Abstract
In plants, low temperature and dehydration activate a set of genes containing C-repeat/dehydration-responsive elements in their promoter. It has been shown previously that the Arabidopsis CBF/DREB1 transcription activators are critical regulators of gene expression in the signal transduction of cold acclimation. Here, we report the isolation of an apparent homolog of the CBF/DREB1 proteins (CBF4) that plays the equivalent role during drought adaptation. In contrast to the three already identified CBF/DREB1 homologs, which are induced under cold stress, CBF4 gene expression is up-regulated by drought stress, but not by low temperature. Overexpression of CBF4 in transgenic Arabidopsis plants results in the activation of C-repeat/dehydration-responsive element containing downstream genes that are involved in cold acclimation and drought adaptation. As a result, the transgenic plants are more tolerant to freezing and drought stress. Because of the physiological similarity between freezing and drought stress, and the sequence and structural similarity of the CBF/DREB1 and the CBF4 proteins, we propose that the plant's response to cold and drought evolved from a common CBF-like transcription factor, first through gene duplication and then through promoter evolution.
Many plants increase their tolerance to freezing after exposure to low nonfreezing temperatures—a phenomenon known as cold acclimation (Hughes and Dunn, 1996; Thomashow, 1998). The major component of this acquired freezing tolerance is the tolerance to dehydration stress caused by extracellular ice formation during the freezing process. The presence of ice lowers the water potential extracellularly and causes water to flow out of cells (Pearce, 1999). Thus, a major cause of freezing damage is the freeze-induced dehydration (Steponkus and Webb, 1992; Thomashow, 1998). Because a plant's ability to survive freeze-induced dehydration is related to its adaptation to drought, it is not surprising that plants respond to low temperature and drought very similarly at the molecular level (Shinozaki and Yamaguchi-Shinozaki, 2000). Many genes, such as RD (responsive to dehydration), ERD (early responsive to dehydration), COR (cold regulated), LTI (low-temperature induced), and KIN (cold inducible), are induced by both low temperature and drought stress (Ingram and Bartels, 1996; Pearce, 1999; Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). The similarity of cold and drought stresses is further demonstrated by experiments showing that mild drought stress can result in increased freezing tolerance in plants (Clavitier and Siminovitch, 1982; Siminovitch and Cloutier, 1983; Guy et al., 1992).
Recently, a major transcriptional regulatory system that controls abscisic acid (ABA) independent gene expression in response to low temperature has been identified (Stockinger et al., 1997; Liu et al., 1998). The system is based on the C-repeat (CRT)/dehydration-responsive element (DRE) cis-acting element and the trans-acting DNA-binding protein CBF/DREB1 (CRT-binding factor or DRE-binding protein). There are three CBF/DREB1 genes present in the Arabidopsis genome arranged in a tandem array within a region of 8.7 kb (Gilmour et al., 1998; Medina et al., 1999). Their expression is induced by low temperature, and they, in turn, activate the expression of many low temperature-responsive genes (Seki et al., 2001). When overexpressed constitutively in Arabidopsis plants, they induce the expression of downstream genes under non-stress conditions, and confer freezing, drought, and salt tolerance to the transgenic plants (Jaglo-Ottosen et al., 1998; Kasuga, et al., 1999; Gilmour et al., 2000). Therefore, this class of genes represents a critical component in the signal transduction of cold acclimation.
Because the CRT/DRE element is sufficient for drought-inducible gene expression (Yamaguchi-Shinozaki and Shinozaki, 1994), it strongly suggests the existence of drought-inducible transcription factors that bind to the element. In an earlier study, two transcription factors, DREB2a and DREB2b, were identified based on their ability to bind the CRT/DRE element in vitro and in yeast (Saccharomyces cerevisiae; Liu et al., 1998). Because they are induced by drought stress and are able to induce the expression of genes that contain the CRT/DRE cis-acting element in protoplast transient assays, they were good candidates to be involved in drought signal transduction. Interestingly, overexpression of the DREB2 cDNA in transgenic plants only caused weak induction of the downstream genes and did not result in obvious phenotypes (Liu et al., 1998). It is proposed that translational modifications are necessary for the activity of those proteins in transgenic plants (Shinozaki and Yamaguchi-Shinozaki, 2000), but this hypothesis remains to be demonstrated experimentally.
In this study, we report the isolation CBF4, a gene coding for a protein that is the closest homolog of CBF/DREB1 proteins in Arabidopsis. The expression of CBF4 is induced rapidly during drought stress and by ABA treatment, but not by cold. The overexpression of CBF4 under the constitutive cauliflower mosaic virus (CaMV) 35S promoter resulted in the expression of cold- and drought-induced genes under non-stress conditions. The transgenic plants are also more tolerant to freezing and drought conditions. Thus, we suggest that CBF4 plays a role in the signal transduction of drought adaptation in Arabidopsis plants. Because of the high sequence homology between CBF4 and the CBF/DREB1 proteins, especially in the DNA-binding domain, downstream gene activation is presumably through the binding of the CRT/DRE element. We propose that the signal transduction of cold acclimation and drought adaptation in pathways involving the CRT/DRE element evolved from a common CBF/DREB1 locus, first through gene duplication and then through the evolution of their promoters.
RESULTS
CBF4 Is the Closest Homolog of CBF/DREB1 Proteins
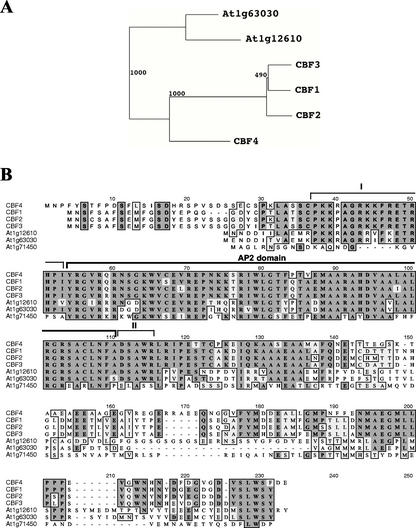
As a part of a functional genomics program on Arabidopsis transcription factors, one protein was recognized as the AP2/ERF protein (At5g51990) most closely related to the three previously described Arabidopsis CBF/DREB1 proteins (CBF1/DREB1b, At4g25490; CBF2/DREB1c, At4g25470; and CBF3/DREB1a, At4g25480; Stockinger et al., 1997; Gilmour et al., 1998). We designated the gene coding for the protein CBF4. The CBF4 open reading frame was initially detected in the sequence of P1 clone MSG15 (GenBank accession no. AB015478), and it was shown by RACE to correspond to an intronless and expressed gene. Phylogenetic relationships between CBF1-4 and At1g63030 and At1g12610, the two most closely related genes to CBF1-4 within the Arabidopsis AP2/ERF family are shown in Figure 1A. CBF4 is the only other member of the AP2/ERF transcription factor family for which substantial sequence similarity with CBF1-3 extends beyond the conserved AP2 domain (Fig. 1B). CBF4 is 224 amino acids in length, shares 63% overall amino acid sequence identity with the three CBF/DREB1 proteins, and 91% to 94% identity within the AP2/ERF DNA-binding domain (Fig. 1B). It has been noted recently that all CBF/DREB1 proteins share common signature motifs that bracket the AP2 domain, and those motifs are found in CBF-like proteins that are conserved across species (Jaglo et al., 2001). Both signature motifs are present in the CBF4 protein (Fig. 1B). In the phylogenetic analysis of the complete Arabidopsis AP2/ERF gene family, which consists of at least 144 members, Atg71450 is next to, but falls outside of, the clade defined by the other six proteins; it is included in the multiple sequence alignment as a control (Fig. 1B).
Figure 1.
Phylogenetic analysis and sequence comparison of CBF4. A, Phylogenetic tree showing the relationships between CBF4, CBF1-3, At1g63030, and At1g12610. The neighbor-joining tree was based on an alignment of the complete protein sequences. Bootstrap values are shown on branches. At1g63030 and At1g12610 are the two most closely related genes to CBF1-4 within the Arabidopsis AP2/ERF family. Addition of other AP2/ERF family members does not change the phylogenetic tree (data not shown). B, Sequence comparison of the proteins CBF4, CBF1-3, At1g63030, At1g12610, and Atg71450. At1g63030 and At1g12610 are the two most closely related genes to CBF1-4 within the Arabidopsis AP2/ERF family. In the phylogenetic analysis of the complete gene family, Atg71450 is next to, but falls outside of, the clade defined by the other six proteins. The predicted AP2 domains and the two signature regions, I and II, previously noted for the CBF1-3 proteins (Jaglo et al., 2001) are also shown.
CBF4 Overexpression Results in Growth Retardation and Activation of COR Genes
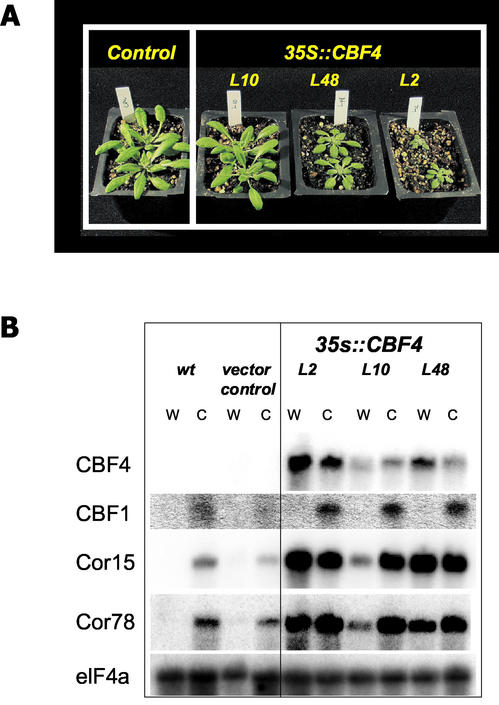
The high degree of sequence similarity between CBF4 and CBF1, 2, and 3 suggested that the proteins were functional homologs. To test this hypothesis, the CBF4 gene was cloned from cDNA, and constitutively overexpressed under the control of the CaMV 35S promoter in transgenic Arabidopsis plants. The effects of CBF4 overexpression on plant growth, COR gene expression, and stress tolerance were tested using the transgenic plants (compared with the published sequence, At5g51990, the clone used for analysis contained a single point mutation that changed amino acid residue 111 from D to G). Thirty independent transgenic lines were identified by kanamycin selection (the selectable marker carried in the transformation vector). Similar to earlier observations of CBF3/DREB1a overexpression using the 35S promoter (Kasuga et al., 1999; Gilmour et al., 2000), the 35S::CBF4 plants showed retarded growth compared with the wild-type controls (Fig. 2A), had shorter petioles and darker green leaves, and the time to flowering was significantly delayed (data not shown). For a detailed characterization, three transgenic lines (L10, L48, and L2) representing the full range of different sizes from minor phenotypic differences (L10) to severe growth retardation (L2) were chosen. Northern analysis indicated that under both normal and cold-acclimated conditions, the transcript levels for CBF4 in the transgenic plants were much greater than those observed for the control plants (wild-type and empty vector control, Fig. 2B). The transcript level for CBF4 did not change significantly upon cold acclimation, neither in the wild type nor in the transgenic plants. The degree of growth retardation (Fig. 2A) correlated with the level of CBF4 expression (Fig. 2B): The highest level of CBF4 expression caused the most severe growth phenotype.
Figure 2.
Growth characteristics of and transcript levels in CBF4-overexpressing transgenic plants. A, Control plants (transformed with empty vector) and CBF4-overexpressing lines L10, L48, and L2 after 4 weeks growth at 22°C. B, Northern analysis of total RNA. Ten micrograms of total RNA prepared from wild-type and empty vector control Arabidopsis plants and from CBF4-overexpressing lines L10, L48, and L2 were blotted and probed with the indicated probes. Plants were either grown at 22°C for 14 to 15 d (W) or at 22°C for 12 to 14 d and then cold treated at 4°C for 7 d (C). eIF4a is a constitutively expressed gene used as a loading control (Metz et al., 1992).
In response to dehydration stress such as cold and drought, wild-type Arabidopsis plants induce the expression of a large number of genes including the COR genes (Seki et al., 2001). It has been shown previously that overexpression of CBF1/DREB1b (Jaglo-Ottosen et al., 1998) or CBF3/DREB1a (Kasuga et al., 1999; Gilmour et al., 2000; Seki et al., 2001) is sufficient to induce the expression of the downstream CRT/DRE element containing COR target genes. To investigate whether the CBF4 transcription factor could also activate the COR genes, expression levels for COR15a and COR78a, known downstream target genes of CBF/DREB1 proteins, were analyzed and shown in Figure 2B. In the control plants (wild type and empty vector control), the COR15a and COR78a transcript levels were low under normal growth conditions, and increased after cold acclimation. Constitutive expression of the CBF4 gene resulted in the expression of both COR15a and COR78a, without a low temperature or drought stimulus (Fig. 2B). Compared with the controls, the COR15a and COR78a transcript levels were significantly higher in all three transgenic lines, and only increased in response to cold in the weakest overexpresser, line L10.
To ensure that the induction of the COR genes was due to CBF4 overexpression rather than the expression of other CBF/DREB1 genes, CBF1/DREB1b gene expression was analyzed as well (the probe used cross-reacts with CBF2 and CBF3). Figure 2B illustrates that the CBF1/DREB1b message was low under normal conditions in all plants, and showed a very similar increase in response to cold in both control and transgenic plants. To further demonstrate the direct involvement of CBF4 in the activation of the COR genes, we used a transient assay developed by Yang et al. (2000) to test CBF4's ability to induce the expression of a COR78a promoter::GUS construct (Horvath et al., 1993). The GUS reporter gene under COR78a promoter was induced 11.2- ± 2.7-fold by CBF4 and 12.3- ± 1.0-fold by CBF3 in this assay (induction level was calculated relative to the empty vector control; data presented were averages of at least eight independent tests). As a control, At4g36900, another AP2 domain transcription factor, was not able to induce the COR78a promoter (0.4- ± 0.1-fold). Taken together, we concluded that the COR gene induction in transgenic plants was a direct consequence of CBF4 expression, and similar to the other CBF/DREB1 genes, CBF4 is a regulator of COR gene expression.
CBF4 Overexpression Results in Plants That Are More Freezing and Drought Tolerant
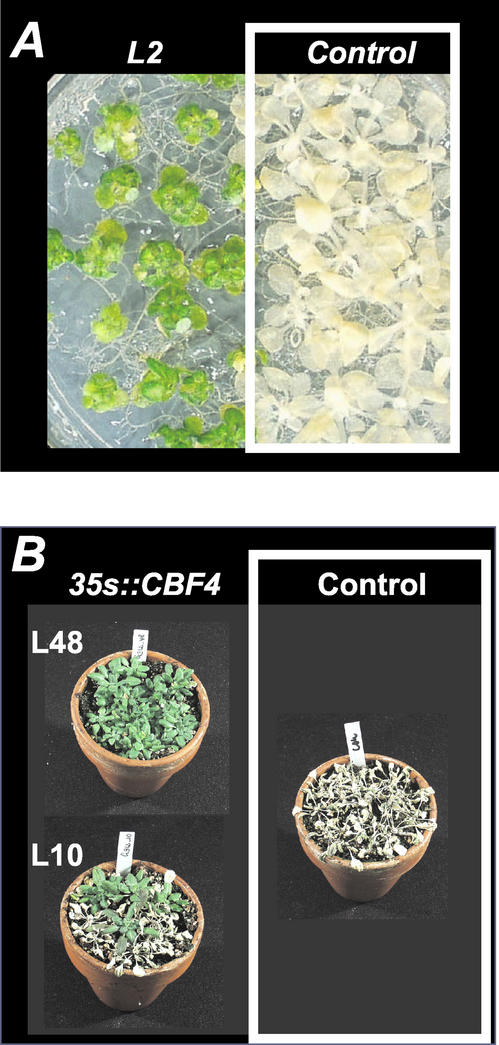
Overexpression of CBF1/DREB1b (Jaglo-Ottosen et al., 1998) or CBF3/DREB1a (Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000) has been reported to increase the freezing, drought, and salt stress tolerance of nonacclimated transgenic plants. To further demonstrate the functionality of the CBF4 gene, we examined the degree of freezing and drought tolerance of the transgenic plants. In a whole plant-freezing assay, 2-week-old plants grown on petri dishes, with or without cold acclimation for 4 d at 4°C, were frozen for 20 h at −15°C (cold-acclimated plants) or −10°C (nonacclimated plants), respectively. Plant survival was scored after 2 d of recovery under normal growth conditions, and representative results are shown in Figure 3A. The constitutive overexpression of the CBF4 gene resulted in an increase in freezing tolerance of the transgenic plants under both cold-acclimating and nonacclimating conditions (Fig. 3A; Table I). Under nonacclimating conditions, only 1% of the wild type plants survived the freezing test compared with 52% to 100% for the different transgenic lines (Table I). Very similar results were obtained after cold acclimation with a 2% survival rate for the wild type, and 54% to 89% for the different transgenic lines (Table I).
Figure 3.
Effects of freezing and drought stress on transgenic plants overexpressing CBF4. A, Transgenic plants from line L2 and empty vector controls were grown side by side on petri dishes for 2 weeks, then frozen at −10°C for 20 h. Photographs were taken 2 d after transfer to 22°C. (B) Plants were grown for 2 weeks with normal watering, withheld from water for 9 d, and rewatered for 4 d before photographs were taken. Control and transgenic plants are shown under the same conditions.
Table I.
Survival rates of transgenic plants under different stress conditions
| CBF4-Overexpressing Lines | Survivala | Totalb | Survivalc |
|---|---|---|---|
| % | |||
| Freezing tolerance (nonacclimated)d | |||
| Control | 1 | 169 | 1 |
| L10 | 44 | 85 | 52 |
| L48 | 29 | 35 | 83 |
| L2 | 44 | 44 | 100 |
| Freezing tolerance (cold acclimated)e | |||
| Control | 4 | 220 | 2 |
| L10 | 50 | 91 | 55 |
| L48 | 15 | 28 | 54 |
| L2 | 73 | 82 | 89 |
| Drought tolerancef | |||
| Control | 3 | 181 | 2 |
| L10 | 44 | 98 | 45 |
| L48 | 123 | 141 | 87 |
No. of survival plants.
Total no. of plants used in each assay.
Percentage of survival plants.
Two-week-old plants grown on petri dishes at 22°C and then frozen at −10°C for 20 h.
Two-week-old plants grown on petri dishes at 22°C, transferred to 4°C for 4 d, and then frozen at −15°C for 20 h.
Two-week-old soil-grown plants withheld water for 9 d, rewatered, and scored 4 d later. Plants were considered dead if all the leaves were brown and there was no regrowth 4 d after rewatering. Water loss during the drought period was similar for all pots independent of the plant size (data not shown).
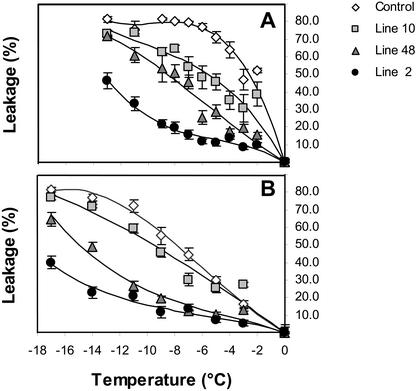
To quantify the degree of added freezing protection, electrolyte leakage experiments were done. Under nonacclimating conditions (Fig. 4A), the EL50 (temperature that causes 50% leakage) value for the control was −2.6°C compared with −5.6°C (L10) and up to −13.3°C (L2) for the 35S::CBF4 plants. The level of freezing tolerance correlated with the level of CBF4 overexpression. Cold acclimation (Fig. 4B) further increased the level of freezing tolerance to −7.9°C for the control, and −9.5°C (L10) and up to −18.7°C (L2) for the transgenic CBF4 plants. Therefore, without cold acclimation, high-level overexpression of CBF4 was sufficient to result in at least the same level of freezing tolerance as cold-acclimated control plants. After cold acclimation, the level of freezing tolerance increased even further in the 35S::CBF4 plants. This result is similar to what has been reported for the overexpression of CBF1/DREB1b (Jaglo-Ottosen et al., 1998) and CBF3/DREB1a genes (Kasuga et al., 1999; Gilmour et al., 2000).
Figure 4.
Quantification of freezing tolerance of transgenic plants overexpressing the CBF4 gene. Using the electrolyte leakage assay, freezing tolerance was measured in plants grown on petri dishes for 2 weeks without (A) and with (B) cold acclimation at 4°C for 7 d. Model curves fitting up to third order linear polynomial trends were determined for each electrolyte leakage experiment. Based on those curves, EL50 (temperature causing 50% ion leakage) values were calculated for nonacclimated (control, −2.6°C; L10, −5.6°C; L48, −8.6°C; and L2, −13.3°C) and cold-acclimated (control, −7.9°C; L10, −9.5°C; L48, −14.9°C; and L2, −18.7°C) conditions.
To assay if the transgenic plants are more drought tolerant than the wild-type controls, 14-d-old plants grown on soil were held without water for 9 d before the plants were watered again (Fig. 3B). Although only 2% of the wild-type plants survived this treatment, the survival rate for the 35S::CBF4 plants was much higher with 45% for L10 and 87% for L48 (Table I; L2 was not included in this test). The increased survival rate 4 d after rewatering illustrates that the overexpression of CBF4 resulted in transgenic plants that are significantly more drought tolerant than the wild-type controls. Again, the level of drought tolerance correlated with the level of CBF4 overexpression, similar to what has been previously reported for CBF3/DREB1a (Kasuga et al., 1999). From these and the other results described above, we concluded that CBF4 is a fully functional fourth member of the CBF/DREB1 family.
CBF4 Is Induced by Drought and ABA
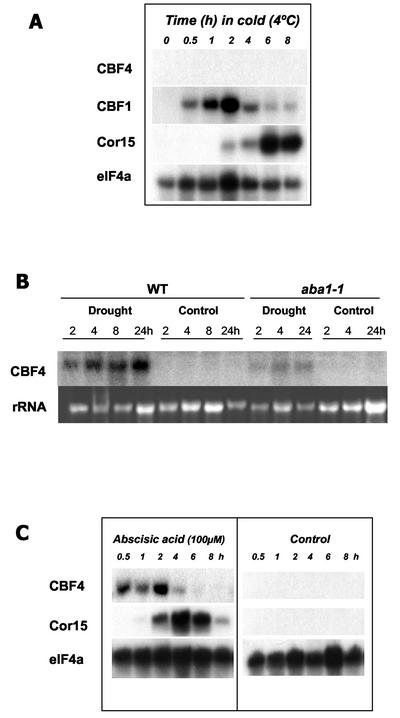
The CBF/DREB1 genes are induced by cold, but not by drought or ABA (Shinwari et al., 1998; Medina et al., 1999). To understand the role CBF4 might play in the signal transduction of dehydration stress response, we studied its gene expression in response to low temperature, drought, and the exogenous application of ABA. RNA from plants subjected to low temperature, drought, or the exogenous application of ABA were blotted and hybridized with probes for CBF4, COR15a, and elF4a (this gene is not responsive to cold, drought, or ABA). Figure 5A showed that, under our experiment condition, CBF4 was not responsive to cold. However, transcripts for CBF4 did accumulate in response to water deficit (Fig. 5B) and exogenous application of ABA (Fig. 5C). To demonstrate that the induction of CBF4 expression is ABA dependent, CBF4 gene expression was analyzed in the ABA-deficient mutant aba1-1 (Koornneef et al., 1982). In aba1-1, the drought induction of CBF4 expression is dramatically reduced (Fig. 5B), indicating that ABA biosynthesis is required for the proper drought-induced induction of CBF4 expression.
Figure 5.
Gene expression analysis of CBF4. Total RNA was prepared from plate-grown Arabidopsis plants subjected to low temperature (4°C; A), dehydration (B), and ABA blotted and probed with indicated probes (C). A, RNA was isolated from plants that were placed in a cold room set at 4°C for the indicated amount of time. B, RNA was isolated from both wild-type and aba1-1 plants placed over desiccant for the indicated amount of time. C, RNA was isolated from plants placed in Gamborgs B5 liquid with 100 μm of ABA for the indicated amount to time. For controls in B and C, the plants were transferred to Gamborgs B5 liquid (see “Materials and Methods”).
DISCUSSION
In this study, we report the isolation of CBF4 as part of a functional genomics research program on transcription factors. The gene encodes a protein that is the closest homolog to the CBF1,2,3/DREB1abc transcriptional activators in Arabidopsis. Although the isolated clone contains a single amino acid change from the published genomic sequence, it appears that this change did not result in a change of its function. The sequence alignment presented in Figure 1 shows that two signature motifs that are conserved among CBF genes across different species (Jaglo et al., 2001) are also present in CBF4. Overexpression of CBF4 using the constitutive CaMV 35S promoter resulted in constitutive expression of COR15a and COR78a, both cold- and drought-inducible and known target genes for CBF1, 2, and 3 (Gilmour et al., 1998; Kasuga et al., 1999). As a result, the transgenic plants are also more tolerant to freezing and drought stress. All of the above phenotypes have been observed for the constitutive overexpression of CBF1 (Jaglo-Ottosen et al., 1998) and CBF3/DREB1a (Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000), indicating that CBF4 is in fact a new and functional member of the CBF/DREB1 family. Interestingly, the level of dehydration tolerance that can be achieved by CBF4 overexpression is very similar for all CBF/DREB1 genes, suggesting that all CBF/DREB1 genes induce the expression of a common set of CRT/DRE target genes.
Unlike that of the other CBF/DREB1 transcription factors (Gilmour et al., 1998; Shinwari et al., 1998; Medina et al., 1999), the expression of CBF4 does not change in response to cold, but is induced by drought stress and ABA treatment (Fig. 5). In an earlier study, two cDNAs encoding AP2 domain transcription factors, DREB2a and b, were identified by a yeast one-hybrid screen (Liu et al., 1998). Both bind to the CRT/DRE element in vitro, and activate expression from the COR78a/RD29a promoter in a transient protoplast assay (Liu et al., 1998). The drought- and salt-responsive expression pattern of the DREB2 genes suggests a role in the activation of the CRT/DRE containing genes in response to those stresses (Nakashima et al., 2000). However, when DREB2 was constitutively overexpressed in Arabidopsis plants, only weak induction of the CRT/DRE-containing target genes was observed, and the plants did not show a significant increase in the level of stress tolerance (Liu et al., 1998). It has been speculated that the activity of the DREB2 proteins is regulated at the posttranscriptional level (Shinozaki and Yamaguchi-Shinozaki, 2000). Here, we show that the overexpression of CBF4 alone is sufficient to provide increased dehydration protection, and we propose that the CBF4 gene also plays a role in the signal transduction of drought adaptation in Arabidopsis plants (Fig. 6). A recent study by Sakuma et al. (2002) showed that CBF4 (DREB1d) expression is induced by salt, but not by drought, cold, or ABA under their experimental conditions. It remains to be investigated whether the different experimental procedures have resulted in these discrepancies.
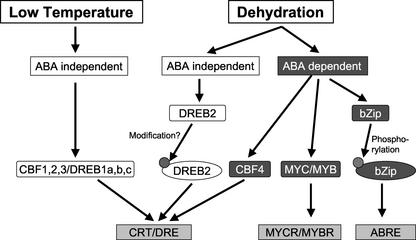
Figure 6.
Simplified model for the regulation of cold- and drought-responsive gene expression.
In Arabidopsis, many genes are induced in response to low temperature and water deficit (Pearce, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong and Zhu, 2001). Previous studies have shown that multiple transcription factors and cis-acting regulatory elements are involved in controlling expression of these genes (Fig. 6). It has been proposed that the ABA-independent cold- and drought-responsive gene expression is regulated by CBF/DREB1 and DREB2 proteins, respectively (Fig. 6; Gilmour et al., 1998; Shinwari et al., 1998; Medina et al., 1999; Nakashima et al., 2000). On the other hand, the ABA-dependent dehydration response involves the ABA-responsive element and Myb/Myc promoter elements recognized by basic-region Leu-zipper (bZIP) and Myb/Myc transcription factors, respectively (Abe et al., 1997; Razik and Quatrano, 1997; Uno et al., 2000; Kim et al., 2001). Here, we show for the first time that the ABA-dependent pathway can also involve the CRT/DRE elements and AP2-type transcription factors (Fig. 6). This observation is in agreement with an earlier study that suggests that the CRT/DRE elements are involved in ABA signal transduction because the ABA-responsive element in the promoter of COR78a/RD29a is not sufficient by itself to elicit an ABA response, and that proper ABA response requires the presence of a region containing the CRT/DRE elements (Yamaguchi-Shinozaki and Shinozaki, 1994). It still remains unclear why the CRT/DRE element by itself does not appear to be sufficient for ABA-dependent gene activation (Yamaguchi-Shinozaki and Shinozaki, 1994).
An analysis of regulatory elements in the 2-kb upstream of the CBF4 gene by PLACE database (Higo et al., 1999; http://www.dna.affrc.go.jp/htdocs/PLACE/) uncovered several putative bZIP- and MYB-/MYC-binding sites. However, because most of the cis-acting elements known to be involved in bZIP or MYB/MYC binding are degenerate 5 to 8 mers, and occur in several thousand Arabidopsis promoters, sequence analysis alone is insufficient to draw any conclusions. A more detailed functional analysis of the CBF4 promoter is needed to identify its key regulatory elements.
CBF1, 2, and 3 are present in the genome as a tandem array on chromosome 4 (Gilmour et al., 1998; Medina et al., 1999), whereas the gene encoding their closest related homolog, CBF4, is present on chromosome 5. Phylogenetic analysis (Fig. 1A) suggests that the ancestor of these four genes underwent a gene duplication event that gave rise to CBF4 and a homolog that subsequently underwent more recent duplication events that produced CBF1, 2, and 3. The fact that the CBF4 promoter region shows little sequence identity with the promoter regions of CBF 1, 2, and 3 (data not shown), which show considerable sequence identity among themselves (Shinwari et al., 1998), suggests that duplications at the CBF1, 2, and 3 locus occurred after differences in regulation were established (or well under way). It has been suggested that one way through which genetic diversity can be generated is by the duplication of genome segments and the subsequent rearrangements (Bancroft, 2000). Promoter scrambling via transposable element insertion and deletion caused rearrangement is shown to be particularly effective way of generating diversity in the promoter regions (Robins and Samuelson, 1992; Kloeckener-Gruissem and Freeling, 1995; Wessler et al., 1995; Britten, 1996; Britten, 1997; Kidwell and Lisch, 1997; Girard and Freeling, 1999). Many traits of agronomic importance have evolved through changes in the expression of regulatory genes (Doebley, 1998; Lukens and Doebley, 2001), and in many cases, those changes were in the promoters rather than the coding regions (Doebley, 1998; Wang et al., 1999; Frary et al., 2000; Zhang et al., 2000). Because the difference between CBF4 and CBF1, 2, and 3 does not appear to have resulted in the selection of fundamentally new functions at the respective loci, we suggest that gene duplication and then promoter evolution have resulted in different CBF loci with conserved protein function, but divergent regulatory elements that are responsive to pertinent environmental cues.
MATERIALS AND METHODS
Plant Growth
For all experiments requiring plate grown plant material, seeds were vapor sterilized with chlorine gas for 90 min, then suspended in a 0.15% (w/v) agarose solution and stratified for 2 to 4 d at 4°C. For selection of transformed plants, seeds were sown to 80% (w/v) Murashige and Skoog supplemented with 0.3% (w/v) Suc and 50 mg L−1 kanamycin. Kanamycin-resistant plants were transplanted to soil after 7 d. All plants were grown under continuous light with a light intensity of approximately 100 μE at 22°C in either environmental control chambers or growth rooms. Cold treatment of plants was done at 4°C under constant illumination from cool-white fluorescent lights (approximately 35 μmol m−2 s−1).
Constructs and Plant Transformation
The 5′ and 3′ ends of CBF4 was determined by RACE following the manufacturer's instructions (Marathon and SMART RACE systems, CLONTECH Laboratories, Palo Alto, CA). The CBF4 gene was amplified by PCR from cDNA using primers 5′-GCACGCGTCGACCATCTTATCCAAAGAAAAAATGAATCC and 5′ GGGAAAGCGGCCGCAACTTATTATCCAGAAAAAGAGCCAAAAAA. The cDNA was derived from a mixed mRNA population from several tissues including shoot, root, flower, rosette leaf, cauline leaf, silique, germinating seed, and a variety of conditions including treatment with auxin (1 μm 2,4-dichlorophenoxyacetic acid), ABA (50 μm), cold (4°C), mannitol (3 m), heat (37°C), sodium chloride (200 mm), pathogen infection (Erysiphe orontii and Fusarium oxysporum), salicylic acid (0.5 mm), and drought. The 720-bp product containing the entire coding region of CBF4 was cloned as an SalI-NotI fragment downstream of the CaMV 35S promoter in a standard binary transformation vector containing a kanamycin resistance selectable marker. The plasmid was introduced into Agrobacterium tumefaciens by electroporation. Arabidopsis plants (Columbia accession) were transformed by the floral dip method (Clough and Bent, 1998). Agrobacteria infiltration of tobacco leaves was carried out essentially the same as described by Yang et al. (2000), except that the CBF genes and the COR78 promoter::GUS constructs were cloned in separate plasmids.
Plant Material and Treatments
Arabidopsis ecotype Columbia was grown in a Percival series 101 (Percival Scientific, Inc., Perry, Iowa) controlled environment chamber at 22°C under continuous illumination of approximately 100 μmol m−2 s−1 for 12 to 14 d. Plants were grown under sterile conditions in petri plates according to Gilmour et al. (1998) with the exception that the seeds were germinated on filter paper for drought, ABA, and control experiments. Cold treatment was preformed by transferring plates to a cold room set at 4°C and were kept under constant illumination from cool-white fluorescent lights (approximately 35 μmol m−2 s−1). For the drought treatment, the filter papers were removed from the plate and placed to dry over desiccant. The ABA treatment was done by transferring the filter papers from the plate and placing them in Gamborgs B5 liquid plus 100 μm ABA. For controls, the plants containing filter papers were transferred to Gamborgs B5 liquid. Tissue was harvested at the indicated times and was immediately frozen in liquid nitrogen before total RNA was extracted.
RNA Hybridization and cDNA Probes
Total RNA was extracted from Arabidopsis plants described by Whitelam et al. (1993) with the exception that the last lithium chloride extraction was omitted. Northern transfers were prepared and hybridized as described by Hajela et al. (1990) and washed at high stringency with the temperature being 60°C instead of 50°C (Stockinger et al., 1997). Full-length cDNAs of CBF4, CBF1, COR15a, and eIF4a were labeled with P-32 with the random primers DNA labeling system (Life Technologies/Gibco-BRL, Cleveland) as directed by the manufacturer.
Stress Tolerance Assays
In the whole-plant freezing assay, 2-week-old plants grown on petri dishes, with or without cold acclimation for 4 d at 4°C, were frozen for 20 h at −15°C (cold-acclimated plants) or −10°C (nonacclimated plants), respectively. Plant survival was scored after 2 d of recovery under normal growth conditions. Electrolyte leakage assays were performed essentially as described (Gilmour et al., 2000) with a minor modification: Instead of leaves, two to four complete 2-week-old plants grown on petri dishes were used per tube. For the drought stress evaluation, pots (10-cm diameter) were filled with a 1:1 (v/v) vermiculite:perlite mix. To ensure even plant growth a thin (1-cm) layer of promix soil (Hummert International, Earth City, MO) was added. Seedlings were grown for 2 weeks with constant watering before the water was withheld. After 9 d without water, all the pots were rewatered simultaneously and the plant regrowth was scored 4 d later.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We thank Pierre Broun, Mark Leibman, and George Chiang at Mendel Biotechnology (Hayward, CA) and Katie Diller at Michigan State University/Plant Research Laboratory (East Lansing) for experimental assistance.
Footnotes
This work was supported in part by the National Science Foundation (grant no. 9983311 to J.Z.Z.), by the Department of Energy (grant no. DEFG0291ER20021 to M.F.T.), and by the Michigan Agricultural Experiment Station (to M.F.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006478.
LITERATURE CITED
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I. Insights into the structural and functional evolution of plant genomes afforded by the nucleotide sequences of chromosomes 2 and 4 of Arabidopsis thaliana. Yeast. 2000;17:1–5. doi: 10.1002/(SICI)1097-0061(200004)17:1<1::AID-YEA3>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci USA. 1996;93:9374–9377. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- Clavitier Y, Siminovitch D. Correlation between cold- and drought-induced frost hardiness in winter wheat and rye varieties. Plant Physiol. 1982;69:256–258. doi: 10.1104/pp.69.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Doebley J. Maize as a model system for investigating the molecular basis of morphological evolution in plants. Symp Soc Exp Biol. 1998;51:127–132. [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Girard L, Freeling M. Regulatory changes as a consequence of transposon insertion. Dev Genet. 1999;25:291–296. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Guy C, Haskell D, Neven L, Klein P, Smelser C. Hydration-state-responsive proteins link cold and drought stress in spinach. Planta. 1992;188:265–270. doi: 10.1007/BF00216823. [DOI] [PubMed] [Google Scholar]
- Hajela RK, Horvath DP, Gilmour SJ, Thomashow MF. Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990;93:1246–1252. doi: 10.1104/pp.93.3.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, McLarney BK, Thomashow MF. Regulation of Arabidopsis thaliana L. (Heyn) cor78 in response to low temperature. Plant Physiol. 1993;103:1047–1053. doi: 10.1104/pp.103.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Dunn MA. The molecular biology of plant acclimation to low temperature. J Exp Bot. 1996;47:291–305. [Google Scholar]
- Ingram J, Bartels D. The molecular basis for dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 2001;25:247–259. doi: 10.1046/j.1365-313x.2001.00947.x. [DOI] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B, Freeling M. Transposon-induced promoter scrambling: a mechanism for the evolution of new alleles. Proc Natl Acad Sci USA. 1995;92:1836–1840. doi: 10.1073/pnas.92.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens L, Doebley J. Molecular evolution of the teosinite branched gene among maize and related grasses. Mol Biol Evol. 2001;18:627–638. doi: 10.1093/oxfordjournals.molbev.a003843. [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz AM, Timmer RT, Browning KS. Sequences for two cDNAs encoding Arabidopsis thaliana eukaryotic protein synthesis initiation factor 4A. Gene. 1992;21:313–314. doi: 10.1016/0378-1119(92)90112-3. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/a:1006321900483. [DOI] [PubMed] [Google Scholar]
- Pearce RS. Molecular analysis of acclimation to cold. Plant Growth Regul. 1999;29:47–76. [Google Scholar]
- Razik MA, Quatrano RS. Effect of the nuclear factors EmBP1 and viviparous1 on the transcription of the Em gene in HeLa nuclear extracts. Plant Cell. 1997;9:1791–1803. doi: 10.1105/tpc.9.10.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins DM, Samuelson LC. Retrotransposons and the evolution of mammalian gene expression. Genetica. 1992;86:191–201. doi: 10.1007/BF00133720. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun. 1998;250:161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- Siminovitch D, Cloutier Y. Drought and freezing tolerance and adaptation in plants: some evidence of near equivalences. Cryobiology. 1983;20:487–503. doi: 10.1016/0011-2240(83)90037-8. [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Webb MS. Freeze-induced dehydration and membrane destabilization in plants. In: Somero G, Osmond B, editors. Water and Life: Comparative Analysis of Water Relationships at the Organismic Cellular and Molecular Level. Berlin: Springer-Verlag; 1992. pp. 338–362. [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998;118:1–8. doi: 10.1104/pp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Stec A, Hey J, Lukens L, Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. Abiotic stress signal transduction in plants: molecular and genetic perspectives. Plant Physiol. 2001;112:152–166. doi: 10.1034/j.1399-3054.2001.1120202.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought low-temperature or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by agro infiltration of tobacco leaves. Plant J. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Chopra S, Peterson T. A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell. 2000;12:2311–2322. doi: 10.1105/tpc.12.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]