Abstract
Each fiber of cotton (Gossypium hirsutum) is a single epidermal cell that rapidly elongates to 2.5 to 3.0 cm from the ovule surface within about 16 d after anthesis. A large number of genes are required for fiber differentiation and development, but so far, little is known about how these genes control and regulate the process of fiber development. To investigate gene expression patterns in fiber, a cDNA, GhTUB1, encoding β-tubulin was isolated from a cotton fiber cDNA library. The analyses of RNA northern-blot hybridization and reverse transcriptase-polymerase chain reaction demonstrated that GhTUB1 transcripts preferentially accumulated at high levels in fiber, at low levels in ovules at the early stage of cotton boll development, and at very low levels in other tissues of cotton. The corresponding GhTUB1 gene including the promoter region was isolated by screening a cotton genomic DNA library. To demonstrate the specificity of the GhTUB1 promoter, the 5′-flanking region including the promoter and 5′-untranslated region was fused with the β-glucuronidase reporter gene. The expression of the reporter chimera was examined in a large number of transgenic cotton plants. Histochemical assays demonstrated that GhTUB1::β-glucuronidase fusion genes were expressed preferentially at high levels in fiber and primary root tip of 1- to 3-d-old seedlings and at low levels in other tissues such as ovule, pollen, seedling cotyledon, and root basal portion. The results suggested that the GhTUB1 gene may play a distinct and required role in fiber development. In addition, the GhTUB1 promoter may have great potential for cotton improvement by genetic engineering.
Upland cotton (Gossypium hirsutum) and Sea Island cotton (Gossypium barbadense) are the most important fiber crops for the textile industry. Although classical breeding has contributed tremendously in terms of quality improvement and yield increase in the twentieth century, further improvements for fiber strength, length, chemical compatibility for dye binding, water absorption, thermal properties, and resistance to wrinkle and shrinkage are needed for textile and other industrial applications. The potential for improving these properties through classical breeding is limited because of requirements of species compatibility and available traits. Genetic engineering will provide novel approaches by introducing target genes of different sources into cotton to overcome the disadvantages of classical breeding (John and Keller, 1996).
Cotton fibers, or seed hairs, are single-cell trichomes that undergo rapid and synchronous elongation during seed development. Fiber development consists of four overlapping stages, initiation, primary cell wall formation, secondary cell wall formation, and maturation (Basra and Malik, 1984). During the initial stages of fiber development, 30% of epidermal cells on the ovule surface begin to enlarge and elongate rapidly. The primary cell wall formation starts at anthesis, and continues up to 19 to 20 d after anthesis (DPA). The primary wall is made up of cellulose, hemicellulose, pectins, proteins, and waxes. The elongating fiber cells are highly vacuolated. A central vacuole forms and occupies most of the cell volume (Basra and Malik, 1984; Kosmidou-Dimitropoulou, 1986). The secondary cell wall formation starts at about 16 DPA, overlapping with the late primary wall formation. During this period (16–40 DPA), rapid cellulose biosynthesis results in massive quantities of cellulose deposited in the secondary cell wall. The maturation of cotton fiber is at 40 to 50 DPA and is associated with changes in mineral content and enzyme levels/activities (Basra and Malik, 1984). Mature fiber is a biological composite of cellulose, water, small quantities of proteins, pectins, hemicellulose, mineral substances, wax, and small amount of organic acids, sugars, and pigments that provides excellent wearability and esthetics (Basra and Malik, 1984; Ryser, 1985; Arthur, 1990).
A number of genes, which are differentially expressed during different stages of fiber development, are required for fiber differentiation and development. So far, only a few of the genes, involved in the biosynthesis of the large numbers of fiber-specific structural proteins, enzymes, polysaccharides, waxes, or lignins, have been identified (John and Crow, 1992; John and Keller, 1995; John, 1996; Kawai et al., 1998; Liu et al., 2000; Ma et al., 1995, 1997; Orford and Timmis, 1998; Song and Allen, 1997). On the basis of their fiber-specific expression, these genes may offer potential for cotton fiber improvement.
Microtubules play important roles in a large number of basic cellular processes and in morphogenesis in higher plants (Kopczak et al., 1992). Microtubules in plants construct four arrays, i.e. preprophase band, mitotic spindle, phragmoplast, and cortical array, which function in the division and elongation of the plant cells. Three of the arrays (preprophase bands, phragmoplasts, and cortical microtubules) are unique to plant cells. Cortical microtubules are thought to provide spatial information to the organization of cellulose microfibrils in plant cells (Whittaker and Triplett, 1999). During cell elongation, highly organized microfibrils of cellulose confine turgor-driven cell expansion to a single major axis of growth (Giddings and Staehelin, 1991; Delmer and Amor, 1995).
Microtubules are one of the major cytoskeleton filaments of eukaryotic cells and mainly consist of tubulin, a heterodimeric protein composed of two highly conserved subunits, α- and β-tubulin. A less abundant form, γ-tubulin, is also found in higher plants (Liu et al., 1994). Both α- and β-tubulin genes are encoded by multigene families in eukaryotes (Cleveland and Sullivan, 1985; Silflow et al., 1987). There are at least six α-tubulin genes and nine β-tubulin genes in Arabidopsis (Kopczak et al., 1992; Snustad et al., 1992) and seven α-tubulin genes and six β-tubulin genes in maize (Zea mays; Montoliu et al., 1989, 1990, 1992; Villemur et al., 1992, 1994), respectively. In cotton, although nine α-tubulin and seven β-tubulin isotypes were identified on immunoblots of two-dimensional gels and the expression level of α-tubulin genes was investigated in fiber cells (Dixon et al., 1994; Whittaker and Triplett, 1999), there is no report on the isolation and characterization of an α- or β-tubulin gene so far. Here, we report the first isolation and characterization of a β-tubulin gene that is predominantly expressed in cotton fibers.
RESULTS
GhTUB1 cDNA and GhTUB1 mRNA Expression Patterns
Poly(A+) RNAs from cotton young fibers of about 8 and 14 DPA, respectively, were used to construct a cotton fiber cDNA library. About 300 cDNA clones were randomly picked and sequenced. Nine clones with potential involvement in cytoskeleton and cell expansion were selected by analyzing the sequence data.
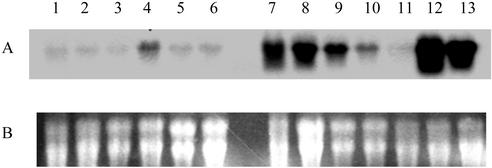
To find cDNA clones preferentially expressed in cotton fibers, the expression patterns of the selected cDNA clones were analyzed by northern-blot hybridization with total RNA isolated from cotton fibers, ovules, anthers, petals, stems, leaves, cotyledons, and roots, using the 3′-untranslated regions (3′-UTRs) of the nine cDNAs as gene-specific probes. The experimental results showed that one cDNA clone was strongly expressed in young fibers of 8 and 14 DPA and was also moderately or weakly expressed in young ovules (seeds) of 4, 8, 14, and 21 DPA (Fig. 1). We subsequently isolated the full-length cDNA by further screening of the fiber cDNA library. The cDNA was 1.6 kb in length and contained an open reading frame (ORF) encoding a β-tubulin polypeptide of 445 amino acids in length that shared a high percentage of homology with known β-tubulin proteins in plants. This is the first full-length β-tubulin gene (GhTUB1; GenBank accession no. AF487511) isolated in cotton.
Figure 1.
Northern analysis of GhTUB1 transcripts in cotton fiber, ovule (seed), petal, anther, leaf, stem, cotyledon, and root. Total RNA (20 μg lane−1) from leaf (1), stem (2), root (3), cotyledon (4), petal (5), anther (6), 4, 8, 14, 21, and 28 DPA ovule (7–11; young seed), and 8 and 14 DPA fiber (12 and 13) was fractionated on a denaturing 1.2% (w/v) agarose gel and transferred to nylon membrane (see “Materials and Methods”). A, Autoradiograph of RNA hybridization; B, loading of total RNA (20 μg lane−1) fractionated on a denaturing 1.2% (w/v) agarose gel.
Isolation and Characterization of the GhTUB1 Gene
Cotton genomic DNA libraries were screened using a GhTUB1 fragment as the probe. Three positive clones were isolated from the genomic libraries, two of which contained the complete GhTUB1 gene (GenBank accession no. AF487511) that was identical to the GhTUB1 cDNA. The isolated GhTUB1 gene contains an intact ORF, a full 3′-downstream sequence, and a shorter fragment of 5′-upstream region from the ORF. On the basis of the sequence of the GhTUB1 gene, we isolated two fragments (0.93 and 1.4 kb, respectively) upstream from the translational start site by Genome Walker PCR, for characterization of the promoter. Comparing the nucleotide and predicted polypeptide sequences of the cotton GhTUB1 gene with those in GenBank, we found that it shared more than 90% homology at the amino acid level and more than 70% homology at the nucleotide level in the gene coding region with the known β-tubulin cDNAs and genes from other plants, such as Arabidopsis, rice (Oryza sativa), maize, soybean (Glycine max), and pea (Pisum sativum; Guiltinan et al., 1987; Liaud et al., 1992; Snustad et al., 1992; Villemur et al., 1994; Koga-Ban et al., 1995).
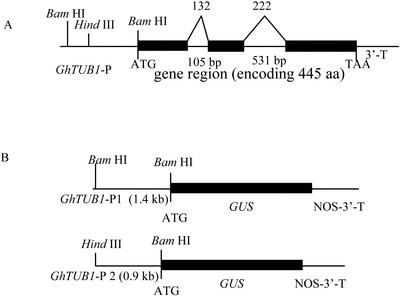
The GhTUB1 gene contained two introns in the ORF (Fig. 2A). The first intron at position 132 is 105 bp long, and the second intron at position 222 is 531 bp in length. Snustad et al. (1992) indicated that the intron positions in Arabidopsis β-tubulin genes are precisely conservative. The GhTUB1 gene showed that the introns are exactly at these conserved and homologous positions, as in the Arabidopsis β-tubulin genes.
Figure 2.
GhTUB1 gene structure and construction of the chimeric genes between the GhTUB1 5′-flanking region and the GUS gene. The physical structure of the GhTUB1 gene was characterized in detail. GhTUB1 encodes a 445-amino acid β-tubulin polypeptide. The translated portions of exons are denoted by black boxes; introns, the 5′-flanking region (including putative promoter and 5′-UTR), and the 3′ terminus are denoted by lines. The lengths of the introns in base pairs are indicated. The number at the boundaries of each exon indicates the codon at which the intron is located. Intron 1 splits codon 132, and intron 2 splits codon 222. The translation initiation and translation termination codons are shown. The length of the 5′-flanking region and cloning sites used for fusion constructs are shown. A, GhTUB1 gene structure; B, GhTUB1::GUS fusion; aa, amino acids.
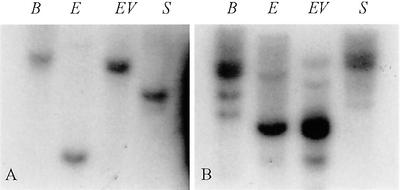
To investigate whether there are other β-tubulin genes that are closely related to GhTUB1, we used 5′-flanking sequences (0.9 kb) from GhTUB1 as gene-specific probe to hybridize with gel blots of cotton genomic DNA digested with four restriction enzymes (BamHI, EcoRI, EcoRV, and SacI). Figure 3A shows that the GhTUB1 probe hybridized to a single band in each of four restriction enzyme digests. This hybridization result suggests that the GhTUB1 gene is a single-copy gene in the cotton genome.
Figure 3.
Genomic DNA Southern-blot analysis. Total genomic DNA (30 μg lane−1) digested with restriction enzymes BamHI (B), EcoRI (E), EcoRV (EV), and SacI (S) and fractionated on a 0.8% (w/v) agarose gel was blotted to nylon membranes and hybridized with 32P-labeled GhTUB1 5′-region gene-specific probe (A) and 32P-labeled GhTUB1 ORF (third exon) probe (B).
In addition, we used the third exon sequence (680-bp fragment) of ORF from the GhTUB1 gene as a probe, and performed DNA Southern-blot analysis to determine the number of homologous β-tubulin genes in cotton. Among the four different restriction enzymes used, one main hybridizing band and two to four weak bands appeared in the hybridization experiments (Fig. 3B). The main band apparently represented the GhTUB1 gene, whereas the weak bands were other homologous cotton β-tubulin genes.
Preferential Expression of GhTUB1::GUS in Fiber
To determine the specificity of the GhTUB1 promoter, two different lengths of the GhTUB1 5′-flanking region including the putative promoter fragment and 5′-UTR before the translational initiation codon ATG were fused with the bacterial glucuronidase (GUS) coding sequence (Fig. 2B).
The GhTUB1::GUS constructs were introduced into cotton by Agrobacterium tumefaciens-mediated transformation. pBI121 vector (cauliflower mosaic virus [CaMV]35S::GUS) was used as a positive control, because CaMV35S promoter is a constitutive promoter active in all tissues of cotton and other plants (Odell et al., 1985; Ow et al., 1987; McCabe and Martinell, 1993). After 12 months of selection on the kanamycin-selective medium (see “Materials and Methods”), 22 and 33 independent kanamycin-resistant lines (5–20 plants each line) for construct pTUB14 (1.4-kb GhTUB1 promoter fragment fused with the GUS gene) and construct pTUB10 (0.9-kb GhTUB1 promoter fragment fused with the GUS gene), respectively, were regenerated. Southern-blot analysis (data not shown) confirmed that 15 of 22 lines for construct pTUB14 and 21 of 33 lines for construct pTUB10 are positive using the GUS gene as a probe, demonstrating GhTUB1::GUS was integrated into cotton genomes. From the 36 independent transgenic cotton lines, 13 were single-transgene insertion sites (single copy) in the cotton genome, and the remaining were two or more insertion sites (two or more copies) in the cotton genome.
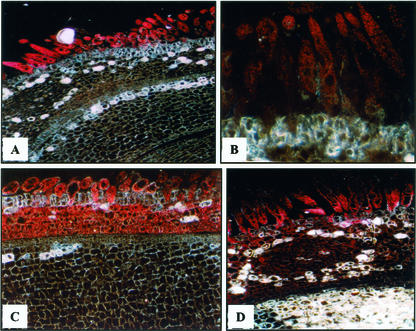
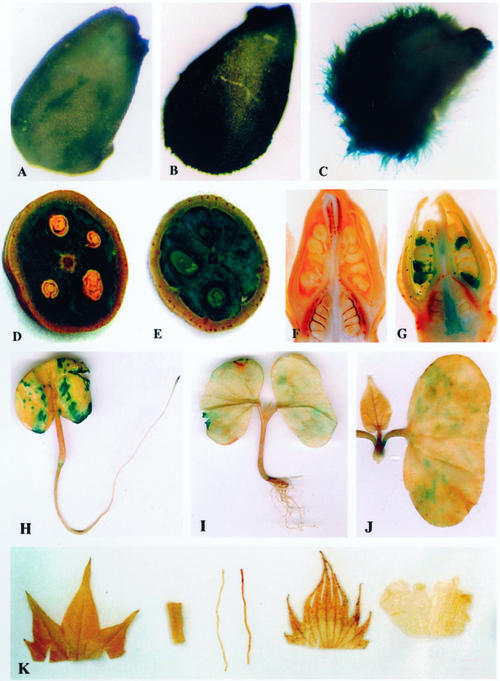
Transgenic cotton plants and their progenies (T1 or T2 heterozygous and homozygous plants) for each independent line were examined with histochemical assays for GUS expression, using non-transformed plants as negative controls. All of the 36 lines (at least five plants for each line) were examined, and we focused on 13 single-insertion lines and studied transgene segregation in heterozygous plants and GUS expression in homozygous plants. The histochemical assay in a large number of transgenic plants (more than 200 plants each generation) showed that GUS expression in the homozygous transgenic plants was not significantly different from that in the heterozygous transgenic plants. For all of the lines with 1.4-kb GhTUB1::GUS fusion, high levels of GUS expression were only shown in the young fibers (Figs. 4, A and B, and 5, A–D). The 0.9-kb GhTUB1 promoter was strongly expressed in the young fibers and in the inner cell layers of the seed coats (Fig. 4, C and D) and was weakly, moderately to strongly expressed in the pollens, ovaries, styles, and young seeds (Fig. 5, E and G), whereas the 1.4-kb GhTUB1 promoter showed weak or no expression in the above tissues except fibers (Figs. 4, A and B, and 5, D and F). The results suggest that some sequence elements found only in the 1.4-kb promoter may be important for fiber-specific expression of the GhTUB1 gene in cotton. However, no GUS activity was detected in leaves, roots, stems, petals, and sepals during different development stages in the transgenic plants (Fig. 5K). For fiber, a higher level of GUS activity was observed in the primary wall formation stage, and then GUS activity gradually declined with fiber development. Non-transformed plants showed no GUS activity in any tissue under the assay conditions used.
Figure 4.
Histochemical localization of GUS gene expression at the early stage of the fiber development in transgenic plants containing the GhTUB1/GUS fusion gene. A through D, Micrographs of 5-μm-thick cross sections of 1 to 2 DPA ovules. A and B, Early and late of 2-DPA ovules (250× and 700×, respectively) of transgenic cotton with 1.4-kb GhTUB1::GUS fusion. High level of GUS activity was only found in the fiber, and very weak GUS staining was seen in the inner cell layers of seed coat in some transgenic lines, but no GUS expression was detected in the outermost cell layer except fiber cells. C and D, One- and 2-DPA ovules (500× and 350×, respectively) of transgenic cotton with 0.9-kb GhTUB1::GUS fusion. Strong GUS activity was observed in the fiber and in the inner cell layers of the seed coat, but no GUS staining was detected in the outermost cell layer (the epidermis) except fiber cells. In all of the transgenic ovules, GUS activity was at an undetectable level in the embryo.
Figure 5.
Histochemical localization of GUS gene expression in transgenic cotton plants containing the GhTUB1/GUS fusion gene. A through C, One-, 2-, and 3-DPA ovules (50×). Strong GUS activity was observed in the fiber. D, Fourteen-DPA boll (cross section) of transgenic cotton with 1.4-kb GhTUB1::GUS fusion. Strong GUS activity was seen in the developing fiber. E, Fourteen-DPA boll (cross section) of transgenic cotton with 0.9-kb GhTUB1::GUS fusion. Strong GUS activity was detected in the developing fiber, and weak GUS staining was also visible in the seed coat and the embryo. F, Flower bud (longitudinal section, 5×) of transgenic cotton with 1.4-kb GhTUB1::GUS fusion. GUS activity was undetectable in the tissues of the bud. G, Flower bud (longitudinal section, 5×) of transgenic cotton with 0.9-kb GhTUB1::GUS fusion. Moderate or strong GUS activity was observed in the pollen, and weak or moderate GUS staining was found in the ovary and style. H through J, Three-, 7-, and 10-d-old seedlings of transgenic cotton with 1.4-kb GhTUB1::GUS fusion. H, Three-day-old cotton seeding. GUS gene was expressed moderately or weakly in the cotyledon and root basal portion, and high level of GUS expression was located in the primary root tip. I, Seven-day-old cotton seedling. Weak GUS activity was observed only in the cotyledon. J, Ten-day-old cotton seedling. GUS activity was decreased to a very low level in the cotyledon, and no GUS expression was detected in the leaf and shoot apex. K, From left to right: leaf, stem, root, sepal, and petal of transgenic cotton plant. No GUS activity was found in all of the tissues.
Expression of GhTUB1::GUS during Early Seedling Development
To examine all possible expression patterns of the GhTUB1 gene during cotton plant development, independent transgenic cotton lines and their progenies were histochemically assayed at different development stages, including seedlings and juvenile and mature plants. Examination of T1 and T2 generations showed there was similar GUS expression patterns in all of the transgenic lines containing the 1.4-kb GhTUB1 promoter fragment, although some variation was observed in the intensity of GUS activity among the lines. Transgenic cotton seeds were soaked in water for 1 d and then germinated in soil. In 2- to 3-d-old seedlings, moderate GUS activity was detected in the two cotyledons and the basal portion of the root, whereas strong GUS staining was seen in the root tip (Fig. 5H). In the 7-d-old seedlings, very weak GUS staining was only found in the basal portion of the primary root in some transgenic lines, whereas GUS activity was at an undetectable level in the lateral root system, including root tips (Fig. 5I). Markedly decreased GUS activity similarly was seen in cotyledons of seedlings in which true leaves had just emerged (Fig. 5I). As the seedlings developed, GUS activity was gradually decreased to very low levels until undetectable in cotyledons of 10-d-old seedlings (Fig. 5J), and no GUS staining was found in the root system. Shoot apices and leaves in the seedlings did not show detectable GUS activity (Fig. 5, I and J). When the seedlings developed to juvenile plants with full-expanded leaves in 25 to 30 d, GUS activity was undetectable in all of the tissues of the juvenile plants under the conditions examined.
By means of gene-specific reverse transcriptase (RT)-PCR (see “Materials and Methods”), we further verified that GhTUB1 transcripts (mRNAs) accumulated at low levels in 3-d-old cotyledons, and no RT-PCR band was seen in the RNA samples from the roots and hypocotyls of 3- and 7-d-old seedlings (data not shown). The level of GhTUB1 RT-PCR products from the 3-d-old cotyledons agreed with expression of the GhTUB1::GUS fusion genes in the cotyledons at the same development stage. GhTUB1 RT-PCR products were at undetectable levels in the 7-d-old cotyledons under the RT-PCR conditions used, suggesting GhTUB1 activity rapidly declined as seedlings developed. However, the GhTUB1 expression in the cotyledons was much lower than in the fibers (Fig. 1). This result, together with the GUS histochemical assay results, suggested that GhTUB1 is expressed at moderate to low levels in the cotyledons, the basal portion and tip of the main root, and GhTUB1 expression is then turned off.
DISCUSSION
The GhTUB1 Is Preferentially Expressed in Cotton Fiber
The transcripts of the GhTUB1 gene exhibited the highest accumulation in young fibers of cotton at 8 DPA, and then there was a gradual decrease until expression levels were undetectable with further development of the fibers. This suggests that the expression of GhTUB1 is specifically regulated at the transcriptional level during cotton fiber and ovule development, as has been described for some other genes (John and Crow, 1992; Ma et al., 1995, 1997; John, 1996; Rinehart et al., 1996; Song and Allen, 1997; Kawai et al., 1998; Shin and Brown, 1999; Whittaker and Triplett, 1999). We also isolated the GhTUB1 gene and characterized the gene structure and expression, focusing on the role of the promoter that may have potential commercial value in the improvement of cotton fiber.
Several other genes with fiber-preferential expression have been characterized. John and Crow (1992) identified the E6 gene that is preferentially expressed in fiber. E6 transcripts were detected throughout the development of the fiber, whereas concentrations of the E6 mRNA and protein are highest during the late primary cell wall and early secondary cell wall synthesis stages. A chimera between the 5′-upstream region of the E6 gene and the coding region of a carrot (Daucus carota) extensin gene demonstrated that the E6 promoter directed the gene expression in a fiber-specific and developmentally regulated fashion. Although further study revealed that E6 gene is not critical to the normal development or structural integrity of cotton fiber (John, 1996), its useful promoter and other regulatory elements have been applied for modification of fiber properties through directing the heterologous gene expression in fiber (John and Keller, 1996). John and colleagues also isolated and characterized two other fiber-specific genes, H6 and FbL2A, that were expressed in a similar manner as E6 (John and Keller, 1995; Rinehart et al., 1996). Analysis of mRNA accumulation for the genes involved in osmoregulation during cell expansion of developing fibers revealed that the gene transcripts accumulated to highest levels during the period of peak expansion (12–15 DPA) and then declined with the onset of secondary cell wall synthesis (Smart et al., 1998). Two discrete patterns of the transcript accumulation of α-tubulin genes were observed in developing cotton fibers. Transcripts of GhTua2/3 and GhTua4 genes increased in abundance from 10 to 20 DPA, whereas GhTua1 and GhTua5 transcripts were abundant only through to 14 DPA, and dropped significantly at 16 DPA with the onset of secondary wall synthesis (Whittaker and Triplett, 1999). GhEXP1, an expansin cDNA isolated from cotton fibers (Orford and Timmis, 1998), is also a fiber-specific gene whose transcripts were most abundant during the elongation phase of fiber growth and decreased gradually after 16 to 20 DPA (Orford and Timmis, 1998; Ruan et al., 2001). Ma and colleagues isolated cotton Ltp3, Ltp6 genes, which were specifically expressed in fibers (Ma et al., 1995, 1997; Hsu et al., 1999; Liu et al., 2000). Expression of these genes can be loosely described as fiber specific, although many “fiber-specific” genes, like GhTUB1, exhibit low levels of expression in other tissues. The fiber-specific expression patterns of these genes may provide important clues about their roles during fiber development.
The GhTUB1 Promoter Directs the Developmentally Regulated and Tissue-Preferential Expression of the Gene
Expression of GUS or other marker genes has been extensively used in assessing activity and tissue specificity of plant promoters. We linked the putative promoter fragments of GhTUB1 to GUS, and introduced the chimeric genes into cotton through A. tumefaciens-mediated gene transfer. GUS activities in different tissues of transgenic plants were detected by histochemical assay. Strong GUS expression was observed in the developing fibers, but no or low levels of GUS activity were detected in other tissues (such as cotyledons, hypocotyls, leaves, stems, roots, ovules, pollens, petals, and sepals) of the stable transgenic cotton plants (T0–T2 generation). This result was consistent with analysis of GhTUB1 mRNA accumulation (Figs. 1, 4, and 5), demonstrating that the 1.4-kb GhTUB1 promoter contains all of the cis elements required for fiber-specific and developmental regulated expression of the gene. Other studies have found that the FbL2A promoter is active during the secondary wall formation, whereas the E6 promoter is more active during the primary wall formation in fiber cell development (John and Crow, 1992; Rinehart et al., 1996). In our observation, the GhTUB1 promoter behaves in a manner similar to the E6 promoter in the developing fibers, with the maximum level of promoter expression occurring in the young fibers at the stage of primary cell wall formation and decreasing gradually thereafter.
It should be noted that GhTUB1 is not expressed in leaf, stem, petal, and sepal trichomes, which, like fibers, elongate by tip growth. Therefore, the β-tubulin isotype encoded by GhTUB1 is not required for trichome growth per se. If GhTUB1 is in fact required for proper microtubule function in developing fibers, it may be because of coevolution with fiber-preferential expression patterns.
The previous studies on plant tubulin genes have demonstrated that promoters direct the developmentally regulated, tissue-specific expression of the genes. In Arabidopsis, the TUA1 promoter is preferentially active in post-mitotic pollen (Carpenter et al., 1992), whereas the TUB1 promoter is predominantly active in epidermal and cortical cells of primary roots, and the TUB8 promoter exhibits high activity in the endodermal and phloem cells of primary roots and in the vascular tissues of leaves, stems, and flowers (Chu et al., 1998). In maize, the tua1 promoter is mainly active in cortex-producing meristematic cells and in pollen, whereas tua3 is active in cells that are differentiating to form vascular bundles in the root and shoot apices (Uribe et al., 1998). Our results, based on both GhTUB1 mRNA and the activity of the GhTUB1 promoter, suggest that GhTUB1 plays a distinct and required role in fiber-preferential expression. The differential expression of the tubulin genes may be linked to processes where microtubules have different functions, suggesting that in plants, as in animals, there are functional differences in the tubulin isotypes.
The results of this study contribute to an understanding of the regulation of gene expression in developing fibers. GhTUB1 may be closely related to processes of cell elongation during fiber development, although we do not know what the precise function of the gene is in fiber cells yet. With the isolation and characterization of a fiber-specific promoter, GhTUB1, we are able to express target gene products in the developing fiber. The GhTUB1 and an additional fiber-specific promoter that we have isolated (X.-B. Li, L. Cai, N.-H. Cheng, and J.-W. Liu, unpublished data) will be useful tools for modifying fiber properties through genetic engineering.
MATERIALS AND METHODS
Collection of Plant Material
Cotton (Gossypium hirsutum cv Coker 312) seeds were surface-sterilized with 70% (v/v) ethanol for 30 to 60 s and 10% (v/v) H2O2 for 30 to 60 min, followed by washing with sterile water. The seeds were germinated on one-half Murashige and Skoog medium in light at 28°C in a culture room. Cotyledons and hypocotyls cut from sterile seedlings were used as transformation explant materials. Cotton cvs DP5415 and Xuzhou142 plants were grown in pots for DNA and RNA extraction.
RNA Isolation and Construction of Cotton cDNA Libraries
Total RNA was extracted from young fibers, ovules, anthers, petals, leaves, cotyledons, and roots of cotton by the following method. Two grams of cotton tissue were frozen in liquid nitrogen and thoroughly homogenized into fine powders. Six milliliters of extraction buffer, consisting of 4 mol L−1 guanidine thiocyanate, 25 mmol L−1 sodium citrate (pH 7.0), 0.5% (w/v) sacrosine, and 100 mmol L−1 β-mercaptoethanol, was added to the homogenized powder in a 50-mL tube. One-tenth volume of 2 mol L−1 sodium acetate, 1 volume of acidic phenol, and 0.2 volume of chloroform was added to each tube, vigorously mixed by vortex, set on ice for 15 min, and then centrifuged at 5,000 rpm for 15 min at 4°C. The upper phase (water phase) was transferred to a new tube, and 100 μL of RNAMATRIX glass beads (Bio 101, Vista, CA) were added to each tube. After shaking (50 rpm) for 15 min at room temperature and 5 min of centrifugation, the pellets were resuspended with 6 mL of 6 mol L−1 guanidine thiocyanate, spun down again, and washed twice with 60% (v/v) ethanol. The pellets were resuspended with 300 μL of RNase-free water and centrifuged for 2 min at 14,000 rpm. The supernatant was transferred to a clean tube, and 2 volumes of ethanol and 0.1 volume of sodium acetate were added to each tube for RNA precipitation. After centrifuging at 14,000 rpm for 30 min, the RNA pellet was washed with 70% (v/v) ethanol. The isolated RNA was dissolved in RNase-free water and stored at −80°C.
Poly(A+) mRNA was prepared from a pool of total RNA isolated from 8 and 14 DPA fibers by using an mRNA purification kit (Qiagen USA, Valencia, CA). Complementary DNA was synthesized and cloned into the EcoRI-XhoI sites of the ZAP Express Vector by using a cDNA synthesis kit according to manufacturer's instruction (Stratagene, La Jolla, CA). Ligation mixture was packaged by using a ZAP-cDNA Gigapack Gold III cloning kit (Stratagene).
Isolation of GhTUB1 cDNA Clone and Northern-Blot Analysis
Three hundred cDNA clones randomly selected from the cotton fiber cDNA library were partially sequenced with the T3 primer (5′-AATTAACCCTCACTAAAGGG-3′) in the ZAP vector (Stratagene). Nine clones (including GhTUB1) with potential involvement in cytoskeleton and cell expansion were selected by analyzing the sequence data and then completely sequenced. For each candidate, a gene-specific 3′-UTR sequence probe was prepared by PCR amplification. The PCR primers for the GhTUB1 probe were GhTUB1-3′L (5′-AAATCTAATGGAATAATTTGGATGT-3′) in −1 to +23 of 3′-UTR from stop codon and GhTUB1-3′R (5′-ACTTAAGGTGTACTTGAAATTACT-3′) in complementary chain +201 to +178 of 3′-UTR from stop codon. RNA samples (20 μg lane−1) from different cotton tissues were separated on 1.2% (w/v) agarose-formaldehyde gels and transferred onto Hybond-N nylon membranes by capillary blotting. Gene-specific probes were labeled with [32P]dCTP by using the random primer method (Prime-a-Gene Labeling System Kit, Promega, Madison, WI) at 28°C for 1 h. RNA northern-blot hybridization was performed at 42°C overnight in RNA hybridization solution (47% [v/v] formamide, 3× SSPE, 1% [w/v] SDS, and 6.5% [v/v] 100× Denhardts) with 32P-labeled gene-specific probes. The membranes were washed three times at 55°C for 15 min in 0.1× SSC and 0.5% (w/v) SDS. After drying briefly, the membranes were exposed to X-film (Eastman Kodak, Rochester, NY) with two intensifying screens at −80°C for 1 to 3 d.
For isolating the full GhTUB1 cDNA, 2 × 105 cDNA clones were screened with a [α-32P]dCTP probe (GhTUB1 cDNA 3′-UTR fragment) generated using a random primer method (Prime-a-Gene Labeling System, Promega). The membrane filters (Hybond N, Amersham Biosciences AB, Uppsala) were hybridized overnight in ExpressHyb solution (BD Biosciences Clontech, Palo Alto, CA) at 68°C and washed with 0.1× SSC and 0.5% (w/v) SDS for 30 to 60 min. The 32P-labeled membranes were exposed to x-ray film at −80°C. Positive cDNA clones were excised with the helper phage.
RT-PCR Analysis
Total RNA samples (2 μg reaction−1) from roots, cotyledons, and hypocotyls of 3- and 7-d-old seedlings and 8 DPA fibers of mature plants were used as RT-PCR templates. The oligonucleotide primers were GhTUB1-6L (5′-GTACCCTCAAGCTCACTACTC-3′) in +638 to +658 of ORF from translational initiation site and GhTUB1-3′R (5′-ACTTAAGGTGTACTTGAAATTACT-3′) in complementary chain +201 to +178 of 3′-UTR from stop codon. The amplified region included the last intron junction for the GhTUB1 cDNA. Potential genomic DNA contamination could be ruled out based on the length of the products, because the genomic products would have been 531 bp longer than the cDNA products. RT-PCR reaction was performed using One-Step RT-PCR kit (Qiagen) according to the manufacturer's instruction. In brief, a 50-μL reaction containing a 2-μg template, 50 pmol of primers, 20 nmol of each dNTP, 1× One-step RT-PCR buffer, 2 μL of enzyme mix, and 4 units of RNase inhibitor was subjected to reverse transcription at 50°C for 30 min, followed by initial PCR activation at 95°C for 15 min, and then 40 PCR cycles at 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min, and a final extension at 72°C for 10 min.
DNA Isolation and Southern-Blot Analysis
Total DNA was extracted and purified from leaves of cotton plants by using the Paterson's method (1993) with some modification. Two grams of leaves were homogenized thoroughly in liquid N2. Twenty milliliters of ice-cold extraction buffer was added to the homogenized tissues in a 50-mL tube and centrifuged at 2,500 rpm for 15 min. After removing the supernatant, 10 mL of lysis buffer was added to each tube. The resuspended pellets were incubated in 65°C for 30 min. Ten milliliters of chloroform was added to each tube, mixed with the samples and centrifuged at 3,500 rpm for 10 min. The supernatant was transferred to a clean tube, and the chloroform extraction was repeated one more time. The supernatant was transferred to a clean tube and 0.6 volume of isopropanol was added to each tube for DNA precipitation. After centrifuging at 3,500 rpm for 30 min, the DNA was washed with 70% (v/v) ethanol. The isolated genomic DNA was dissolved in sterile water and stored at −20°C.
Total genomic DNA from cotton leaves was digested with restriction enzymes, separated on agarose gels, and transferred onto Hybond-N nylon membranes by capillary blotting. DNA Southern blots were hybridized overnight in ExpressHyb solution (BD Biosciences Clontech) at 68°C with [32P]DNA probes prepared by random labeling (Prime-a-Gene Labeling System, Promega). After hybridization, the blots were washed at 68°C in 0.1× SSC, 0.5% (w/v) SDS for 30 to 60 min. The 32P-labeled membranes were exposed to x-ray film at −80°C for 1 to 3 d.
Construction and Screening of Cotton Genomic Libraries
For construction of the genomic library, DNA was partially digested with BamHI or EcoRI, and the DNA fragments (4–12 kb) were cloned in the BamHI or EcoRI site of ZAP express vector (Stratagene) according to the manufacturer's instruction. The ligation mixture was packaged using a Gigapack Gold packaging kit (Stratagene). About 3 × 106 clones were screened with an [α-32P]dCTP probe (GhTUB1 cDNA 3′-UTR fragment) generated using a random primer method (Prime-a-Gene Labeling System, Promega). The membrane filters (Hybond N, Amersham Biosciences AB) were hybridized overnight in ExpressHyb solution (BD Biosciences Clontech) at 68°C and washed with 0.1× SSC and 0.5% (w/v) SDS for 30 to 60 min. The 32P-labeled membranes were exposed to x-ray film at −80°C. Positive clones were excised with the helper phage, and phagemid DNA was isolated for GhTUB1 gene sequence analysis.
Construction of Genome Walker Libraries and Genome Walker PCR
Genome Walker libraries were constructed by using Universal Genome Walker kit according to manufacturer's instruction (BD Biosciences Clontech). Cotton genomic DNA (2.5–5 μg) in each reaction was digested at 37°C overnight with a restriction enzyme. Five enzymes (DraI, EcoRV, PvuII, ScaI, and StuI) were used in five reactions, respectively. After purification with phenol and chloroform extraction and ethanol precipitation, the digested DNA was ligated to Genome Walker adapters (5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT-3′) at 16°C overnight. Primers for PCR-based DNA walking in Genome Walker Libraries were gene-specific GhTUB1-P1 (5′-TATCTGATTGCCGCATTGGCCACCTTG-3′) and GhTUB1-P2 (5′-GGATGTGAAGGATTTCTCTCATTTTCTC-3′) in complementary chain +48 to +22 and +22 to −6 of ORF from translational initiation site, adapter sequence AP1 (5′-GTAATACGACTCACTATAGGGC-3′) and AP2 (5′-ACTATAGGGCACGCGTGGT-3′). Two Genome Walker PCR reactions were carried out successively using Advantage-HF PCR Kit, which is a KlenTaq-based system with Pfu-like high fidelity and efficiency in the amplification of DNA template, and Advantage 2 PCR Kit (BD Biosciences Clontech). GhTUB1-P1 and AP1 were used in primary PCR, and GhTUB1-P2 and AP2 were used in secondary (nested) PCR. In a 50-μL PCR mix, 1 μL of each Genome Walker DNA library was used as templates in the primary PCR, and 2 μL of primary PCR products was used as templates in secondary PCR. The PCR was started at 95°C for 1 min, followed by 35 cycles consisting of 95°C for 15 s and 68°C for 4 min, and a final extension at 68°C for 6 min. Target PCR products were expected to overlap GhTUB1 5′-UTR (59 bp) for identifying the GhTUB1 promoter sequence.
DNA Sequence Analysis
GhTUB1 genomic and cDNA clone sequencing was conducted by ABI Prism 377 DNA Sequencer (Applied Biosystems, Foster City, CA) following the protocol provided by the manufacturer, except that several degenerate GhTUB1-specific primers were used. Sequences were analyzed using DNA analysis program (DNAStar software).
GUS Reporter Constructs and Cotton Transformation
A HindIII site and a BamHI site were introduced at the 5′-end and 3′-end of the GhTUB1 5′-upstream region (including the putative promoter fragment and 5′-UTR before translational initiation codon ATG), respectively, by PCR. The HindIII/BamHI fragment (0.93 kb) of GhTUB1 5′-upstream region was subcloned into pGEM-T vector (Promega). Plasmid DNA containing the GhTUB1 5′-upstream fragment was digested with HindIII and BamHI, and subcloned into the HindIII/BamHI sites of the pBI121 vector, to replace the CaMV 35S promoter and generate the chimeric GhTUB1::GUS construct (pTUB10). Another BamHI/BamHI fragment (1.4 kb) from the GhTUB1 5′-upstream region was similarly subcloned into the BamHI site of the pBI101 vector to regenerate the chimeric GhTUB1::GUS construct (pTUB14; Fig. 3B).
Cotton cv Coker 312 cotyledon and hypocotyl explants were transformed with chimeric GhTUB1::GUS fusion genes using Agrobacterium tumefaciens-mediated DNA transfer. After cutting into 5- × 5-mm pieces or 5-mm segments, the explants were immersed in A. tumefaciens (strain LBA4404) suspension (OD = 0.2–0.4) for 15 min. The infected explants were transferred onto callus-induced medium (Murashige and Skoog salts + B5 vitamins, complemented with 0.1 mg L−1 2,4-dichlorophenoxyacetic acid, 0.1 mg L−1 kinetin, 0.75 g L−1 MgCl2, 30 g L−1 Glc, and 2.0 g L−1 phytogel, pH 6.4) at 24°C for 2 d of cocultivation. After being washed with liquid Murashige and Skoog medium, the explants were cultured on selection medium (callus-induced medium with 50 mg L−1 kanamycin and 200 mg L−1 cefetoxime) in the light at 28°C for selecting transformants. After being subcultured for about 3 months, the kanamycin-resistant calli were transferred onto differentiation medium (DM; Murashige and Skoog salts and B5 vitamins, complemented with 19 g L−1 KNO3, 0.75 g L−1 MgCl2, 30 g L−1 Glc, and 3 g L−1 phytogel, pH 6.4). The somatic embryos begin to be formed when the calli were subcultured on DM medium for about 3 to 5 months. The somatic embryos were further developed to maturation on the DM medium, and then were transferred onto GM medium (one-half Murashige and Skoog with 0.01 mg L−1 naphthalene-acetic acid) for germination. A total of 325 regenerated transgenic cotton plants (T0 generation) from 36 independent transformed lines were transplanted in soil for maturation. To identify the homozygous transgenic progeny plants (T1 and T2 generations), the T2 or T3 seeds from each transgenic progeny plant were germinated on selection medium (one-half Murashige and Skoog inorganic salts with 200 mg L−1 kanamycin, without Suc) for assaying the ratio of kanamycin-resistant seedlings to sensitive ones, and then the putative homozygous plants were further confirmed by Southern blot, PCR, and histochemical assay.
Histochemical Assay of GUS Gene Expression
Histochemical assays for GUS activity in transgenic cotton plants were conducted according to the protocol described previously by Jefferson et al. (1987) with some modifications. Fresh tissues from the plants were incubated in 5-bromo-4-chloro-3-indolylglucuronide (X-gluc) solution consisting of 0.1 mol L−1 sodium phosphate (pH 7.0), 10 mmol L−1 EDTA, 0.5 mmol L−1 potassium ferrocyanide, and 0.5 mmol L−1 potassium ferricyanide, and 0.1% (w/v) X-gluc (BD Biosciences Clontech) for 4 to 8 h. The stained plant materials were then cleared and fixed by rinsing with 100% and 70% (v/v) ethanol successively, and the samples were examined and photographed directly or under a microscope.
After the 1 to 3 DPA ovules were incubated in X-gluc solution, the stained ovules were fixed with 2.5% (v/v) glutaraldehyde in 0.1 mol L−1 sodium phosphate buffer (pH 7.2) overnight at room temperature, dehydrated through conventional ethanol series, and embedded in plastics according to manufacturer's instruction (Historesin Embedding Kit, Leica, Wetzlar, Germany). Sections (5–7 μm thick) were made with a Leica microtome. The ovule cross sections were examined and photographed under a microscope.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
ACKNOWLEDGMENT
We thank Dr Wei-Cai Yang for his valuable suggestions and comments.
Footnotes
This work was supported by Delta and Pine Land Co. and by the National Science and Technology Board (Republic of Singapore).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005538.
LITERATURE CITED
- Arthur JC. Cotton. In: Kroschwitz JI, editor. Polymers: Fibers and Textile: A Compendium. New York: Wiley; 1990. pp. 119–141. [Google Scholar]
- Basra AS, Malik CP. Development of the cotton fiber. Int Rev Cytol. 1984;89:65–113. [Google Scholar]
- Carpenter JL, Ploense SE, Snustad DP, Silflow CD. Preferential expression of an α-tubulin gene of Arabidopsis in pollen. Plant Cell. 1992;4:557–571. doi: 10.1105/tpc.4.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B, Wilson TJ, McCune-Zierath C, Snustad DP, Carter JV. Two β-tubulin genes, TUB1 and TUB8, of Arabidopsis exhibit largely nonoverlapping patterns of expression. Plant Mol Biol. 1998;37:785–790. doi: 10.1023/a:1006047129410. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Sullivan KF. Molecular biology and genetics of tubulin. Annu Rev Biochem. 1985;54:331–365. doi: 10.1146/annurev.bi.54.070185.001555. [DOI] [PubMed] [Google Scholar]
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DC, Seagull RW, Triplett BA. Changes in the accumulation of α- and β-tubulin isotypes during cotton fiber development. Plant Physiol. 1994;105:1347–1353. doi: 10.1104/pp.105.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Staehelin LA. Microtubule-mediated control of microfibril deposition: a re-examination of the hypothesis. In: Lloyd CW, editor. The Cytoskeletal Basis of Plant Growth and Form. London: Academic Press; 1991. pp. 85–99. [Google Scholar]
- Guiltinan MJ, Ma DP, Barker RF, Bustos MM, Cyr RJ, Yadegari R, Fosket DE. The isolation, characterization and sequence of two divergent β-tubulin genes from soybean (Glycine max L.) Plant Mol Biol. 1987;10:171–184. doi: 10.1007/BF00016154. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Creech RG, Jenkins JN, Ma DP. Analysis of promoter activity of cotton lipid transfer protein gene Ltp6 in transgenic tobacco plants. Plant Sci. 1999;143:63–70. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME. Structural characterization of genes corresponding to cotton fiber mRNA, E6: reduced E6 protein in transgenic plants by antisense gene. Plant Mol Biol. 1996;30:297–306. doi: 10.1007/BF00020115. [DOI] [PubMed] [Google Scholar]
- John ME, Crow LJ. Gene expression in cotton fiber: cloning of the mRNAs. Proc Natl Acad Sci USA. 1992;89:5769–5773. doi: 10.1073/pnas.89.13.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME, Keller G. Characterization of mRNA for a proline-rich protein of cotton fiber. Plant Physiol. 1995;108:669–676. doi: 10.1104/pp.108.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME, Keller G. Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc Natl Acad Sci USA. 1996;93:12768–12773. doi: 10.1073/pnas.93.23.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Aotsuka S, Uchimiya H. Isolation of a cotton CAP gene: a homologue of adenylyl cyclase-associated protein highly expressed during fiber elongation. Plant Cell Physiol. 1998;39:1380–1383. doi: 10.1093/oxfordjournals.pcp.a029346. [DOI] [PubMed] [Google Scholar]
- Koga-Ban Y, Niki T, Nagamura Y, Sasaki T, Minobe Y. cDNA sequences of three kinds of beta-tubulins from rice. DNA Res. 1995;2:21–26. doi: 10.1093/dnares/2.1.21. [DOI] [PubMed] [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis thaliana contains at least six expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidou-Dimitropoulou K. Hormonal influences in fiber development. In: Mauney JR, Stewart JM, editors. Cotton Physiology. Memphis, TN: The Cotton Foundation; 1986. pp. 361–373. [Google Scholar]
- Liaud MF, Brinkmann H, Cerff R. The β-tubulin gene family of pea: primary structures, genomic organization and intron-dependent evolution of genes. Plant Mol Biol. 1992;18:639–651. doi: 10.1007/BF00020007. [DOI] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. γ-Tubulin in Arabidopsis: gene sequence, immunoblot, and immunofluorescence studies. Plant Cell. 1994;6:303–314. doi: 10.1105/tpc.6.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Creech RG, Jenkins JN, Ma DP. Cloning and promoter analysis of the cotton lipid transfer protein gene Lpt3. Biochim Biophys Acta. 2000;1487:106–111. [PubMed] [Google Scholar]
- Ma DP, Liu HC, Tan H, Creech RG, Jenkins JN, Chang YF. Cloning and characterization of a cotton lipid transfer protein gene specifically expressed in fiber cells. Biochim Biophys Acta. 1997;1344:111–114. doi: 10.1016/s0005-2760(96)00166-x. [DOI] [PubMed] [Google Scholar]
- Ma DP, Tan H, Si Y, Greech RG, Jenkins JN. Differential expression of a lipid transfer protein gene in cotton fiber. Biochim Biophys Acta. 1995;1257:81–84. doi: 10.1016/0005-2760(95)00077-p. [DOI] [PubMed] [Google Scholar]
- McCabe DE, Martinell BJ. Transformation of elite cotton cultivars via particle bombardment of meristems. Biotechnology. 1993;11:596–598. [Google Scholar]
- Montoliu L, Puigdomenech P, Rigau J. The Tubα3 gene from Zea mays: structure and expression in dividing plant tissues. Gene. 1990;94:201–207. doi: 10.1016/0378-1119(90)90388-8. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomenech P. A tandem of α-tubulin genes preferentially expressed in radicular tissues from Zea mays. Plant Mol Biol. 1989;193:427–438. doi: 10.1007/BF00015650. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomenech P. Analysis by PCR of the number of homologous genomic sequences to α-tubulin in maize. Plant Sci. 1992;84:179–185. [Google Scholar]
- Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Orford SJ, Timmis JN. Specific expression of an expansin gene during elongation of cotton fibers. Biochim Biophys Acta. 1998;1398:342–346. doi: 10.1016/s0167-4781(98)00065-7. [DOI] [PubMed] [Google Scholar]
- Ow DW, Jacobs JD, Howell SH. Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc Natl Acad Sci USA. 1987;84:4870–4974. doi: 10.1073/pnas.84.14.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep. 1993;11:122–127. [Google Scholar]
- Rinehart JA, Peterson MW, John ME. Tissue-specific and developmental regulation of cotton gene FbL2A: demonstration of promoter activity in transgenic plants. Plant Physiol. 1996;112:1331–1341. doi: 10.1104/pp.112.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U. Cell wall biosynthesis in differentiating cotton fibers. Eur J Cell Biol. 1985;39:236–256. [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. The control of single-cell cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell. 2001;13:47–60. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Brown RM. GTPase activity and biochemical characterization of a recombinant cotton fiber annexin. Plant Physiol. 1999;119:925–934. doi: 10.1104/pp.119.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Oppenheimer DG, Kopczak SD, Ploense SE, Ludwig SR, Haas NA, Snustad DP. Plant tubulin genes: structure and differential expression during development. Dev Genet. 1987;8:435–460. [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wikkins TA. Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol. 1998;116:1539–1549. doi: 10.1104/pp.116.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Allen RD. Identification of a cotton fiber-specific acyl carrier protein cDNA by differential display. Biochim Biophys Acta. 1997;1351:305–312. doi: 10.1016/s0167-4781(96)00218-7. [DOI] [PubMed] [Google Scholar]
- Uribe X, Torres MA, Capellades M, Puigdomenech P, Rigau J. Maize α-tubulin genes are expressed according to specific patterns of cell differentiation. Plant Mol Biol. 1998;37:1069–1078. doi: 10.1023/a:1006067710312. [DOI] [PubMed] [Google Scholar]
- Villemur R, Haas NA, Joyce CM, Snustad DP, Silflow CD. Characterization of four new β-tubulin genes and their expression during male flower development in maize (Zea mays L.) Plant Mol Biol. 1994;24:295–315. doi: 10.1007/BF00020169. [DOI] [PubMed] [Google Scholar]
- Villemur R, Joyce CM, Haas NA, Goddard RH, Kopczak SD, Hussey PJ, Snustad DP, Silflow CD. α-Tubulin gene family of maize (Zea mays L.): evidence for two ancient α-tubulin genes in plants. J Mol Biol. 1992;227:81–96. doi: 10.1016/0022-2836(92)90683-b. [DOI] [PubMed] [Google Scholar]
- Whittaker DJ, Triplett BA. Gene-specific changes in α-tubulin transcript accumulation in developing cotton fibers. Plant Physiol. 1999;121:181–188. doi: 10.1104/pp.121.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]