Abstract
Selenium (Se) phytovolatilization, the process by which plants metabolize various inorganic or organic species of Se (e.g. selenate, selenite, and Se-methionine [Met]) into gaseous Se forms (e.g. dimethylselenide), is a potentially important means of removing Se from contaminated environments. Before attempting to genetically enhance the efficiency of Se phytovolatilization, it is essential to elucidate the enzymatic pathway involved and to identify its rate-limiting steps. The present research tested the hypothesis that S-adenosyl-l-Met:l-Met S-methyltransferase (MMT) is the enzyme responsible for the methylation of Se-Met to Se-methyl Se-Met (SeMM). To this end, we identified and characterized an Arabidopsis T-DNA mutant knockout for MMT. The lack of MMT in the Arabidopsis T-DNA mutant plant resulted in an almost complete loss in its capacity for Se volatilization. Using chemical complementation with SeMM, the presumed enzymatic product of MMT, we restored the capacity of the MMT mutant to produce volatile Se. Overexpressing MMT from Arabidopsis in Escherichia coli, which is not known to have MMT activity, produced up to 10 times more volatile Se than the untransformed strain when both were supplied with Se-Met. Thus, our results provide in vivo evidence that MMT is the key enzyme catalyzing the methylation of Se-Met to SeMM.
The trace element selenium (Se) is an essential micronutrient with important benefits for animal and human nutrition; however, at high doses, Se is toxic (Wilber, 1980; Van Vleet and Ferrans, 1992; Lemly, 1997). Major sources of Se pollution are agricultural drainage from seleniferous soils and industrial wastewater. Se pollution is a worldwide problem and there is a tremendous demand for the cleanup of Se-contaminated soil and water. Phytoremediation, the use of plants to remove, stabilize, or detoxify pollutants, is a highly promising solution to counter the Se problem (Bañuelos et al., 1995; Salt et al., 1998; Terry and Zayed, 1998).
Se volatilization, the process by which gaseous Se forms are produced from inorganic or organic Se compounds (Lewis et al., 1966; Zieve and Peterson, 1984; Duckart et al., 1992; Terry et al., 1992, 2000), is particularly attractive for the phytoremediation of Se-contaminated environments because it completely removes Se from the local food chain (Atkinson et al., 1990; Terry and Zayed, 1998). The major volatile Se form produced by plants and microbes is dimethylselenide (DMSe; Lewis et al., 1974). DMSe is 600 to 700 times less toxic than selenate or selenite, two Se species that are commonly present in polluted areas (McConnell and Portman, 1952; Ganther et al., 1966; Wilber, 1980).
The formation of DMSe in many plants is thought to proceed via the sulfur (S) assimilation pathway (Terry et al., 2000). To enhance the efficiency of Se volatilization by plants, it is essential that we fully elucidate the biochemical pathway involved in Se assimilation and volatilization. Once we have determined the rate-limiting steps in the pathway, it should be possible to enhance the efficiency of Se volatilization by overexpressing the genes encoding key enzymes. Our recent research using Indian mustard (Brassica juncea) plants demonstrated that the uptake and activation of selenate are important rate-limiting steps (de Souza et al., 1998; Pilon-Smits et al., 1999). Overexpression of the gene encoding ATP sulfurylase resulted in transgenic plants that exhibited increased reduction of activated selenate to organic Se forms; this showed that the rate-limiting step of selenate reduction had been overcome (Pilon-Smits et al., 1999). However, Terry et al. (2000) proposed that other steps further along the Se assimilation pathway may also be limiting for Se volatilization (e.g. the synthesis and methylation of Se-Met; Fig. 1). With respect to rate limitation in the later steps of the pathway, de Souza et al. (2000) concluded that the production of selenonium compounds from Se-Met was rate limiting, rather than the conversion of selenonium compounds to DMSe.
Figure 1.
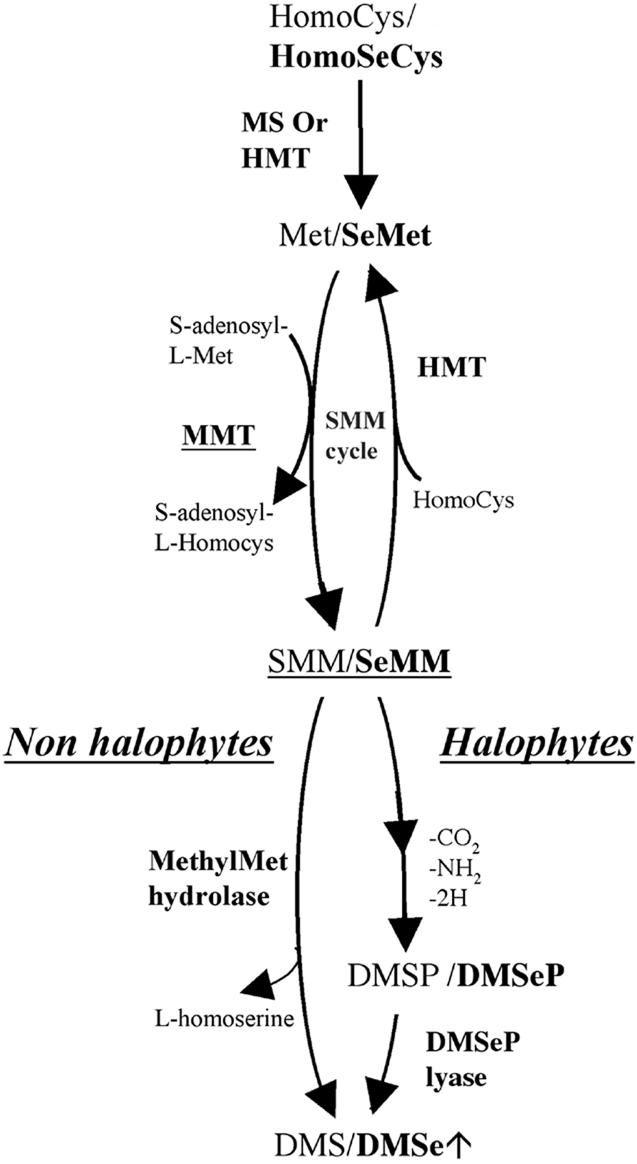
Biosynthesis of dimethylsulfide (DMS) and DMSe in plants. Biosynthesis of DMSe is thought to occur via the S volatilization pathway.
The synthesis of Se-Met is carried out by two possible enzymes, Met synthase or homo-Cys S-methyltransferase. By analogy with the S assimilation pathway, Terry et al. (2000) proposed that Se-Met is methylated to Se-methyl Se-Met (SeMM) by the S-adenosyl-l-Met:Met-S-methyltransferase (MMT; E.C. 2.1.1.12). The key role played by MMT in the production of sulfonium compounds has been well characterized. MMT catalyzes the transfer of a methyl group from S-adenosyl-l-Met to l-Met, resulting in the production of S-adenosyl-l-homo-Cys and S-methyl-l-Met (SMM; James et al., 1995; Pimenta et al., 1998; Bourgis et al., 1999). cDNA clones for MMT have been isolated from barley (Hordeum vulgare; Pimenta et al., 1998), Wollastonia biflora, maize (Zea mays), and Arabidopsis (Bourgis et al., 1999).
Assuming that Se-Met is methylated to SeMM, the next step in the Se assimilation and volatilization pathway is the formation of DMSe from SeMM. Two possible pathways have been suggested (Fig. 1). In non-halophytes, the production of DMSe is thought to occur directly from the hydrolysis of SeMM, whereas in halophytes, SeMM may be converted to dimethylselenoniopropionate (DMSeP; Terry et al., 2000). The latter hypothesis is consistent with the pathway of DMS production in halophyte plants, where the major precursors of DMS are the sulfonium compounds SMM and dimethylsulfoniopropionate (Dacey et al., 1987; Mudd and Datko, 1990; Rennenberg, 1991; Hanson et al., 1994, 1997; Kocsis et al., 1998).
In members of the Brassicaceae, e.g. Arabidopsis and Indian mustard, which are non-halophytes, SeMM is more likely to be the precursor of DMSe rather than DMSeP. Lewis et al. (1974) demonstrated that Se-methyl Se-Met was the source of DMSe production in cabbage (Brassica oleracea) leaves. This idea is supported by the fact that DMSeP was not detected in Se-Met-supplied Indian mustard plants, although they were capable of taking up, assimilating, and volatilizing Se at high rates when supplied with DMSeP (de Souza et al., 2000).
In Arabidopsis, MMT is most likely to be encoded by a single copy gene (Bourgis et al., 1999). To determine if MMT plays a role in Se volatilization, we adopted a reverse genetics approach. We identified an Arabidopsis T-DNA mutant knockout for MMT, which showed a dramatically reduced Se volatilization rate compared with wild type (WT). We then tested the capability of the mutant to volatilize Se by chemical complementation with SeMM. To confirm the view that MMT is a critical enzyme in Se volatilization using a different approach, we generated a recombinant Escherichia coli strain overexpressing the MMT from Arabidopsis to see whether volatile Se production can be substantially enhanced in an organism that does not have this enzymatic (MMT) capability (Thanbichler et al., 1998).
RESULTS
MMT Expression Is Up-Regulated in the Presence of Selenate
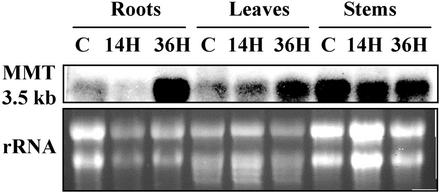
An indication that MMT might be involved in the production of volatile Se from selenate was obtained by northern analysis. Total RNA was extracted from roots, leaves, and stems of Indian mustard (treated with 100 μm selenate for 14 and 36 h) and hybridized with an MMT-specific probe corresponding to the entire MMT cDNA. Indian mustard (which we use as a model plant in our research on phytoremediation) is a species of Brassicaceae that is closely related to Arabidopsis (BrassicaDB, http://ukcrop.net/brassica.html#brassicadb).
One band of 3.5 kb corresponding to the size of the Arabidopsis MMT mRNA was revealed. For each lane, the MMT hybridization signal was compared with the amount of ribosomal RNA 18S and 25S visualized by ethidium bromide. We observed an up-regulation of the MMT expression in roots and leaves after 36 h of selenate treatment. The up-regulation was higher in roots than in leaves. We did not observe any up-regulation in stems (Fig. 2).
Figure 2.
Expression analysis of MMT in roots, leaves, and stems of Indian mustard in the presence of 100 μm selenate. Ten micrograms of total RNA were loaded for each sample and hybridized with an MMT-specific probe. MMT expression was up-regulated in roots and leaves (R-36H, l-36H) in the presence of selenate after 36 h but not in stems (St-36H). No up-regulation was detected in roots, leaves, and stems (R-14H, L-14H, and St-14H) after only 14 h. Untreated tissue from roots (R-C), leaves (L-C), and stems (St-C) served as control. The ethidium bromide-stained 18S and 25 S ribosomal RNA show the relative amount of RNA loaded in each lane.
Isolation of the T-DNA Mutant Disrupted for Its MMT Gene
Because the entire Arabidopsis genome was recently completely sequenced (Arabidopsis Genome Initiative, 2000), we were able to confirm that the gene for MMT is single copy. Using the BLAST program (Altschul et al., 1997), we identified only one bacterial artificial chromosome (BAC; clone K21G20, accession no. AB025612) containing the MMT gene.
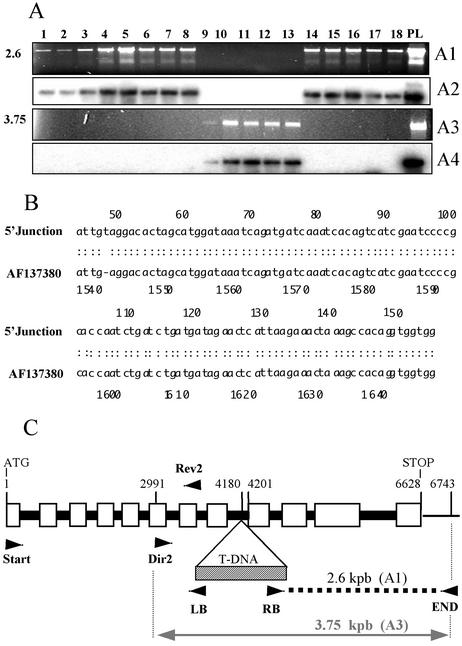
Using a PCR-based screen, we identified one line disrupted for the MMT gene in the Feldmann collection of Arabidopsis T-DNA mutants (Arabidopsis Biological Resource Center [ABRC], Columbus, OH). This mutant was designated Δmmt. A junction MMT/T-DNA was detected with the combination of primers RB-F and MMT-END (Fig. 3A). The similarity search (BLAST) for the generated PCR fragment RB-END that corresponds to the junction gave alignments with both the MMT mRNA (AF137380) and the BAC K21G20 (AB025612; Fig. 3B). This enabled us to locate the T-DNA insertion in the eighth intron (nucleotide 4,180, from the ATG codon), 22 bases upstream of the junction with the ninth exon (nucleotide 4,201; Fig. 3C). It was previously reported that T-DNA insertions that have occurred in introns of Arabidopsis lead to a complete disruption of the affected gene (Krysan et al., 1999; Papi et al., 2000). Homozygous descendants were isolated from the progeny of the Δmmt parental line and identified by PCR (Fig. 3A).
Figure 3.
Identification of the T-DNA mutant disrupted for the MMT and characterization of the T-DNA insertion. A, The MMT/T-DNA junction (2.6 kb) was amplified by PCR (primers RB-F and MMT-End) and identified by Southern blotting in the parental line Δmmt (PL) and the descendants 1 through 8 and 14 through 18, ethidium bromide gel (A1), and hybridization with a specific MMT probe (A2). PCR with the primers MMT-Dir2 and MMT-End generated the intact genomic MMT fragment (3.75 kb) in the PL and in the descendants 9 through 13, but not in descendants 1 through 8 and 14 through 18, ethidium bromide gel (A3), and hybridization (A4) as for A2. PL is heterozygous for the insertion, descendants 1 through 8 and 14 through 18 are homozygous, and descendants 9 through 13 did not inherit it. B, Sequence alignment of the T-DNA/MMT junction (PCR-fragment RB-F/MMT-END) with the MMT mRNA (AF137380; shown is the 5′ region of the alignment). C, Map of the T-DNA insertion. The T-DNA insertion is located at the end of intron 8 (nucleotide 4,180). The T-DNA/MMT junction (A1) is represented by the bold dotted line. The intact MMT-Dir2-MMT-End fragment (A3) is represented by the gray arrowed line. The small arrows represent the location of the primers used. The figure is not to scale. The black rectangles represent the introns and the white ones represent the exons.
Genetic and Molecular Characterization of the Mutant
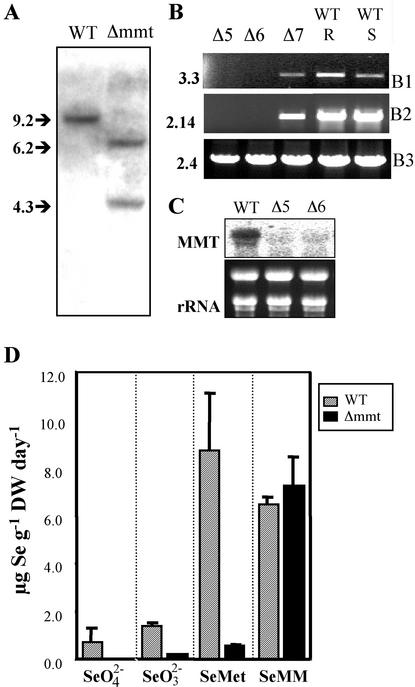
To confirm the T-DNA insertion in the MMT gene, we performed a Southern analysis. Genomic DNA of the Arabidopsis WT (ecotype Wassilewskija) and one homozygous descendant Δmmt was cleaved with the restriction enzyme NheI and hybridized with the MMT-specific probe that was used previously for the identification of the mutant Δmmt. We detected a variation in the restriction patterns (Fig. 4A). A single 9.2-kb fragment was observed in the WT, whereas the mutant Δmmt showed two fragments of 6.2 and 4.3 kb in size. This pattern is expected with regard to the location of the T-DNA insertion and the T-DNA restriction map (sequence available at ABRC). To determine how the expression of MMT gene in Δmmt was affected by the T-DNA insertion, reverse transcriptase (RT)-PCR and northern analysis were performed. Using RT-PCR, no MMT fragment was amplified in the two homozygous descendants, Δmmt-5 and Δmmt-6, with two sets of specific MMT primers, MMT-Start/MMT-End and MMT-Dir2/MMT-End (Fig. 4B). Total RNA from Δmmt-5 and Δmmt-6 was probed with a specific MMT probe corresponding to the MMT cDNA, and no hybridization signal was detected (Fig. 4C). Therefore, the T-DNA line Δmmt is knockout.
Figure 4.
Genetic and physiological characterization of the mutant Δmmt. A, The T-DNA insertion in MMT gene generated a variation in restriction patterns between Arabidopsis WT and Δmmt. The genomic DNA was cleaved with the endonuclease NheI and hybridized with an MMT-specific probe (size in kb). B, RT-PCR analysis of the MMT gene. Arabidopsis WT roots (WT R) and shoots (WT S) and three different Δmmt descendants (D5, D6 [both homozygous], and D7 [heterozygous]). Primers MMT-Start and MMT-End (B1) or MMT-Dir2 and MMT-End (B2) were used to detect MMT mRNA. No MMT mRNA expression was found in Δ5 and Δ6, whereas Δ7, WTR, and WTS expressed the MMT gene. mRNA for Met synthase was used as internal standard (B3). PCRs were performed with 35 cycles and product size is in kb. C, Northern analysis of the MMT gene showed no expression of MMT mRNA in the two Δmmt homozygous descendants Δ5 and Δ 6. Twenty micrograms of total RNA were loaded for each sample. Ethidium bromide-stained RNA is shown as a control of loading and RNA intactness. D, Se volatilization from Arabidopsis WT and the MMT knockout mutant Δmmt, treated with 20 μm selenate (SeO42−), selenite (SeO32−), or Se-Met or 7 μm SeMM. The Se volatilization capacity of the knockout mutant was reduced by up to 16-fold compared with the WT when plants were supplied with Se-Met. Disruption of the MMT gene led to a substantial loss of DMSe production. The capacity of the mutant to volatilize Se was restored when SeMM was supplied.
Physiological Characterization of the Mutant Δmmt
Se Volatilization
To determine the effect of the inactivation of the MMT gene on the production of volatile Se, we measured Se volatilization (μg Se volatilized g−1 dry weight d−1) in the knockout mutant, Δmmt, and in the WT. The experimental procedure is described in detail in “Materials and Methods.” In the knockout plant, the production of volatile Se was dramatically reduced compared with the WT (Fig. 4D). When SeO42−, SeO32−, or Se-Met was supplied, the total Se volatilized by Δmmt accounted for only 7%, 13%, and 6%, respectively, of the Se volatilized by the WT.
Because the gene for MMT is a single-copy gene, the residual amounts of volatile Se detected in the mutant are not due to the activity of an MMT isoform. The residual amount of Se volatilized in the mutant when selenate or selenite is supplied could be due to the production of hydrogen selenide. It is possible in plants, as in bacteria, that H2S (or H2Se) is formed from sulfide (S2−; or selenide [Se2−]) before its incorporation in Cys (or seleno-Cys). H2Se is volatile and could easily be absorbed in the trap solution and oxidized to SeO42−. This view is also supported by research with animals showing that the inhibition of the Se methylation is accompanied by an accumulation of H2Se (Sayato et al., 1997). On the other hand, the residual amount observed when Se-Met is supplied could be due to the activity of another amino S-methyltransferase (Farooqui et al., 1985) catalyzing a nonspecific methylation of Se-Met at high Km and/or low Vmax.
Se Accumulation
To determine if the loss of Se volatilization in the knockout plants was accompanied by a higher Se accumulation in the mutant, we measured the concentration of total Se (μg Se g−1 dry weight) in the tissue of Δmmt and WT (Table I). The plant material was the same as that used to measure Se volatilization rates (Fig. 4D). For each form of supplied Se (20 μm selenate, or 20 μm selenite, or 20 μm Se-Met, or 7 μm SeMM), we calculated the difference between Δmmt and WT; more Se was accumulated in the mutant than in the WT (Table I).
Table I.
Se accumulation (μg Se g−1 dry wt) in the mutant Δmmt and the WT
| Lines | 20 μm Selenate | 20 μm Selenite | 20 μm SeMet | 7 μm SeMM |
|---|---|---|---|---|
| Δmmt | 224 ± 13 | 452 ± 59 | 351 ± 21 | 148 ± 20 |
| WT | 218 ± 2.6 | 321 ± 45 | 320 ± 26 | 111 ± 10 |
| Δmmt − WT | 6 | 131 | 31 | 36 |
The difference in Se accumulation (Δmmt − WT) shows that after 8 d of Se treatment, Δmmt accumulates more Se. Shown are the averages and sd of three replicates.
Se Tolerance
The root length root of 12-d-old seedlings (see “Materials and Methods”) was measured to compare the tolerance of mutant and WT plants with selenate and Se-Met. In the absence of Se, there was no significant difference in root length between the WT and the mutant. In the presence of 25 μm selenate, the WT roots are slightly longer than the mutant. In the presence of 10 or 20 μm Se-Met, the roots of the WT were 3 times longer than the mutant (Table II), showing that the WT was substantially more tolerant to high levels of Se than the mutant.
Table II.
Root length of Δmmt and the WT in absence or presence of Se
| Lines | No Selenium | 25 μm Selenate | 10 μm SeMet | 20 μm SeMet |
|---|---|---|---|---|
| mm | ||||
| WT | 40 ± 3.64 | 32.6 ± 5.3 | 17.7 ± 6.47 | 6.8 ± 4.35 |
| Δmmt | 43 ± 5.1 | 24.1 ± 3.6 | 6.1 ± 4.28 | 2.2 ± 2 |
| Length root ratio (WT:Δmmt) | 0.93 | 1.35 | 2.9 | 3.1 |
Root length of 12-d seedlings (average and sd of 20 plants). For more accuracy, measurements were taken using a stereomicroscope for SeMettreated plants.
Chemical Complementation of Δmmt
To confirm that the absence of Se volatilization in the mutant was due to the disruption of the MMT gene, we performed a chemical complementation experiment with 7 μm SeMM, the putative product of MMT enzyme. The SeMM was synthesized as l-seleno-Met Se-methylsulfonium bromide by methylation of Se-Met with methanol in the presence of sulfuric acid according to the method described for the synthesis of SMM by Floyd and Lavine (1954). The chemical structure of the product was confirmed by NMR, and the yield was determined by atomic absorption. The Se volatilization experiment was performed as described previously. Treatment with SeMM restored the ability of the mutant to produce volatile Se (Fig. 4D), demonstrating that SeMM is the product of the MMT enzyme and the substrate for DMSe production. Both the WT and the mutant plants accumulated an average of 130 μg Se g−1 dry weight, showing that the supplied SeMM was taken up and metabolized.
Se Volatilization from E. coli Overexpressing the MMT Gene from Arabidopsis
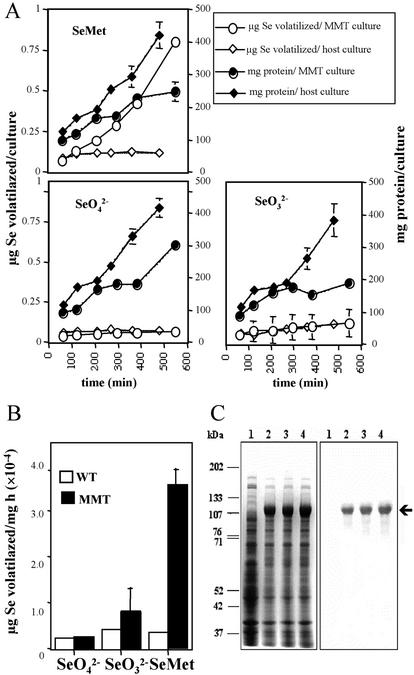
To correlate the rate of Se volatilization with the expression of the MMT protein, we measured the production of volatile Se from E. coli cultures overexpressing the recombinant plant MMT in the presence of SeO42−, SeO32−, or Se-Met. The plant cDNA from Arabidopsis (ecotype Columbia) was cloned by RT-PCR and heterologously expressed in strain BL21(DE3) as a 10His-tag fusion protein using the pET16b vector derivative pET16MMT. The MMT cDNA was sequenced using different oligonucleotides listed in Table I. No mismatches were detected between our sequence and the MMT sequence in the BAC K21G20 (AB025612). During the logarithmic growth phase, which was monitored by the increase in total cell protein, the MMT cultures showed an exponential production of volatile Se when supplied with Se-Met. Only a low threshold level of volatilization was observed from the untransformed host (Fig. 5A). Se volatilization was not time dependent when either strain was supplied with SeO42− or SeO32−. Volatilization rates (Fig. 5B) were calculated from the last time point measured in the logarithmic growth phase. The MMT cultures showed up to a 10-fold higher Se-volatilization from Se-Met compared with the untransformed host strain. When supplied with SeO32−, the MMT cultures volatilized Se at twice the host strain rate. There was no essential difference between the rates obtained from SeO42− in the MMT and host cultures. The MMT cultures volatilized Se from Se-Met at a 5-fold higher rate than from SeO32−, whereas Se volatilization from SeO42− accounted for only one-third of the rate obtained with SeO32−. In the host cultures, the rate was independent of the Se form supplied.
Figure 5.
Kinetics of Se volatilization from recombinant E. coli overexpressing the MMT gene from Arabidopsis. A, Kinetics of Se volatilization and cell protein content in E. coli BL21(DE3) overexpressing MMT from Arabidopsis (MMT culture) and the untransformed host culture supplied with different Se compounds (50 μm SeO42−, SeO32−, or Se-Met). B, Se volatilization rates from Se-Met were up to 10-fold higher in MMT cultures compared with the host cultures. Rates were calculated from the last time point measured in the exponential growth phase (see A). C, Overexpression of the recombinant MMT from Arabidopsis in E. coli. SDS-PAGE and western blot showing the presence of the 120-kD MMT from Arabidopsis (arrow) as a 10His-fusion protein expressed in E. coli BL21(DE3) cultures treated with different Se-compounds (50 μm SeO42−, SeO32−, or Se-Met) to measure Se volatilization. Lanes: 1, total protein from the untransformed host E. coli BL21(DE3); 2, total protein from E. coli BL21(DE3) transformed with plasmid pETMMT for the overexpression of the 10His-MMT, SeO42− treatment; 3, same as 2, SeO32− treatment; 4, same as 2, Se-Met treatment.
Because the growth of the MMT culture is slower than the untransformed E. coli, the total amount of protein was also lower in the MMT culture (Fig. 5A). This could be due to the fact that the MMT culture has to produce a large amount of the recombinant MMT protein, which has a negative effect on cell division.
The expression of recombinant MMT was confirmed by SDS-PAGE and western-blot analysis (Fig. 5C). Protein extracts from the MMT cultures that were used for volatilization contained a major polypeptide with an overproduction level of 10% of 15% of the total protein. The molecular mass, as judged by SDS-PAGE, was between 107 and 133 kD, which is in agreement with an expected size of 120 kD (117 kD MMT + 3 kD 10His fusion) derived from the DNA sequence. Specific antibodies raised against the MMT from W. biflora recognized the overexpressed protein as the recombinant MMT.
DISCUSSION
Our results provide in vivo evidence that MMT is the key enzyme catalyzing the methylation of Se-Met to SeMM in Arabidopsis (a non-halophytic plant). This conclusion is based on the following. First, we demonstrated that an Arabidopsis T-DNA mutant, knockout for MMT, has a vastly decreased ability to volatilize Se; the mutant volatilized 93%, 87%, and 94% less than WT when supplied with selenate, selenite, and Se-Met, respectively (Fig. 4D). Second, we showed that this mutant could be chemically complemented; the ability to volatilize Se was restored by supplying SeMM, the putative enzymatic product of MMT (Fig. 4D). Third, E. coli overexpressing MMT cDNA from Arabidopsis produced up to 10-fold higher levels of volatile Se compared with the untransformed host strain BL21(DE3) when supplied with Se-Met. Because E. coli is not known to have MMT or the ability to synthesize SMM (Thanbichler et al., 1998), this substantial increase in Se volatilization upon the introduction of the Arabidopsis MMT cDNA emphasizes the importance of MMT for Se volatilization in plants. Thus, these three experimental observations show that the MMT is a key enzyme, playing a major role in the production of volatile Se in plants, and that Met S-methyltransferase could also be designated Se-Met Se-methyltransferase.
Because of its potential importance to phytoremediation, a major goal of our research is to determine the enzymatic steps that are rate limiting for Se volatilization. Our results show that the amount of Se volatilized increased progressively when we supplied Se as selenite compared with selenate, and as Se-Met compared with selenite (all three Se forms being supplied at 20 μm, Fig. 4D). Even when supplying only 7 μm SeMM, the amount of volatilized Se was almost as high as that with 20 μm Se-Met (Fig. 4D).
The form of Se supplied may influence volatilization in two ways: (a) It may affect volatilization rate directly, and/or (b) it may affect volatilization rate indirectly by altering the extent of Se uptake and accumulation by the plant, the rate of volatilization being dependent on Se concentration in plant tissues (Terry et al., 2000). To assess the effect of uptake/accumulation on volatilization, we recalculated the data in terms of the rate of volatilization per unit of Se accumulated by plant tissues (Fig. 6). This also allows us to correct for the fact that we supplied SeMM at 7 μm Se rather than 20 μm as in selenate, selenite, and Se-Met. When the data are reexpressed as volatilization per unit Se accumulated (Fig. 6), they show that volatilization increased 1.3-fold when we supplied Se to WT Arabidopsis plants as selenite compared with selenate, 6.3-fold with Se-Met versus selenite, and 2-fold with SeMM versus Se-Met. These results show that we obtained progressively greater increases in Se volatilization (per unit of Se accumulated) because we supplied forms of Se that were downstream in the Se/S assimilation pathway. Thus, there are not only rate-limiting steps between the conversion of selenate to selenite (as demonstrated earlier, Terry et al., 2000) and between selenite and Se-Met, but also between Se-Met and SeMM. The importance of MMT as a rate-limiting step is further emphasized by the fact that we obtained a 29-fold increase in volatilization rate when we supplied SeMM rather than Se-Met to the MMT knockout mutant (Fig. 6), thereby completely overcoming the absence of the MMT enzyme.
Figure 6.
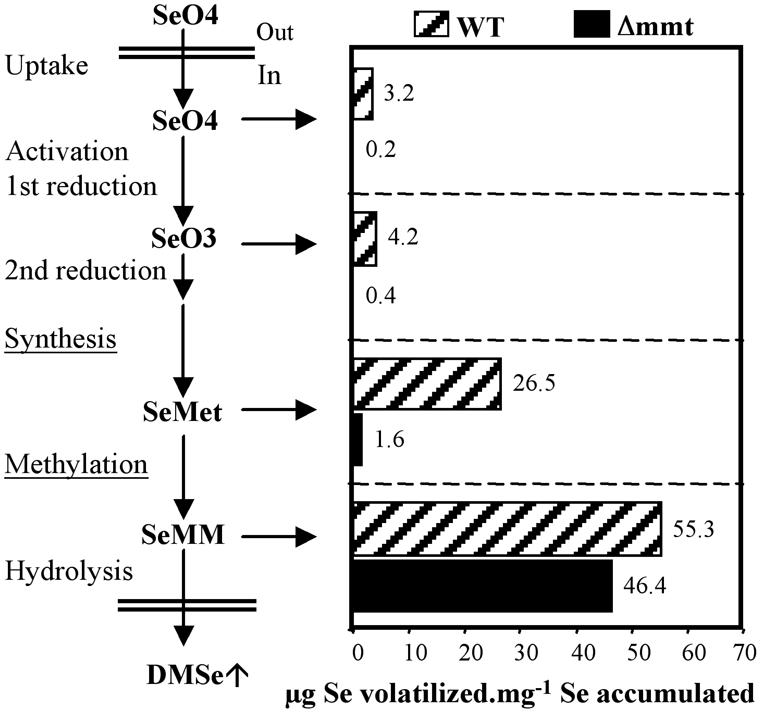
Recalculation of Se volatilization data (Fig. 4D) from Arabidopsis WT, and the mutant Δmmt, as the amount (μg) of Se volatilized per mg Se accumulated (i.e. the amount of Se accumulated = amount of Se in plant tissue + the amount of Se volatilized).
The Se accumulation data showed that the mutant (which does not volatilize Se) accumulated more Se than the WT (Table I). When supplied with SeMM, the mutant still accumulated more Se than the WT. This result suggests that the part of SeMM that is not directly hydrolyzed to give DMSe could be converted back to Se-Met by the homo-Cys S-methyltransferase, the enzyme that converts SMM to Met (Thanbichler et al., 1998; Ranocha et al., 2000). In the WT, but not in the mutant, the newly synthesized Se-Met is methylated to SeMM by the MMT and this compound is then converted to DMSe.
The use of the root length as a measure of the tolerance to Se showed that the mutant, Δmmt, is more affected by the presence of toxic concentrations of Se than the WT. This difference in tolerance between the mutant and the WT is more evident in the presence of Se-Met (Table II). The decreased tolerance of the mutant shows that the loss of Se volatilization capability has an effect on Se tolerance, suggesting that Se volatilization might be used by plants as a mechanism of detoxification.
Our results have implications with respect to other aspects of Se physiology. Studies have shown that the rate of Se volatilization is much greater in root tissue compared with shoot tissue (Terry et al., 2000). In the absence of selenate, our results show that Indian mustard shoots have the highest level of MMT expression; however, after induction by selenate, roots and shoots had the same level of expression (Fig. 2). Thus, the higher rates of DMSe production from roots compared with shoots that we observed earlier (Zayed and Terry, 1994; Terry et al., 2000) does not appear to be due to a greater MMT expression in root tissue. Alternatively, roots may volatilize Se at higher rates than shoots because there is a larger SeMM pool in roots than shoots. Bourgis et al. (1999) demonstrated that SMM (the chemical analog of SeMM) produced in shoots is transported to other organs via the phloem. Ranocha et al. (2001) recently showed that the SMM cycle occurs throughout Arabidopsis tissues and that the SMM pool is largest in roots. Thus, assuming that SeMM, like SMM, is translocated from shoots to roots, the pool of SeMM in roots may be substantially increased, leading to an increased volatilization.
A second implication of our research relates to the question as to whether microbes are involved in Se volatilization by plants (Terry et al., 2000). Because all of our experiments with Arabidopsis plants were performed axenically, it is clear that plants have all the enzymes necessary to volatilize Se from each of the supplied Se forms (selenate, selenite, Se-Met, or SeMM). Thus, our results confirm those of de Souza et al. (1999a, 1999b) who showed that although rhizosphere bacteria facilitate the uptake of selenate into root tissue, they do not appear to be responsible for the production of volatile Se.
In conclusion, the present research shows that MMT is the enzyme responsible for the methylation of Se-Met to SeMM and that MMT is an important rate-limiting enzyme in Se phytovolatilization. Although the Arabidopsis mutant knockout for the MMT gene lacks the SMM cycle, it is viable and does not present a particular phenotype except that it exhibited a slight decrease of its fertility (some silics are small and less seeds are produced than the WT; data not shown). This suggests that the mutant under the culture conditions used in our experiments was able to overcome the absence of the SMM cycle.
The role of the SMM cycle in plants has not yet been completely elucidated. Mudd and Datko (1990) suggested earlier that the SMM cycle serves to prevent depletion of free Met. Because plants lack the negative feedback to regulate the S-adenosyl-Met pool, Ranocha et al. (2001) proposed recently that the SMM cycle is the main mechanism to control the S-adenosyl-Met level in plants. Thus, the MMT mutant may serve in the future to elucidate certain aspects of the SMM cycle and its role in plants.
MATERIALS AND METHODS
Identification and Isolation of the T-DNA Line Disrupted for the MMT
Two Arabidopsis T-DNA mutant collections (Wassilewskija ecotype Ws-2; and T. Jack, ecotype Columbia Col-6, gl1-1; Kenneth A. Feldmann [Department of Plant Sciences, University of Arizona, Tucson]), were provided by ABRC. Each collection contains 6,000 independent lines and is organized in pools of DNA according to a model that allows the identification of the subpool of 10 lines, which contains the tagged line of interest.
A PCR-based screen was performed with two sets of MMT-specific primers in combination with oligonucleotides specific for the T-DNA left border (LB) or right border (RB; Table III). The PCR products obtained were hybridized with a mixture of two probes specific of the MMT gene that correspond to PCR fragments amplified with the primer sets MMT-Start/MMT-Rev2 and MMT-Dir2/MMT-END (Table III). Genomic DNA was extracted from each line of the subpool of 10 and PCR was performed with the set of primers generating the MMT/T-DNA junction. The MMT disrupted line was screened for its kanamycin resistance. Resistant plants were transferred to soil and allowed to self-fertilize. Genomic DNA was extracted from individual plants. Homozygous descendants were identified by PCR with the primers bounding the T-DNA insertion (MMT-Dir2/MMT-END) and primers specific of the MMT/T-DNA junction (RB-F/MMT-End).
Table III.
List of primers used in this study
| Name of Primers | Sequences |
|---|---|
| MMT-STARTab | 5′ TTCACGATAATGGCGGACCTCTCCTC 3′ |
| MMT-ENDa | 5′ AGAAACAACAGAACAAAGGAACGCAGAC 3′ |
| MMT-REV2a | 5′ TTGCCACAGCCCTTGATGGAAACAC 3′ |
| MMT-DIR2a | 5′ GCTGATGAGAAGATTCCATTCCTAGCC 3′ |
| RB-Fa | 5′ GCTCAGGATCCGATTGTCGTTTCCCGCCTT 3′ |
| LB-Fa | 5′ GATGCAATCGATATCAGCCAATTTTAGAC 3′ |
| RB-Ja | 5′ TCGGGCCTAACTTTTGGTG 3′ |
| LB-Ja | 5′ GAAGTTTCTCATCTAAGCCCCCATTT 3′ |
| MS-ATGc | 5′ AATGGCTTCACACATTGTTGGATAC 3′ |
| MS-STOPc | 5′ ACCATATGCATCAAATCTACCTTCG 3′ |
| T7d | 5′ TAATACGACTCACTATAGGG 3′ |
| MMT-fwdd | 5′ CACACAATGACGATAATGGCGGACCTCTCCTCTG 3′ |
| MMT-REVd | 5′ GTGTGTCATATGGCACGTTTCAGTTAGCAAGGACG 3′ |
| MMT-STOPb | 5′ TGGCACGTTTCAGTTAGCAAGGACG 3′ |
| MMT-REV1d | 5′ GAGGAATTTCTCCATTCGCTGAGT 3′ |
| MMT-DIR1d | 5′ GCTGAGCTTTCAGTCACTGAGATT 3′ |
Primers used for the isolation of the T-DNA mutant disrupted for the MMT (bold indicates primers detecting the insertion).
Primers used for cloning of the Arabidopsis MMT cDNA.
Primers specific of the internal standard (Met synthase) used in the RT-PCR experiment.
Primers used for sequencing of the MMT cDNA.
Plasmid Construction
Plasmid pET16MMT, a pET16b (Novagen, Madison, WI) derivative, was used for the overexpression of the recombinant MMT from Arabidopsis (ecotype Columbia) in Escherichia coli. It contains a 3,231-bp NdeI-fragment generated by PCR from plasmid pTOPOMMT using the oligonucleotides MMT-fwd and MMT-rev. pTOPOMMT harbors the MMT cDNA of Arabidopsis isolated by RT-PCR using the oligonucleotides MMT-Start and MMT-Stop. The PCR product was subsequently cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA).
Nucleic Acid Procedures
Principal methods used for the cloning of DNA, plasmid isolation from bacteria, restriction, Southern blotting, agarose gel electrophoresis, and PCR were performed according to Sambrock et al. (1989). The amplification of DNA for cloning was carried out using Turbo Pfu DNA polymerase (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Amplitaq DNA polymerase (Perkin-Elmer Applied Biosystems, Foster City, CA) was used for the PCR screen of the T-DNA mutants. Annealing temperatures for oligonucleotides were calculated depending on their G:C content and inserted mismatches. DNA sequencing was carried out by the University of California Sequencing Facility (Berkeley). Genomic DNA was isolated from Arabidopsis using the cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1990). Total RNA was extracted from mature plants utilizing Trizol reagent (Life Technologies/Gibco-BRL, Cleveland). RNA electrophoresis, northern blotting, hybridization, and washing were performed as described by Hwang and Herrin (1994). MMT-DNA probes were labeled with [32P]dCTP by random priming (Ready to go Kit, Amersham, Buckinghamshire, UK). RT-PCR was performed using RT Superscript II (Life Technologies/Gibco-BRL) and Turbo Pfu DNA polymerase (Stratagene) or Amplitaq DNA polymerase (Perkin-Elmer Applied Biosystems) as described in the user instructions for Superscript II RT. All primers used are listed in Table III.
Se Volatilization and Se Accumulation from Arabidopsis
Arabidopsis was grown in liquid cultures at 22°C under permanent light using a modified protocol described by Xiang and Oliver (1998). Twenty sterilized seeds were used to inoculate 500-mL flasks containing 200 mL of medium (full-strength Murashige and Skoog salts [Sigma, St. Louis] and 2% [w/v] Glc, pH 5.8). Two weeks after inoculation, the cultures were treated with 20 μm selenate (NaSeO4), 20 μm selenite (NaSeO3), 20 μm l-Se-Met, or 7 μm SeMM. Three replicate cultures were used for each treatment. Se volatilization was measured using the method of de Souza et al. (1998), which we modified as follows. One week after Se treatment, the liquid cultures were transferred to 500-mL gas washing bottles (Fisher Scientific, Loughborough, Leicestershire, UK) that were connected by Teflon tubing to 500-mL washing bottles with fritted discs containing the alkaline peroxide trap solution. A continuous air flow (1.5 L min−1) was passed through the flasks by applying suction at the outlet, whereas the incoming air was bubbled into the liquid culture after passing through a 0.22-μm syringe filter. The setup was placed in the greenhouse under standard conditions. As a negative control, Se volatilization from the different Se compounds was measured in flasks without plant material. After the volatile Se was collected for 24 h, the trap solutions were treated according to de Souza et al. (1998). The plant tissue was washed thoroughly with distilled water, dried (55°C, 3 d), ground, and digested as described in the USEPA Methods 7742 (USEPA, 1994) and 3050B (USEPA, 1996). The Se content in trap solutions and the acid-digested plant tissues were analyzed by vapor-generation atomic absorption spectroscopy (AA975, Varian, Palo Alto, CA).
Se Tolerance
Twelve-day-old seedlings were grown on solid media (one-half-strength Murashige and Skoog [Sigma]); 1% (w/v) Glc, pH 5.8; 0.4% (w/v) phytagar [Life Technologies/Gibco-BRL]) in the presence of 25 μm selenate or 10 or 20 μm Se-Met, under long day conditions (16 h light/8 h dark) and at 22°C.
Se Volatilization from E. coli
E. coli strain BL21(DE3) (Novagen) was transformed with plasmid pET16MMT according to Hanahan (1985). Transformants were grown at 37°C in Luria-Bertani medium (Sambrock et al., 1989) containing 100 mg L−1 ampicillin, and growth was monitored by measuring the A595. At an absorbance of 0.6, the expression of the 10His-MMT was induced by adding 0.5 mm isopropyl-β-d-thiogalactoside. The untransformed E. coli strain BL21(DE3) was grown in Luria-Bertani to the same density. Both cultures were divided into 200-mL aliquots, which were transferred to sterile 500-mL gas washing bottles (Fisher Scientific) and treated with 50 μm NaSeO4, 50 μm NaSeO3, or 50 μm Sel-Met. There were three replicates for each treatment. The bottles were connected to the trap solution as illustrated above and placed in a laminar flow hood at room temperature. At appropriate intervals during the exponential growth phase of the bacteria, aliquots were withdrawn from the trap solution to determine the Se content as described above. At the same time intervals, the absorbance (595 nm) and protein content of the cultures were monitored.
Protein Methods
Bacterial protein extracts for SDS-PAGE were prepared by cell lysis with 1% (w/v) SDS at 95°C in the presence of 10 mm dithiothreitol, subsequently separated by SDS-PAGE on 7.5% (w/v) gels (Laemmli, 1970), and either stained with Coomassie Brilliant Blue or transferred to polyvinylidene difluoride membranes according to Michov (German patent no. 4127546, cited in Michov, 1996). Polyclonal antibodies (1:8,000 [v/v] dilution) raised against MMT from Wollastonia biflora (James et al., 1995) were used to detect the recombinant MMT overexpressed in E. coli. Immunoprecipitates were visualized by alkaline phosphatase-conjugated goat:anti-rabbit immunoglobulins and 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate (Boehringer Mannheim/Roche, Basel) staining. For protein determination in E. coli, cultures cells were washed with water and treated according to de Souza and Yoch (1995).
ACKNOWLEDGMENTS
The authors thank the ABRC (Columbus, OH) for providing the T-DNA mutant collections, and Prof. Andrew D. Hanson (Horticultural Science Department, University of Florida, Gainesville) for providing the MMT antibodies.
Footnotes
This work was supported by the Torrey Mesa Research Institute, Syngenta Research and Technology (San Diego).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001693.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Atkinson R, Aschmann SM, Hasegawa D, Thompson-Eagle ET, Frankenberger WT., Jr Kinetics of the atmospherically important reactions of dimethylselenide. Environ Sci Technol. 1990;24:1326–1332. [Google Scholar]
- Bañuelos GS, Terry N, Zayed A, Wu L. Managing high soil selenium in soil with phytoremediation. In: Schuman GE, Vance GF, editors. Selenium: Mining, Reclamation, and Environmental Impact. Proceedings of the 12th Annual Meeting of the American Society for the Surface Mining and Reclamation, Gillette, WY. Princeton, WV: American Society of Surface Mining and Reclamation; 1995. pp. 394–405. [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Taraczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL et al. S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell. 1999;11:1485–1497. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacey JWH, King GM, Wakeham SG. Factors controlling emission of dimethylsulfide from salt marshes. Nature. 1987;330:643–645. [Google Scholar]
- de Souza MP, Chu D, Zhao M, Zayed AM, Ruzin SE, Schichnes D, Terry N. Rhizosphere bacteria enhance selenium accumulation and volatilization by Indian mustard. Plant Physiol. 1999a;119:565–573. doi: 10.1104/pp.119.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza MP, Huang CPA, Chee N, Terry N. Rhizosphere bacteria enhance the accumulation of selenium and mercury in wetland plants. Planta. 1999b;209:259–263. doi: 10.1007/s004250050630. [DOI] [PubMed] [Google Scholar]
- de Souza MP, Lytle CM, Mulholland MM, Otte ML, Terry N. Selenium assimilation and volatilization from dimethylselenoniopropionate by Indian mustard. Plant Physiol. 2000;122:1281–1288. doi: 10.1104/pp.122.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza MP, Pilon-Smits EAH, Lytle M, Hwang S, Tai J, Honma TSU, Yeh L, Terry N. Rate-limiting steps in selenium assimilation and volatilization by Indian mustard. Plant Physiol. 1998;117:1487–1494. doi: 10.1104/pp.117.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza MP, Yoch DC. Purification and characterization of dimethylsulfoniopropionate lyase from an Alcaligenes-like dimethylsulfide-producing marine isolate. Appl Environ Microbiol. 1995;61:21–26. doi: 10.1128/aem.61.1.21-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Duckart EC, Waldron LJ, Doner HE. Selenium uptake and volatilization from plants growing in soil. Soil Sci. 1992;153:94–99. [Google Scholar]
- Farooqui JZ, Tuck M, Paik WK. Purification and characterization of enzymes from Euglena gracilis that methylate methionine and arginine residues of cytochrome c. J Biol Chem. 1985;260:537–545. [PubMed] [Google Scholar]
- Floyd NF, Lavine TF. Isolation of methionine from protein hydrolysates as methylsulfonium salt. J Biol Chem. 1954;207:119–123. [PubMed] [Google Scholar]
- Ganther HE, Levander OA, Saumann CA. Dietary control of selenium volatilization in the rat. J Nutr. 1966;88:55–60. doi: 10.1093/jn/88.1.55. [DOI] [PubMed] [Google Scholar]
- Hanahan D. . In DNA Cloning, Vol I. IRL Press, Oxford, pp 109–135. 1985. Techniques for transformation of Escherichia coli [Google Scholar]
- Hanson AD, Rivoal J, Paquet L, Gage DA. Biosynthesis of 3-dimethylsulfoniopropionate by Wollastonia biflora (L.) DC. Plant Physiol. 1994;105:103–110. doi: 10.1104/pp.105.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Trossat C, Nolte KD, Gage DA. 3-Dimethylsulfoniopropionate biosynthesis in higher plants. In: Cram WJ, editor. Sulfur Metabolism in Higher Plants, Molecular, Ecophysiological and Nutritional Aspects, Third Workshop, Newcastle upon Tyne. Leiden, The Netherlands: Backhuys Publishers; 1997. pp. 147–154. [Google Scholar]
- Hwang S, Herrin DL. Control of lhc gene transcription by the circadian clock in Chlamydomonas reinhardtii. Plant Mol Biol. 1994;26:557–569. doi: 10.1007/BF00013743. [DOI] [PubMed] [Google Scholar]
- James F, Nolte KD, Hanson A. Purification and properties of S-adenosyl-l-methionine:l-methionine S-methyltransferase from Wollastonia biflora leaves. J Biol Chem. 1995;270:22344–22350. doi: 10.1074/jbc.270.38.22344. [DOI] [PubMed] [Google Scholar]
- Kocsis MG, Nolte KD, Rhodes D, Shen TL, Gage DA, Hanson AD. Dimethylsulfoniopropionate biosynthesis in Spartina alterniflora: evidence that S-methylmethionine and dimethylsulfoniopropylamine are intermediates. Plant Physiol. 1998;117:273–281. doi: 10.1104/pp.117.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemly AD. Environmental implications of excessive selenium: a review. Biomed Environ Sci. 1997;10:415–435. [PubMed] [Google Scholar]
- Lewis BG, Johnson CM, Broyer TC. Volatile selenium in higher plants. The production of dimethylselenide in cabbage leaves by enzymatic cleavage of Se-methyl selenomethionine selenonium salt. Plant Soil. 1974;40:107–118. [Google Scholar]
- Lewis BG, Johnson CM, Delwiche CC. Release of volatile selenium compounds by plants: collection procedures and preliminary observations. J Agric Food Chem. 1966;14:638–640. [Google Scholar]
- McConnell KP, Portman OW. Toxicity of dimethylselenide in the rat and mouse. Proc Soc Exp Biol Med. 1952;79:230–231. doi: 10.3181/00379727-79-19333. [DOI] [PubMed] [Google Scholar]
- Michov B. Blotting der proteine. In: Gruyter W, editor. Elektrophorese: Theorie und Praxis. New York. 1996. pp. 270–281. [Google Scholar]
- Mudd SH, Datko AH. The S-methylmethionine cycle in Lemna paucicostata. Plant Physiol. 1990;93:623–630. doi: 10.1104/pp.93.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev. 2000;14:28–33. [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Hwang S, Lytle CM, Zhu Y, Tai JC, Bravo RC, Chen Y, Leustek T, Terry N. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol. 1999;119:123–132. doi: 10.1104/pp.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta MJ, Kaneta T, Larondelle Y, Dohmae N, Kamiya Y. S-Adenosyl-l-methionine:l-methionine S-methyltransferase from germinating barley: purification and localization. Plant Physiol. 1998;118:431–438. doi: 10.1104/pp.118.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, Bourgis F, Ziemak MJ, Rhodes D, Gage DA, Hanson AD. Characterization and functional expression of cDNAs encoding methionine-sensitive and -insensitive homocysteine S-methyltransferases from Arabidopsis. J Biol Chem. 2000;275:15962–15968. doi: 10.1074/jbc.M001116200. [DOI] [PubMed] [Google Scholar]
- Ranocha P, McNeil SD, Ziemak MJ, Li C, Tarczynski MC, Hanson AD. The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 2001;25:575–584. doi: 10.1046/j.1365-313x.2001.00988.x. [DOI] [PubMed] [Google Scholar]
- Rennenberg H. The significance of higher plants in the emission of sulfur compounds from terrestrial ecosystems. In: Sharkey TD, Holland EA, Mooney HA, editors. Trace Gas Emission by Plants. San Diego: Academic Press; 1991. pp. 217–260. [Google Scholar]
- Salt DE, Smith RD, Raskin I. Phytoremediation. In: Jones RL, Somerville CR, Walbot V, editors. Annual Review of Plant Physiology and Plant Molecular Biology. Palo Alto, CA: Annual Reviews Inc.; 1998. pp. 643–668. [DOI] [PubMed] [Google Scholar]
- Sambrock J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sayato Y, Nakamuro K, Hasegawa T. Selenium methylation and toxicity mechanism of selenocystine. Yakugaku Zasshi. 1997;117:665–672. doi: 10.1248/yakushi1947.117.10-11_665. [DOI] [PubMed] [Google Scholar]
- Terry N, Carlson C, Raab TK, Zayed A. Rates of selenium volatilization among crop species. J Environ Qual. 1992;21:341–344. [Google Scholar]
- Terry N, Zayed A. Phytoremediation of selenium. In: Frankenberger WT, Engberg R, editors. Environmental Chemistry of Selenium. New York: Marcel Dekker Inc.; 1998. pp. 633–656. [Google Scholar]
- Terry N, Zayed AM, de Souza MP, Tarun AS. Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:401–432. doi: 10.1146/annurev.arplant.51.1.401. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Neuhierl B, Bock A. S-Methylmethionine metabolism in Escherichia coli. J Bacteriol. 1998;181:662–665. doi: 10.1128/jb.181.2.662-665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA. Methods for Chemical Analysis of Water and Wastes. Washington, DC: USEPA; 1994. Selenium (atomic absorption, borohydride reduction), Method 7742. [Google Scholar]
- USEPA. Methods for Chemical Analysis of Water and Wastes. Washington, DC: USEPA; 1996. Acid digestion of sediments, sludges, and soils, Method 3050B. [Google Scholar]
- von Vleet JF, Ferrans VJ. Etiological factors and pathologic alterations in selenium vitamin E deficiency and excess in animals and humans. Biol Trace Element Res. 1992;33:1–21. doi: 10.1007/BF02783988. [DOI] [PubMed] [Google Scholar]
- Wilber CG. Toxicology of selenium: a review. Clin Toxicol. 1980;17:171–230. doi: 10.3109/15563658008985076. [DOI] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ. Glutathione metabolic genes coordinately respond to heavy metal and jasmonic acid in Arabidopsis. Plant Cell. 1998;10:1539–1550. doi: 10.1105/tpc.10.9.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed AM, Terry N. Selenium volatilization in roots and shoots: effects of shoot removal and sulfate level. J Plant Physiol. 1994;143:8–14. [Google Scholar]
- Zieve R, Peterson PJ. Volatilization of selenium from plants and soil. Sci Total Environ. 1984;32:197–202. [Google Scholar]