM5 RNA (Pneuli et al., 1994a). The results from both TM29 and TM5 transgenic plants suggest that tomato SEP orthologues may both normally limit parthenocarpic fruit development. This function has not been found for the Arabidopsis SEP genes (Honma and Goto, 2001; Pelaz et al., 2000, 2001a, 2001b). The basis for these different requirements is not clear, but might be attributed to differences in fruit structure and fruit development processes between tomato and Arabidopsis. A precedent for such differences is seen with the floral organ identity gene PI (PISTILLATA), which is associated with parthenocarpic fruit development in apple (Malus domestica) but not in Arabidopsis (Yao et al., 2001).
TM29 Controls the Maintenance of Floral Meristem Identity
A remarkable phenotype of the TM29 transformants was the eventual emergence of ectopic shoots from the fruit, which eventually produced both leaves and flowers. This phenotype suggested that the floral meristems have undergone reversion by acquiring shoot meristem identity. In comparison, in the sep1/2/3 triple mutants, floral meristems have only lost determinacy but not undergone floral reversion because only new flowers, but not shoots, are found to replace the fourth whorl of carpels (Pelaz et al., 2000). The TM29 phenotype is more similar to the phenotypes of the Arabidopsis leafy-6 and ag-1 mutants grown under short-day conditions (Okamuro et al., 1996; Mizukami and Ma, 1997), and FBP2 cosuppressed petunia flowers (Angenent et al., 1994). A significant difference between the ectopic shoot in TM29 transgenic flowers and the ectopic inflorescence described previously is the observation that the TM29 ectopic shoot initiated leaf-like organs as well as flowers. This suggests that the reversion has gone a step further from floral meristem to vegetative growth.
The ectopic shoot produced by TM29 transgenic flowers can be compared with the floral reversion seen in Impatiens balsamina. Under noninductive flowering conditions, flower development in I. balsamina reverts dramatically to vegetative growth, producing leaves until inductive conditions are imposed despite the continued expression of floral meristem identity genes (Pouteau et al., 1997, 1998b). The molecular control of floral reversion in I. balsamina is still largely unclear. Analyses of the transcription patterns of floral meristem identity genes Imp-FLO, Imp-FIM, and Imp-SQUA, orthologues of snapdragon FLORICAULA, FIMBRIATA, and SQUAMOSA, did not provide an adequate explanation for this phenomenon (Pouteau et al., 1997, 1998a, 1998b).
In Arabidopsis, floral reversion seems to be regulated by photoperiod and the plant hormone gibberellin. In the ag and leafy mutants, floral reversion occurs only under short days, a noninductive condition for flowering, which can be inhibited by exogenous application of gibberellins (Okamuro et al., 1996). Our results with tomato suggest that TM29 may be required for the maintenance of floral meristem identity. Because floral reversion has not been reported in tomato, a photoperiod insensitive plant, the control of this characteristic is less well understood. We investigated the effects of photoperiod and gibberellin on the tomato transgenic phenotypes. The transgenic phenotypes were found to be consistent for 8- and 16-h photoperiods. Similarly, there was no indication of GA control on floral reversion in tomato (C. Ampomah-Dwamena, B. Veit, and J.-L. Yao, unpublished data). Our results suggest floral reversion may be controlled differently in tomato and may not be influenced by photoperiod or GA.
In summary, TM29 is a homolog of SEP genes judging by sequence, expression, and function analyses. Like the SEP genes, TM29 is involved in the control of floral organ development. The evidence presented also suggests that TM29 may play an important role in the maintenance of floral meristem identity and fruit development in tomato. Further studies identifying TM29-interacting genes will help unravel the mechanisms controlling the maintenance of flowering.
MATERIALS AND METHODS
cDNA Library Screening and DNA Sequence Analysis
A primary cDNA library was constructed with mRNA extracted from young tomato (Lycopersicon esculentum Mill. cv UC82B) fruit (1–7 DPA) using the λ-Uni-Zap XR vector (Stratagene, La Jolla, CA) as described by Kvarnheden et al. (2000). Degenerate primers based on the conserved protein sequences MGRGKV/I and LCDAEV in the MADS domain were used to amplify a 145-bp MADS box fragment (Yao et al., 1999). The DNA fragment containing a mixture of amplified MADS box sequences was used further to screen the library at low stringency. Sequencing revealed five different clones; one of them was named TM29. The full-length cDNA of TM29 was sequenced and confirmed to be a MADS box gene. Alignment of predicted protein sequences was performed with the GCG program (version 9, Genetics Computer Group, Madison, WI). Phylogenetic relationships were established using ClustalW with the neighbor-joining method (Saitou and Nei, 1987; Thompson et al., 1994) and plotted with the TREEVIEW program (Page, 1996). Only the corresponding sequences in the MADS box, intergenic region, and K box were used in the analyses.
DNA-Blot Hybridization
Genomic DNA was isolated from young tomato leaf tissues as described by Doyle and Doyle (1990). DNA (20 μg) was digested with EcoRI, HindIII, and XbaI in separate reactions. The digests were separated on 1% (w/v) agarose gel and transferred onto Hybond N+ membrane (Amersham, Buckinghamshire, UK). A 0.7-kb fragment from the 3′ end of TM29 cDNA was labeled and used as a probe. Hybridization and washing of blots were as described previously (Church and Gilbert, 1984). Hybridization signals were visualized with the Storm 840 Phospho-Imaging system (Alphatech, Arlington, VA) and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
RNA-Blot Hybridization
Total RNA was isolated from selected tissues using Trizol reagent (Gibco-BRL, Gaithersburg, MD) according to the manufacturer's instructions. For RNA-blot analysis, RNA (10 μg) was denatured in RNA sample buffer (2.2 m formaldehyde, 50% [v/v] deionized formamide, 50 mm MOPS [pH 7.0], and 1 mm EDTA) and electrophoresed on 1% (w/v) agarose gel in 1× Tris-acetate EDTA buffer. For detection of specific transcripts, sense or antisense probes were generated by in vitro transcription using T3 and T7 RNA polymerases (Roche Diagnostics, Mannheim, Germany) respectively. For use as template, a 0.7-kb PCR fragment was amplified with gene-specific primers: ITM-03 (5′-GCA ATT AAC CCT CAC TAA AGG G GGT ACC AAA AGT GCA GCT-3′) and ITM-04 (5′-GCT AAT ACG ACT CAC TAT AGG GGG TTC ACA ACG TTC ACC T-3′). These had T3 and T7 promoter sequences (underlined) at their 5′ ends, respectively. Hybridization and washing conditions were the same as described for the Southern-blot hybridization (Church and Gilbert, 1984).
To assess the expression of other MADS box genes in the transgenic plants, 3′ cDNA fragments of TM5 and TAG1 (Pneuli et al., 1994a, 1994b) were used as probes to detect their transcripts in selected transgenic plants. The MADS box sequences were not included in any of the probes used to reduce cross hybridizations to other MADS box genes. To reprobe blots, previous probes were stripped with 0.1% (w/v) SDS at 100°C for 5 min.
RNA in Situ Hybridization
The methods for labeling RNA probes, tissue preparation, and in situ hybridization were as described by Jackson (1992). The 0.7-kb PCR fragment of TM29, described above, was used as template for generating digoxigenin-labeled sense and antisense probes with T3 and T7 RNA polymerases, respectively. Immunological detection was performed with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indoyl phosphate toluidine salt (Roche Diagnostics) as described by Coen et al. (1990).
Binary Vector Construction and Plant Transformations
To generate transgenic plants with down-regulated TM29 expression for phenotypic analysis, the full-length cDNA of TM29 was cloned between the cauliflower mosaic virus 35S RNA promoter and the octopine synthase gene 3′-untranslated region in pART7 plasmid (Gleave, 1992) in the sense or antisense orientation. The 35S-cDNA-octopine synthase fragment of each construct was then inserted into the NotI site in the T-DNA region of pART69 (J.-L. Yao, unpublished data). The binary vector, pART69, is a derivative of pART27 (Gleave, 1992) and contains the NPTII and uidA genes. These vectors containing the TM29 sequences were transformed into Agrobacterium tumefaciens strain LBA4404 cells by electroporation and used in tomato transformation experiments.
Tomato transformation experiments were carried out using the cv Microtom as described by Meissner et al. (1997). Transgenic shoots were rooted on Murashige and Skoog basal medium (Gibco-BRL) supplemented with 1 mg L−1 indoyl butyric acid and 50 mg L−1 kanamycin. Rooted plants were transferred to soil in the greenhouse.
Plant Growth Conditions
Transgenic and WT plants were grown in the glasshouse, usually under 16 h of light supplemented by incandescent lamps (150–180 μmol m−2 s−1) at a temperature of 23°C ± 1°C. For short-day treatment, plants were placed in a growth chamber, with 8 h of light supplied by cool-white flourescent bulbs (120 μmol m−2 s−1) and a temperature of 24°C ± 0.5°C.
PCR
To confirm the presence of the T-DNA constructs in the transformed tomato plants, PCR was performed with Taq DNA polymerase (Roche Diagnostics). A 35S promoter sense primer, P35S-1 (5′-GTC ACT TCA TCA AAA GGA CAG-3′), was used in combination with TM29-specific primer ITM-04 to amplify a 1.45-kb DNA fragment from the sense-transformed plants. As an internal control, this primer combination did not give any fragment from the antisense lines or the non-transgenic plants. P35S-1 and ITM-03 amplified a 1.06-kb DNA fragment from the antisense-transformed plants. PCR conditions were as follows: initial denaturation at 95°C for 2 min, 25 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min plus a final extension at 72°C for 5 min.
Scanning Electron Microscopic Analyses
Plant samples were fixed in a 50% (w/v) ethanol, 0.9 m glacial acetic acid and 3.7% (w/v) formaldehyde for 15 h and dried in a BalTec CPD 030 critical point drier (BalTec, Balzers, Liechtenstein, Germany). Samples were dissected under a stereomicroscope by removing some parts to reveal the organs to be examined. These were mounted onto stubs and coated with gold in a Polaron E5100 sputter coater (Philips, Eindhoven, The Netherlands). Specimens were examined in a 505 scanning electron microscope (Philips) at 15 kV.
Tissue Preparation and Staining
To observe the early developmental stages of ectopic inflorescence, 8-μm tissue sections of ovary at 0 to 6 DPA were prepared from the AS/45 transgenic line, which has a severe phenotype, using the method described previously by Jackson (1992). For staining, tissues were dewaxed in Histoclear (National Diagnostics, Atlanta), rehydrated through serial dilutions of ethanol, and allowed to dry as described by O'Brien and McCully (1981). Tissues were stained in 0.01% (w/v) toluidine blue (pH 4.5) and photographed using a Vanox AHT3 light microscope (Olympus, Tokyo).
ACKNOWLEDGMENTS
We are grateful to Anders Kvarnheden for the tomato cDNA library, Julie Nichols for growing plants in the greenhouse, Anna Henderson for electron microscopy, Martin Heffer for assistance with photography, and Bart-Jan Janssen for critical reading of the manuscript.
Footnotes
This work was supported by the New Zealand Foundation for Research, Science, and Technology (contract no. CO6411).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005223.
LITERATURE CITED
- Allen KD, Sussex IM. Falsiflora and anantha control early stages of floral meristem development in tomato (Lycopersicon esculentum Mill.) Planta. 1996;200:254–264. [Google Scholar]
- Angenent G, Franken J, Busscher M, Van Dijken A, Van Went J, Dons H, Van Tunen A. A novel class of MADS box genes is involved in ovule development in Petunia. Plant Cell. 1995;7:1569–1582. doi: 10.1105/tpc.7.10.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, Weiss D, van Tunen AJ. Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant J. 1994;5:33–44. doi: 10.1046/j.1365-313x.1994.5010033.x. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E, Meyerowitz E. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliot R, Murphy G, Carpenter R. floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Davies B, Eagea-Cortines M, de Andrade S, Saedler H, Sommer H. Multiple interactions amongst floral homeotic proteins. EMBO J. 1996;15:4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Flanagan C, Ma H. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsis flowers. Plant Mol Biol. 1994;26:581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- Gleave A. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Immink R, Hannapel D, Ferrario S, Busscher M, Franken J, Campagne L, Angenent G. A Petunia MADS box gene involved in the transition from vegetative to reproductive development. Development. 1999;126:5117–5126. doi: 10.1242/dev.126.22.5117. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Jackson D. In situ hybridisation in plants. In: Gurr S, McPherson M, Bowles D, editors. Molecular Plant Pathology: A Practical Approach. Oxford: IRL Press; 1992. pp. 163–174. [Google Scholar]
- Janssen B-J, Lund L, Sinha N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998;117:771–786. doi: 10.1104/pp.117.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, An G. Isolation and characterization of a rice MADS box gene belonging to the AGL2 gene family. Mol Cell. 1997;7:45–51. [PubMed] [Google Scholar]
- Kang H, Jang S, Chung J, Cho Y, An G. Characterization of two rice MADS box genes that control flowering time. Mol Cell. 1997;7:559–566. [PubMed] [Google Scholar]
- Kotilainen M, Elomaa P, Uimari A, Albert VA, Deyue Y, Teeri TH. GRCD1, an AGL2-like MADS Box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell. 2000;12:1893–1902. doi: 10.1105/tpc.12.10.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvarnheden A, Yao J-L, Zhan X, O'Brien I, Morris BAM. Isolation of three distinct CycD3 genes expressed during fruit development in tomato. J Exp Bot. 2000;51:1–9. doi: 10.1093/jexbot/51.352.1789. [DOI] [PubMed] [Google Scholar]
- Llop-Tous I, Dominguez-Puigjaner E, Palomer X, Vendrell M. Characterisation of two divergent endo-B-1,4-glucanase cDNA clones highly expressed in the nonclimacteric strawberry fruit. Plant Physiol. 1999;119:1415–1421. doi: 10.1104/pp.119.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Angosto T, Gomez P, Payan C, Capel J, Huijser P, Salinas J, Martinez-Zapater J. Tomato flower abnormalities induced by low temperatures are associated with changes of expression of MADS-box genes. Plant Physiol. 1998;117:91–100. doi: 10.1104/pp.117.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanfosky MF. Molecular characterisation of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex Plant Reprod. 1998;11:22–28. [Google Scholar]
- Mazzucato A, Taddei A, Soressi G. The parthenocarpic fruit (pat) mutant of tomato (Lycopersicum esculentum Mill.) sets seedless fruits and has aberrant anther and ovule development. Development. 1998;125:107–114. doi: 10.1242/dev.125.1.107. [DOI] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A. A new model system for tomato genetics. Plant J. 1997;12:1465–1472. [Google Scholar]
- Mizukami Y, Ma H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell. 1992;71:119–131. doi: 10.1016/0092-8674(92)90271-d. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Mol Biol. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TP, McCully ME. The Study of Plant Structure. Principles and Selected Methods. Victoria, Australia: Termarcarphi Pty Ltd.; 1981. [Google Scholar]
- Okamuro JK, den Boer BGW, Lotys-Prass C, Szeto W, Jofuku KD. Flowers into shoots: photo and hormonal control of a meristem identity switch in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:13831–13836. doi: 10.1073/pnas.93.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi S, Crosby W, Yanofsky MF. APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 2001a;26:385–394. doi: 10.1046/j.1365-313x.2001.2641042.x. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Tapia-Lopez R, Alvarez-Buylla E, Yanofsky M. Conversion of leaves into petals in Arabidopsis. Curr Biol. 2001b;11:182–184. doi: 10.1016/s0960-9822(01)00024-0. [DOI] [PubMed] [Google Scholar]
- Pneuli L, Hareven D, Broday L, Hurwitz C. The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell. 1994a;6:175–186. doi: 10.1105/tpc.6.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pneuli L, Hareven D, Rounsley S, Yanfosky M, Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell. 1994b;6:163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S, Nicholls D, Tooke F, Coen E, Battey N. The induction and maintenance of flowering in Impatiens. Development. 1997;124:3343–3351. doi: 10.1242/dev.124.17.3343. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Nicholls D, Tooke F, Coen E, Battey N. Transcription pattern of a FIM homologue in Impatiens during floral development and reversion. Plant J. 1998a;14:235–246. doi: 10.1046/j.1365-313x.1998.00114.x. [DOI] [PubMed] [Google Scholar]
- Pouteau S, Tooke F, Battey N. Quantitative control of inflorescence formation in Impatiens balsamina. Plant Physiol. 1998b;118:1191–1201. doi: 10.1104/pp.118.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Theres K. Genetic control of branching in Arabidopsis and tomato. Curr Opin Biol. 1999;2:51–55. doi: 10.1016/s1369-5266(99)80010-7. [DOI] [PubMed] [Google Scholar]
- Sekhar K, Sawhney V. A scanning electron microscope study of the development and surface features of floral organs of tomato (Lycopersicon esculentum) Can J Bot. 1984;62:2403–2413. [Google Scholar]
- Sekhar K, Sawhney V. Ontogenic study of the fusion of floral organs in the normal and “solanifolia” mutant of tomato (Lycopersicon esculentum) Can J Bot. 1987;65:215–221. [Google Scholar]
- Smyth D. A reverse trend-MADS functions revealed. Trends Plant Sci. 2000;5:315–317. doi: 10.1016/s1360-1385(00)01690-3. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A, Luo M, Chaudhury A, Koltunow A. Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development. 2001;128:2321–2331. doi: 10.1242/dev.128.12.2321. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Yanofsky M, Ma H, Bowman J, Drews G, Feldmann K, Meyerowitz E. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–349. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yao J-L, Dong Y-H, Kvarnheden A, Morris B. Seven MADS-box genes in apple are expressed in different parts of the fruit. J Am Soc Hortic Sci. 1999;124:8–13. [Google Scholar]
- Yao J-L, Dong Y-H, Morris B. Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proc Natl Acad Sci USA. 2001;98:1306–1311. doi: 10.1073/pnas.031502498. [DOI] [PMC free article] [PubMed] [Google Scholar]