Abstract
The little success of breeding approaches toward the improvement of salt tolerance in crop species is thought to be attributable to the quantitative nature of most, if not all the processes implicated. Hence, the identification of some of the quantitative trait loci (QTL) that contribute to natural variation in salt tolerance should be instrumental in eventually manipulating the perception of salinity and the corresponding responses. A good choice to reach this goal is the plant model system Arabidopsis, whose complete genome sequence is now available. Aiming to analyze natural variability in salt tolerance, we have compared the ability of 102 wild-type races (named ecotypes or accessions) of Arabidopsis to germinate on 250 mm NaCl, finding a wide range of variation among them. Accessions displaying extremely different responses to NaCl were intercrossed, and the phenotypes found in their F2 progenies suggested that natural variation in NaCl tolerance during germination was under polygenic controls. Genetic distances calculated on the basis of variations in repeat number at 22 microsatellites, were analyzed in a group of either extremely salt-tolerant or extremely salt-sensitive accessions. We found that most but not all accessions with similar responses to NaCl are phylogenetically related. NaCl tolerance was also studied in 100 recombinant inbred lines derived from a cross between the Columbia-4 and Landsberg erecta accessions. We detected 11 QTL harboring naturally occurring alleles that contribute to natural variation in NaCl tolerance in Arabidopsis, six at the germination and five at the vegetative growth stages, respectively. At least five of these QTL are likely to represent loci not yet described by their relationship with salt stress.
A major factor impairing worldwide agricultural productivity is salinity, which is believed to affect nearly one-fifth of the world's irrigated land and causes 107 irrigated hectares to be abandoned each year (Boyer, 1982; Szaboles, 1987; Flowers and Yeo, 1995; Nelson et al., 1999). To solve the problems caused by salinity in agricultural areas, some engineering-based approaches have been applied, such as increased irrigation with water of high quality or soil drainage. Because these expensive solutions are not always practical, the study of plant salt tolerance, with a view to identifying and eventually manipulating the genes involved in salt perception and responses, seems to be a more promising approach.
Plant salt tolerance is a complex trait, which is considered by many authors to be polygenic and hence difficult to dissect and manipulate. The variety of adaptive mechanisms that plants have evolved to cope with salt stress (McCue and Hanson, 1990) makes it difficult to choose of a single trait as a target for manipulation aimed at significantly improving plant salt tolerance. This might explain the lack of success of breeding programs developed with the aim of obtaining crop varieties able to tolerate salt stress while remaining productive in salinized lands (Flowers and Yeo, 1995). Moreover, the series of crosses necessary to complete a breeding program aimed at the introgression of genes responsible for desirable traits is time consuming, and it is not always possible to find wild relatives of a given crop species displaying the traits of interest (Barkla et al., 1999).
Genetic engineering of salt tolerance has been attempted using mutational and transgenic approaches, one of which involves the transfer into model systems, such as tobacco (Nicotiana tabacum) or Arabidopsis, of transgenes designed to constitutively express genes previously known to be involved in salt tolerance in other plants or unicellular organisms. Only marginal success in increasing salt tolerance has been obtained in this way, and the technology is still not widespread in crop plants (Nelson et al., 1999). The only exceptions are the recently obtained transgenic tomato (Lycopersicon esculentum) and canola (Brassica napus) lines, which overexpress the AtNHX1 gene from Arabidopsis, coding for a vacuolar Na+/H+ antiporter (Zhang and Blumwald, 2001; Zhang et al., 2001). These transgenic lines were able to grow and produce almost normal fruits in the presence of 200 mm NaCl, a concentration that inhibits wild-type plant growth.
One strategy for studying and manipulating plant salt tolerance that has received little attention is the analysis of natural variability in a model system such as Arabidopsis. This approach is now feasible because of the availability of high-density genetic maps, which include the positions of hundreds of molecular markers, together with the development of powerful software to map quantitative trait loci (QTL). QTL analysis can be carried out in different primary mapping populations, such as F2, recombinant inbred lines (RILs), doubled haploid lines, and backcross inbred lines (Yano, 2001). Among these, RILs have many advantages when used to map QTL, because they are permanent populations that can be indefinitely amplified, and the new markers that are mapped can immediately be integrated into their genetic map (Alonso-Blanco and Koornneef, 2000).
In recent years, a large number of studies have been performed with the aim of identifying QTL that control agronomic traits such as biotic and abiotic stress resistance, productivity, and earliness in rice (Oryza sativa; McCouch and Doerge, 1995), salt tolerance during vegetative growth in barley (Hordeum vulgare; Mano and Takeda, 1997) and rice (Koyama et al., 2001), some morphological features and earliness in cauliflower (Brassica oleracea; Lan and Paterson, 2000), and salt tolerance during germination and vegetative growth (Foolad, 1999), fruit size (Frary et al., 2000), and soluble solids contents (Fridman et al., 2000) in tomato. In the last two cases, the genes at the QTL detected have been cloned, which demonstrates that the QTL approach not only gives a relatively wide genome interval but also facilitates the identification of the genes responsible for the trait under study. In Arabidopsis, QTL have been identified that control seed dormancy (Van der Schaar et al., 1997), seed size (Alonso-Blanco et al., 1999), seed soluble oligosaccharides and storability (Bentsink et al., 2000), and flowering time (Kowalski et al., 1994; Clarke et al., 1995; Alonso-Blanco et al., 1998; Juenger et al., 2000). The cloning of the CRY2 gene of Arabidopsis, encoding the blue-light receptor cryptochrome 2, using a QTL approach has recently been reported (El-Assal et al., 2001).
In the present work, we analyze NaCl tolerance in 102 accessions and 100 RILs of Arabidopsis, with the aim of identifying loci that control natural variations in salt tolerance during the germination and vegetative growth stages. This experimental approach will hopefully provide valuable insight into the identification of genes whose eventual manipulation could improve plant salt tolerance.
RESULTS
Natural Variability of NaCl Tolerance among Arabidopsis Accessions
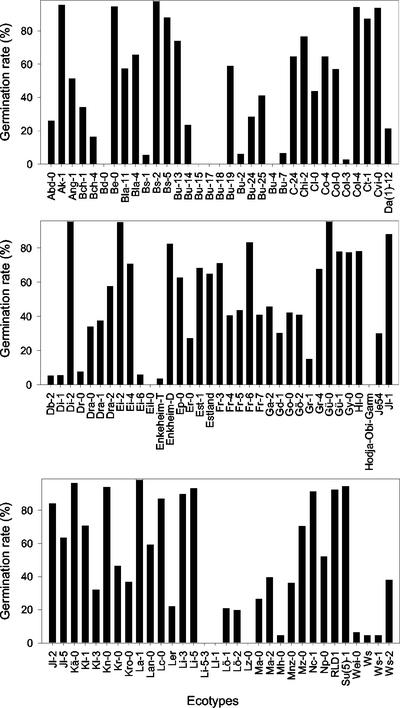
We first analyzed NaCl tolerance in a sample of 102 Arabidopsis wild-type strains or accessions (ecotypes), obtained from the Nottingham Arabidopsis Stock Centre (NASC), all but 12 of which belonged to the Arabidopsis Information Service collection (Röbbelen, 1965; Kranz, 1978). Germination of approximately 100% was obtained when seeds from these accessions were sown on non-supplemented medium. To normalize germination percentages, we referred the scores obtained on salt-supplemented media to those obtained on non-supplemented media. On the basis of the emergence of the radicle through the seed coat as a criterion for discriminating germinated from ungerminated seeds, we found strong variations in the ability of accession seeds to germinate on 250 mm NaCl medium (Fig. 1). Germination levels above 90% were obtained for 16 accessions: Ak-1 (N938), Be-0 (N964), Bs-2 (N998), Bs-5 (N1000), Columbia-4 (Col-4; N1090), Cvi-0 (N1096), Di-2 (N1110), Ei-2 (N1124), Gü-0 (N1212), Kä-0 (N1266), Kn-0 (N1286), La-1 (N1302), Li-5 (N1320), Nc-1 (N1388), RLD1 (N913), and Su(5)-1 (N930). Absolutely no germination was observed in nine accessions: Bd-0 (N962), Bu-4 (N1012), Bu-15 (N1034), Bu-17 (N1036), Bu-18 (N1038), Hodja-Obi-Garm (N922), Li-5–3 (N1324), Ll-1 (N1340), and Lz-0 (N1354). The remaining 77 accessions showed germination percentages ranging from 4.5 to 88.
Figure 1.
Germination rates of different Arabidopsis accessions sown on growth media supplemented with 250 mm NaCl, determined 14 d after sowing.
The accessions showing more than 90% germination on 250 mm NaCl medium were later tested on 300 and 350 mm NaCl. It was seen that N938 (Ak-1) and N1302 (La-1) showed the highest germination levels (94.8% and 93.2%, respectively, on 350 mm NaCl) and these were selected as the most salt tolerant. Accessions unable to germinate on 250 mm NaCl medium were sown on lower salt concentrations (50, 100, and 150 mm NaCl), N1038 (Bu-18), and N922 (Hodja-Obi-Garm) being chosen as the most salt sensitive, because they displayed the lowest germination values (68.9% and 76.7%, respectively, on 150 mm NaCl). We could not find any correlation between the salt tolerance of the selected accessions and the scarce information available on the environmental conditions of their habitats.
The most salt-tolerant accessions (N938 [Ak-1] and N1302 [La-1]) were crossed with the most salt-sensitive ones (N1038 [Bu-18] and N922 [Hodja-Obi-Garm]) to ascertain whether or not their salt tolerance at germination is a monogenic trait. F2 seeds were obtained by selfing F1 individuals grown on non-supplemented medium. Germination of 100% was obtained when F2 seeds from each cross were sown on non-supplemented medium. When additional F2 progeny was sown on 250 mm NaCl, the phenotypic segregation observed did not fit a monogenic transmission pattern, suggesting that the salt tolerance displayed at germination by these accessions was under polygenic control, in agreement with the wide range of variations in salt tolerance found among the accessions (data not shown).
To study whether or not salt tolerance during seed germination and vegetative growth were related, we determined the effect of NaCl on the growth of accessions displaying extremely different responses to NaCl at germination. The N938 (Ak-1), N1302 (La-1), N1038 (Bu-18), and N922 (Hodja-Obi-Garm) accessions were sown on growth medium supplemented with 50 mm NaCl, a concentration already shown to permit the germination and growth of other accessions (Quesada et al., 2000). The fresh and dry weights of stressed and non-stressed plants were determined as described in “Materials and Methods.” We found that the presence of NaCl decreased both dry and fresh weights in the accessions selected as the most salt tolerant at germination (14.5% and 31% fresh weight loss, and 7.5% and 27.2% dry weight loss for N1302 [La-1] and N938 [Ak-1], respectively). In contrast, a 14.7% and 7.3% increase in fresh weight and 0% and 0.9% reduction in dry weight were found for the most salt-sensitive ones (N1038 [Bu-18] and N922 [Hodja-Obi-Garm], respectively). These results suggest that the genetic controls of NaCl tolerance during germination and vegetative growth are independent in Arabidopsis.
Microsatellite Length Variation among Accessions
To investigate the phylogenetic relationships between the accessions selected as the most salt-tolerant and -sensitive during germination, we analyzed variations in repeat number at 22 polymorphic microsatellites in a sample of 11 wild-type strains: the five most salt tolerant (Ak-1 [N938], Bs-2 [N998], Estland [N911], Gü-0 [N1212], and La-1[N1302]) and the five most salt sensitive (Bd-0 [N962], Bu-17 [N1036], Bu-18 [N1038], Hodja-Obi-Garm [N922], and Li-5–3 [N1324]), together with Col-0 as the reference accession used to transform microsatellite length in repeat number for each microsatellite allele (see “Materials and Methods”). As Table I shows, the average number of repeats ranged from 8.55 (nga1145 and nga162) to 43.05 (AthGENEA), whereas the number of alleles per locus ranged from 2 (nga1145) to 10 (nga6, AM4, and nga1139), with an average of 6.45. The most frequent motif analyzed was (GA)n. In addition to the number of alleles per locus, we also estimated gene diversity (expected heterozygosity) to determine the level of microsatellite polymorphism (see “Materials and Methods”). The gene diversity ranged from 0.18 (nga1145) to 0.98 (AM4 and nga6), with an average of 0.79 for the 22 microsatellites. This value is the same as that obtained by Innan et al. (1997), who analyzed a sample of 20 microsatellites in a population of 42 accessions of Arabidopsis while investigating the recent evolutionary history of this species.
Table I.
Variation found at the Arabidopsis microsatellites analyzed
| Locus | Chromosome | Motif Repeated | Accession
|
No. of Alleles | Average | Variance | Gene Diversity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Col | Bd-0 | Bu-17 | Bu-18 | Li-5–3 | Hodja | La-1 | Ak-1 | Estland | Gü-0 | Bs-2 | |||||||
| AthACS | 1 | A | 36 | 32 | 37 | 33/37 | 38 | 33 | 37 | 38 | 30 | 30 | 40 | 7 | 35.09 | 3.37 | 0.92 |
| AthZFPG | 1 | AG | 16 | 8 | 12 | 7/9 | 8 | 9 | 46 | 19 | 7 | 35 | 19 | 8 | 17.00 | 12.67 | 0.95 |
| T27k12Sp6 | 1 | AT | 12 | 11 | 20 | 11/35 | N.D. | 11 | 28 | 38 | 13 | 36 | 38 | 8 | 23.00 | 11.49 | 0.94 |
| AthGENEA | 1 | A | 39 | 52 | 37/54 | 39 | 44 | 42 | 47 | 40 | 52 | 34 | 39 | 9 | 43.05 | 6.23 | 0.93 |
| nga111 | 1 | GA | 16 | 23 | 16/44 | 23/30 | 25 | 30 | 22 | 22 | 23 | 25 | 24 | 7 | 24.23 | 6.02 | 0.92 |
| nga1145 | 2 | GA | 14 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 2 | 8.55 | 1.80 | 0.18 |
| nga1126 | 2 | AG | 14 | 16 | 14 | 18/25 | 19 | 18 | 17 | 14 | 15 | 19 | 14 | 7 | 16.50 | 2.77 | 0.87 |
| nga361 | 2 | AG | 14 | 15 | 11 | 13/22 | 21 | 13 | 12 | 13 | 19 | 11 | 19 | 8 | 15.05 | 3.69 | 0.93 |
| nga168 | 2 | GA | 25 | 24 | 17 | 13/17 | 18 | 13 | 18 | 17 | 20 | 17 | 17 | 6 | 18.27 | 3.60 | 0.83 |
| AthCHIB | 3 | AT | 14 | 6/22 | 12 | 0/6 | 17 | 6 | 12 | 14 | 12 | 6 | 15 | 7 | 11.36 | 5.11 | 0.88 |
| nga162 | 3 | GA | 17 | 6 | 7 | 6 | 9 | 5 | 11 | 9 | 9 | 5 | 10 | 7 | 8.55 | 3.46 | 0.91 |
| AthGAPAab | 3 | CTT | 10 | 10 | 10 | 10/14 | 10 | 14 | 10 | 13 | 10 | 10 | 10 | 3 | 10.82 | 1.59 | 0.41 |
| nga6 | 3 | GA | 31 | 20 | 14 | 18/39 | 14 | 39 | 17 | 16 | 43 | 24 | 25 | 10 | 24.68 | 10.44 | 0.98 |
| nga12 | 4 | GA | 14 | 8/14 | 8 | 8 | 14 | 8 | 8 | 14 | 8 | 14 | 8 | 2 | 10.45 | 3.04 | 0.53 |
| nga1111 | 4 | GA | 16 | 17 | 16 | 26 | 22 | 25 | 16 | 16 | 18 | 17 | 9 | 7 | 18.00 | 4.31 | 0.87 |
| AM4 | 4 | AT | 19 | 12 | 7/17 | 16 | 15 | 23 | 65 | 5 | 7 | 9 | 9 | 10 | 17.45 | 16.60 | 0.98 |
| nga1139 | 4 | GA | 20 | 15 | 14/28 | 18/30 | 16 | 29 | 15 | 21 | 24 | 18 | 15 | 10 | 19.82 | 5.31 | 0.95 |
| nga1107 | 4 | AG | 27 | 16 | 16 | 17 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 3 | 17.09 | 3.29 | 0.35 |
| AthCTR1 | 5 | AG | 16 | 9 | 8 | 11 | 16 | 10 | 11 | 9 | 9 | 7 | 8 | 6 | 10.36 | 2.99 | 0.89 |
| nga139 | 5 | AG | 29 | 22 | 5 | 6 | 6 | 35 | 7 | 13 | 22 | 21 | 13 | 8 | 16.27 | 10.20 | 0.95 |
| AthPHYC | 5 | ATT; GAA | 17 | 22 | 17 | 17/22 | 22 | 17 | 22 | 22 | 22 | 22 | 22 | 2 | 20.41 | 2.42 | 0.48 |
| MBK5 | 5 | CTT | 15 | 7 | 13 | 13/15 | 13 | 15 | 13 | 17 | 7 | 7 | 22 | 5 | 13.00 | 4.15 | 0.83 |
N.D., Not determined.
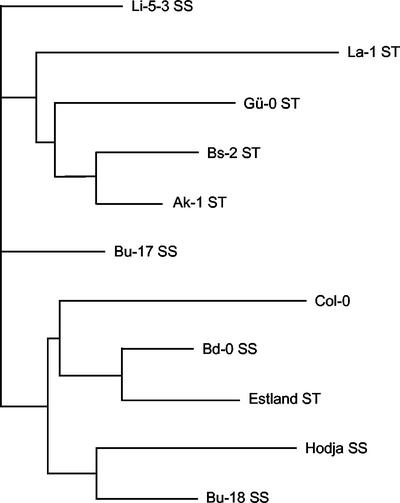
Several distance matrices were obtained with the Microsat program, calculated on the basis of different genetic distance measurements (see “Materials and Methods”) and a consensus phylogenetic tree was constructed from resampled data using the neighbor-joining method. Most, but not all, the accessions with a similar salt tolerance at germination could be grouped in the same clade, indicating that the genetic distance is shorter between strains with similar salt stress responses than between those with different responses (Fig. 2).
Figure 2.
Phylogenetic tree of accessions displaying extremely different responses to NaCl. Extremely salt-sensitive and -tolerant accessions are indicated as SS and ST, respectively. The phylogram was constructed using the NEIGHBOR program included in the PHYLIP 3.5c package, from a distance matrix calculated by the Microsat 1.5d program on the basis of the number of repeats found in 22 polymorphic microsatellites, using the neighbor-joining algorithm and the absolute distance (DAD) parameter.
QTL Analysis
The above results suggested but did not demonstrate that natural variation in NaCl tolerance at the germination stage of Arabidopsis is a polygenic trait. To ascertain the existence of the corresponding QTL and to eventually determine their map positions, we analyzed a sample of 100 RILs derived from a cross between the Col-4 and Landsberg erecta (Ler-0) accessions (Lister and Dean, 1993).
Effects of NaCl on RIL Germination
To analyze the effects of salinity on RIL germination, two to four progenies per RIL were studied (see “Materials and Methods”), sowing 100 seeds from each progeny on growth medium supplemented with 250 mm NaCl. Considering as germinated those seeds in which the radicle had emerged through the seed coat, germination was scored at 24-h intervals from the 3rd d after sowing until the score remained unchanged for three consecutive days. Control sowings were always performed in parallel in non-supplemented media, to normalize germination percentages by referring scores obtained on salt-supplemented media to those obtained on non-supplemented media. Hence, the germination rate for each RIL was the average of the different progenies studied.
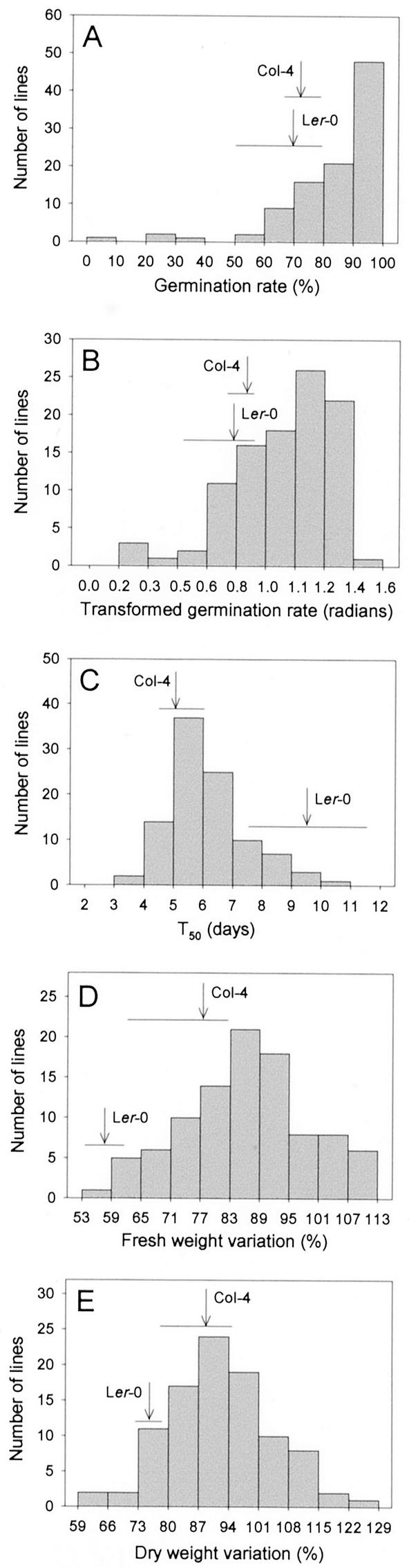
RIL germination on salt-supplemented media was analyzed by using two parameters: the time in days to reach a 50% germination (T50; Foolad, 1999) and the germination rate after 15 d of exposure to 250 mm NaCl. We calculated the T50 for all of the RILs, the only exception being the N1970 line, whose germination rate was only 1.7% on 250 mm NaCl. With regard to the germination rates determined 15 d after sowing, we obtained a wide spectrum of values, ranging from 1.7% (N1970) to 98.5% (N1927 and N1999), most being above 70%. In contrast, differences between the parental accessions Col-4 and Ler-0 were very small (Fig. 3A). Taken together, these results indicate transgressive variation in both directions as a consequence of the effects of alleles that putatively increase and reduce salt tolerance in both parental lines. The distribution fit to normality was determined by χ2 analysis (χ2 = 42.8; P = 3.6 × 10−7; freedom degrees [FD] = 7; Fig. 3A), and the angular (arcsin) transformation was considered the best transformation that normalized the parameter. In this way, normality was improved in the distribution of transformed data (χ2 = 13.027; P = 0.071; FD = 7; Fig. 3B). The frequency distribution of the times to reach a 50% germination (T50) was normal as determined by a χ2 test (χ2 = 15.082; P = 0.035; FD = 7), the differences between the parental accessions being significant and displaying transgression in both directions (Fig. 3C).
Figure 3.
Frequency distribution of the germination rates of the RILs 15 d after sowing (A), their angular (arcsin) transformation (B), and their variation with time estimated from the T50 values (C), on media supplemented with 250 mm NaCl. Frequency distributions are also shown for the variation with time of the fresh (D) and dry weight (E), of plants grown on 50 mm NaCl. Arrows indicate the values corresponding to the parental accessions and the horizontal bars their standard variation.
Effects of NaCl on RIL Vegetative Growth
To study RIL salt tolerance during developmental stages other than germination, sowings were made in medium supplemented with 50 mm NaCl. The fresh and dry weights were determined for each RIL as described in “Materials and Methods,” from the values obtained on NaCl-supplemented and non-supplemented media.
We found great variability in the response of RILs during vegetative growth to the moderated salt stress caused by a medium supplemented with 50 mm NaCl, the distribution of the variation in fresh and dry weight being continuous and normal, as determined by χ2 analyses [(χ2 = 9.455; P = 0.222; FD = 7) and (χ2 = 6.121; P = 0.047; FD = 7) for fresh and dry weight, respectively] (Fig. 3, D and E). Differences between the parental accessions were small for dry weight and higher for fresh weight. Hence, both parental lines carry alleles that increase and reduce salt tolerance, in agreement with the transgression observed.
Correlation Analysis between the Traits Analyzed
Significant correlation was found between the traits analyzed to study salt tolerance at germination, as well as between the traits studied to analyze the response to moderate salt stress during vegetative growth. No significant correlation was found for any other pair of traits (Table II).
Table II.
Correlation coefficients between the traits analyzed
| Germination Ratea | Transformed Germination Ratea | T50 | Variation in Fresh Wt | |
|---|---|---|---|---|
| Transformed germination ratea | 0.9721b | |||
| T50 | −0.6615b | −0.6698b | ||
| Variation in fresh wt | −0.1079 | −0.1418 | 0.0895 | |
| Variation in dry wt | −0.1500 | −0.1988 | 0.2512 | 0.6831b |
Determined 15 d after sowing.
α = 0.01.
QTL Mapping
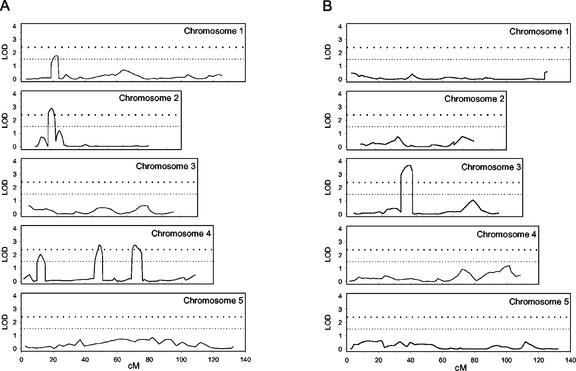
The phenotypic values of the 100 RILs analyzed and the data of the molecular markers of the Arabidopsis genetic map were used for QTL analysis using the MapQTL 4.0 program (Van Ooijen and Maliepaard, 1995), as described in “Materials and Methods.” Regarding the response to salinity at germination, six QTL were detected. Five of these QTL were identified from the transformed values of the germination rates obtained 15 d after sowing, mapping on chromosomes 1, 2, and 4. The QTL on chromosomes 2 (19.1 centiMorgans [cM]) and 4 (48.7 and 72.2 cM) are major QTL (logarithm of odds [LOD] > 2.4), whereas the remaining two are minor QTL (LOD < 2.4; Fig. 4A; Table III). With regard to the time course of germination rates estimated from the T50 values, only a major QTL (LOD = 3.18) was detected, on chromosome 3 (Table III; Fig. 4B).
Figure 4.
QTL contributing to NaCl tolerance at the germination stage of Arabidopsis, as determined with the MapQTL 4.0 program. One hundred RILs were analyzed by determining on growth media supplemented with 250 mm NaCl the angular (arcsin) transformation of the germination rate, 15 d after sowing (A) and the variation with time of the germination rate (B; T50). LOD threshold levels for major (2.4) and minor (1.5) QTL are indicated by dotted lines.
Table III.
QTL involved in NaCl tolerance during seed germination and vegetative growth in Arabidopsis
| Trait | Chromosome | Map Positiona | LOD | Confidence Interval | Percentage of Variance Explained | Additive Effectb |
|---|---|---|---|---|---|---|
| cM | cM | |||||
| Transformed | 1 | 20.9 | 1.72 | 13–33c | 5.7d | −0.07 |
| germination rate | 2 | 19.1 | 2.65 | 13–24e | 8.6d | 0.13 |
| 4 | 11.9 | 2.04 | 7–20c | 6.8d | −0.08 | |
| 4 | 48.7 | 2.75 | 36–57e | 9.3d | 0.10 | |
| 4 | 72.2 | 2.56 | 61–102e | 8.6d | −0.09 | |
| 31.7f | ||||||
| T50 | 3 | 36.3 | 3.18 | 13–48e | 13.7f | −0.50 |
| Variation in fresh wt | 1 | 125.4 | 2.21 | 116–125c | 6.8d | −3.22 |
| 4 | 54.5 | 3.8 | 46–65e | 12.2d | −4.21 | |
| 5 | 37.8 | 4.33 | 34–56e | 14.1d | 5.05 | |
| 5 | 77.3 | 1.95 | 69–88c | 5.1d | −3.39 | |
| 5 | 96.9 | 3.06 | 88–116e | 9.6d | 4.40 | |
| 38.4f |
Maximum likelihood map position.
Additive effect determined as indicated in “Materials and Methods.”
Positions given correspond to 1-LOD confidence intervals.
Percentage of variance explained by each QTL detected for a given trait.
Positions given correspond to 2-LOD confidence intervals.
Percentage of variance explained by all of the QTL detected for a given trait.
Two LOD support intervals as 95% confidence intervals and one LOD support intervals as 90% confidence were established for the major and minor QTL, respectively, the two LOD confidence intervals ranging from 11 to 41 cM and the one LOD confidence intervals from 9 to 20 cM.
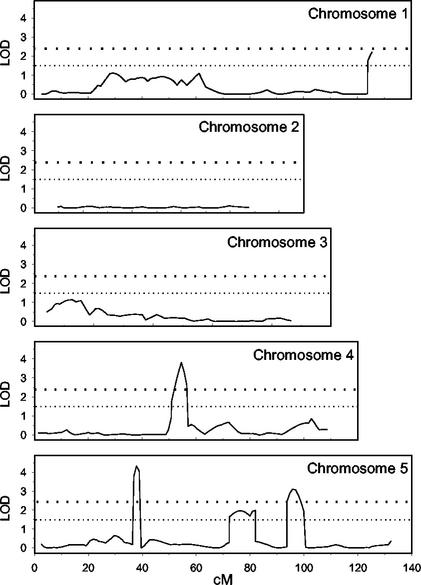
Analysis of the variation in fresh weight on 50 mm NaCl-supplemented media allowed us to identify five QTL involved in salt tolerance during vegetative growth: three major QTL located on chromosomes 4 (54.5 cM) and 5 (37.8 and 96.9 cM) and two minor QTL on chromosomes 1 (125.4 cM) and 5 (77.3 cM; Fig. 5). Their confidence intervals ranged from 19 to 28 cM and from 9 to 19 cM for the major and minor QTL, respectively. No QTL was detected from the dry weight data obtained.
Figure 5.
QTL contributing to NaCl tolerance during vegetative growth in Arabidopsis. One hundred RILs derived from a cross between the Col-4 and Ler-0 accessions were analyzed by determining for each of them the ratio between the fresh weight of the aerial part of the plants grown on media supplemented with 50 mm NaCl and the fresh weight obtained on non-supplemented media, 21 d after sowing. The analysis of the data was performed as described in Figure 4.
Role of the QTL Detected in Salt Tolerance during Germination and Vegetative Growth
We determined the additive effect of each identified QTL, considering the effect as positive when the mean value of a trait in the plants carrying the Col-4 alleles was higher than that of the Ler-0 alleles (Table III). On the contrary, a negative QTL effect was assumed when the mean value of a trait in the plants of the Ler-0 genotypic group was higher than that of the Col-4 group. Therefore, the phenotypic value of the trait under study is increased by Col-4 alleles in the first case and by Ler-0 in the second one. Compared with the Col-4 alleles, Ler-0 alleles increase salt tolerance at four and three loci at germination (chromosomes 1, 3, and 4) and vegetative growth (chromosomes 1, 4, and 5), respectively, whereas Col-4 alleles positively contribute to salt tolerance at the remaining four loci (two of them controlling germination and the other two involved in vegetative growth). The existence of alleles increasing and reducing salt tolerance in Col-4 and Ler-0 is in agreement with the transgression detected in the RILs.
We also determined the phenotypic variation explained for each trait by the QTL detected. For the transformed germination rate, the proportion of the total variation explained by the QTL is 31.7%. With regard to the QTL identified from the T50 values obtained, the identified proportion of the total variation that it explains is 13.7% (Table III).
The proportion of the total phenotypic variation explained by the QTL detected for vegetative growth variation in fresh weight was 38.4%, ranging from 5.1% to 14.1% (Table III). As expected, QTL with a large effect explained more phenotypic variation than those with a small effect.
The map positions of the QTL detected for germination were not coincident with those obtained for the QTL involved in salt response during vegetative growth, suggesting that the mechanisms controlling salt tolerance at both stages are different. This result is in agreement with that obtained when we analyzed salt tolerance during vegetative growth among the Arabidopsis accessions.
Candidate Genes
With the aim of identifying candidate genes for the QTL detected in this work, we compared their map positions with those of the genes previously described and considered likely to be relevant for salt tolerance in Arabidopsis. We considered (a) genes whose mutations cause a salt response different from that of their wild-type ancestors during the vegetative growth (SOS1-SOS3 [Liu and Zhu, 1998; Zhu et al., 1998; Liu et al., 2000; Shi et al., 2000] and PFS1 [Tsugane et al., 1999]) or during germination (SAÑ1-SAÑ4 [Quesada et al., 2000] and rss [Werner and Finkelstein, 1995]); (b) genes required for ABA perception (ABI1-ABI5; Giraudat et al., 1992; Leung et al., 1994, 1997; Meyer et al., 1994; Finkelstein et al., 1998; Finkelstein and Lynch, 2000), modulation of the ABA signal (ERA1 [Cutler et al., 1996] and ABH1 [Hugouvieux et al., 2001]), or ABA biosynthesis (ABA1-ABA3; Koornneef et al., 1982; Léon-Kloosterziel et al., 1996; Xiong et al., 2001) because this plant hormone mediates abiotic stress responses; and (c) genes involved in the response to other environmental stresses such as freezing (SFR1-SRF7 [Thorlby et al., 1999], ESK1 [Xin and Browse, 1998], and CHS1 [Hugly et al., 1990]) or drought (ERD and RD [Taji et al., 1999]).
The results of our search for candidate genes are summarized in Table IV. It is of note that some of the QTL detected map close to genes involved in ABA responses (ABI1, ABI2, and ABI3), biosynthesis (ABA3) or modulation (ABH1). It has previously been reported that abi and aba mutants are more insensitive to salt stress than their wild-type ancestors (Werner and Finkelstein, 1995; Léon-Kloosteitz et al., 1996; Quesada et al., 2000). Genes involved in other environmental responses such as drought (RD26, RD29A, RD29B, and DREB1) or freezing (SFR5 and CBF1) could potentially correspond to some QTL detected in this work (Table IV). Another significant colocalization is the map position of the QTL on chromosome 5 (69–88 cM), which is involved in the variation in fresh weight, and that of the SOS2 gene, whose product is a Ser/Thr protein kinase required for salt tolerance during vegetative growth in Arabidopsis (Liu et al., 2000). For the remaining QTL detected, no obvious candidate gene could be assigned, so that they probably represent new genes not yet assessed for their role in salt tolerance.
Table IV.
Co-ocurrence of QTL positions and candidate genes
| Trait | Chromosome | Map Position (Interval) | Candidate Genes (Map Position) | References |
|---|---|---|---|---|
| cM | ||||
| Transformed germination rate | 1 | 20.9 (13–33) | SFR5 (19.8) ABA3 (25) | Thorlby et al. (1999),Xiong et al. (2001) |
| 2 | 19.1 (13–24) | ABH1 (24) | Hugovieux et al. (2001) | |
| 4 | 72.2 (61–102) | RD26 (72.8) ABI1 (72.9) CBF1 (74) DREB1A&C (74) | Taji et al. (1999),Meyer et al. (1994) Jaglo-Ottosen et al. (1998) Liu et al. (1998) | |
| T50 | 3 | 36.3 (13–48) | ABI3 (38) | Giraudat et al. (1992) |
| Variation in fresh wt | 5 | 77.3 (69–88) | SOS2 (70) | Liu et al. (2000) |
| 5 | 96.9 (88–116) | RD29A (104.7) RD29B (104.7) ABI2 (112) | Horvath et al. (1993),Nordin et al. (1993),Leung et al. (1997) | |
DISCUSSION
As a result of environmental adaptation, Arabidopsis shows a wide spectrum of intraspecific variability in traits such as flowering time (Kowalski et al., 1994; Alonso-Blanco et al., 1998), circadian rhythms (Swarup et al., 1999), and seed size (Alonso-Blanco et al., 1999), which suggests that this plant species may be useful as a source of potential genetic resources.
In this work, we present results obtained from the analysis of natural variations in NaCl tolerance in the model plant Arabidopsis. We first compared the ability of 102 wild-type strains to germinate in saline conditions. The broad spectrum of germination percentages obtained on 250 mm NaCl medium can be interpreted as continuous variation among Arabidopsis races, suggesting, but not demonstrating, that such natural variability is controlled by QTL. We isolated wild-type strains displaying extremely different germination responses to NaCl and the results from their intercrosses indicated that the studied trait was likely to be under polygenic control.
We studied salt tolerance during vegetative growth in the wild-type races selected as extremely salt tolerant or extremely salt sensitive at germination by sowing them on 50 mm NaCl medium. The results obtained when the responses to salt stress during germination and vegetative growth were compared suggested that the genetic controls underlying NaCl tolerance in Arabidopsis are different, because the most tolerant accessions to NaCl at germination were the most sensitive to this salt during vegetative growth. Similar results have been previously reported for other plant species such as soybean (Glycine max; Abel and Mackenzie, 1963), wheat (Triticum aestivum; Kumar et al., 1983), alfalfa (Medicago sativa; Johnson et al., 1992), barley (Mano and Takeda, 1997), and tomato (Foolad, 1999).
We analyzed the phylogenetic relationships between the wild-type strains with extremely different salt tolerance at germination, studying variation in the repeat number at 22 polymorphic microsatellites in these wild-type races. The average gene diversity value obtained (0.79) was the same as that reported by Innan et al. (1997), who studied variation in repeat number at 20 microsatellite loci in 42 accessions. This polymorphism level is also comparable with that described in previous analyses performed in Arabidopsis (Todokoro et al., 1995). The genetic distances obtained indicated that most accessions with similar salt tolerance at germination are phylogenetically related, because they can be grouped in the same clade.
We determined that the RILs used in this work (Lister and Dean, 1993) show moderate variability in their germination rates in 250 mm NaCl. These results agree with those obtained by Van der Schaar et al. (1997), who studied seed dormancy in these RILs, also finding a moderate variability. We found very small differences between the parental accessions (Col-4 and Ler-0) in their germination rates determined 15 d after sowing on 250 mm NaCl, in contrast with the significant differences observed between both wild-type strains in morphological features such as bearing, silique size, or flowering time. Differences between Col-4 and Ler-0 were more pronounced when it came to variations in the time needed to reach T50 on 250 mm NaCl. No significant differences were found in the T50 values reached by the RILs and their parental accessions when sown on non-supplemented media (data not shown), indicating that the T50 differences between Col-4 and Ler-0 are attributable to a different salt stress response.
A significant correlation was found between germination rates and T50 values in the presence of NaCl: RILs with the highest germination rates also yielded the lowest T50 values. With regard to their salt tolerance during vegetative growth, differences found between the parental accessions Col-4 and Ler-0 were higher than those found when we studied the germination rate on NaCl. No correlation was found between the salinity responses of the RILs during germination and vegetative growth, in agreement with the results obtained with the accessions showing extremely different salt tolerance. It is to be noted that the frequency distribution of the traits analyzed in the RILs is continuous and normal, supporting the polygenic nature of salt tolerance in plants (Lindsey and Jones, 1989).
The use of interval analysis and the multiple-QTL model mapping method (MQM; Jansen, 1994) allowed us to identify 11 genomic regions containing loci involved in the response to salt stress in Arabidopsis. This number of loci is similar to that detected by Mano and Takeda (1997) in barley and by Foolad (1999) in tomato. The percentage of variance explained by each QTL found in our work is similar to that reported by Foolad (1999) in tomato: more than 10% and from 5% to 10% for each major and minor QTL, respectively.
When we analyzed salt tolerance at germination, six QTL were detected, five of them contributing to the germination rate and the remaining one to its variation with time estimated from the T50 values. Despite the significant correlation found between T50 and germination rate values, the map positions of T50 and percentage germination QTL were different. The map position of the T50 QTL detected in our work (36.3 cM) is very close to the location of the ABI3 gene (38 cM), which encodes a transcriptional regulator that participates in the transduction of the ABA signal in seeds (Giraudat et al., 1992). Likewise, the QTL on chromosomes 1 (20.9 cM), 2 (19.1 cM), and 4 (72.2 cM) map very close to the genes ABA3 (encoding a molybdenum cofactor sulfurase involved in the last step of ABA biosynthesis [Xiong et al., 2001]), ABH1 (encoding a mRNA cap binding protein required to modulate ABA signaling [Hugovieux et al., 2001]), and ABI1 (encoding a protein phosphatase that participates in ABA signaling [Leung et al., 1994]), respectively.
Because it is well documented that ABA plays a major role in the response to osmotic stress during germination (Begum et al., 1992; Groot and Karssen, 1992; Ni and Bradford, 1993), some of the QTL identified in this work are likely to be related to osmotic stress. In fact, abi and aba mutants are able to germinate better than their wild-type ancestors in saline conditions (Werner and Finkelstein, 1995; Léon-Kloosteitz et al., 1996; Quesada et al., 2000). Significantly, Mano and Takeda (1997) found that the major QTL controlling ABA response at germination in barley mapped very close to that of QTL involved in salt tolerance at the same developmental stage, supporting a role for ABA-related genes in the control of salt responses during germination. Taken together, these results indicate that natural allelic variations in genes involved in ABA signaling or biosynthesis are good candidates to account for the differences in salt responses between Ler-0 and Col-4, the parental accessions of the RILs studied here. Nevertheless, we cannot rule out that other environmental stress response genes that colocalize with the QTL mentioned above (such as SFR5, RD26, CBF1, or DREB1) might be responsible for the differences between Ler-0 and Col-4 regarding salt tolerance during the germination stage.
No candidate genes could be assigned to the remaining QTL detected during germination on the basis of the comparison of their location with that of genes previously mapped and reported to be related with salt stress. Nevertheless, the existence of wild-type strains exhibiting differences in salt response greater than that of Col-4 and Ler-0, as found in our accession analysis, suggests that additional loci may be involved in the control of salt tolerance at germination in Arabidopsis. Detection of these loci would require the analysis of RIL mapping populations derived from crosses including accessions displaying large differences in their responses to salinity.
With regard to the vegetative growth analyses, five QTL were detected from the data for variations in fresh weight. The location of the QTL on chromosome 5 (77.3 cM) is close to that of the SOS2 gene, whose product is a Ser/Thr protein kinase required for K+ nutrition and NaCl tolerance during vegetative growth in Arabidopsis (Liu et al., 2000). Another QTL on chromosome 5 (96.9 cM) maps very close to some genes whose expression is induced in response to osmotic stress, such as RD29A (Horvath et al., 1993) and RD29B (Nordin et al., 1993), both encoding hydrophilic proteins, or whose product is involved in the ABA signal transduction pathway, such as the ABI2 gene (Leung et al., 1997).
The map positions of the QTL found to be involved in salt responses during vegetative growth are different from those of the QTL involved in the same response at germination. This is in agreement with the correlation analysis of our data and with the results obtained in the study of accessions, reinforcing the hypothesis of different genetic controls regulating salt tolerance in different developmental stages in Arabidopsis.
In the work presented here, we have analyzed natural variations in NaCl tolerance in a wide sample of wild-type strains and RILs. Our results indicate that NaCl tolerance in Arabidopsis is a quantitative trait under polygenic control. This study allowed us to identify genomic regions involved in the responses to NaCl at germination and during vegetative growth. Results obtained from the accessions and the RILs indicate the existence of different genetic controls acting on the responses to NaCl during germination and vegetative growth. At least five of the 11 genomic regions identified are likely to represent new loci not yet described by its relationship with salt tolerance. Further analyses involving KCl and mannitol will be required to determine whether these QTL affecting NaCl tolerance are ion specific or are associated to osmotic stress rather than sodium toxicity.
The information reported in this work will contribute to the identification and eventual manipulation of Arabidopsis genes involved in natural variation in salinity responses. This might also be applied to agronomic species, in which their orthologs could be identified and eventually manipulated to increase their yield under saline conditions.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The list of studied Arabidopsis accessions, whose seeds were provided by the NASC, includes the following: NW20, N904, N906, N908, N911, N913, N915, N917, N920, N921, N922, N924, N930, N932, N933, N938, N950, N956, N960, N962, N964, N976, N984, N996, N998, N1000, N1008, N1012, N1018, N1026, N1032, N1034, N1036, N1038, N1040, N1050, N1052, N1074, N1076, N1082, N1090, N1092, N1094, N1096, N1104, N1108, N1110, N1114, N1116, N1118, N1120, N1124, N1126, N1130, N1132, N1140, N1142, N1150, N1170, N1172, N1174, N1176, N1178, N1182, N1184, N1194, N1196, N1198, N1204, N1212, N1214, N1216, N1228, N1248, N1250, N1256, N1266, N1276, N1280, N1286, N1296, N1300, N1302, N1304, N1306, N1316, N1320, N1324, N1340, N1346, N1348, N1354, N1356, N1358, N1366, N1370, N1382, N1388, N1396, N1601, N2223, and N3110.
F8 seeds of a set of 100 RILs developed by Lister and Dean (1993), derived from a cross between the Arabidopsis accessions Ler-0 and Col-4, were analyzed. These seeds were provided by the NASC with the following accession numbers: N1900, N1901, N1903 to N1908, N1910 to N1971, N1973 to N1999, N4664, and N4686.
Sterile (in 150-mm petri dishes containing 100 mL of agar medium) and non-sterile (in pots containing a 1:1:1 [v/v] mixture of perlite, vermiculite, and sphagnum moss) cultures were performed at 20°C ± 1°C, 60% to 70% relative humidity, and continuous illumination of 7,000 lux, as described in Ponce et al. (1998).
Detection of Microsatellite Variation
DNA isolation and PCR amplifications were performed as described in Ponce et al. (1999). This high-throughput method is based on multiplex PCR amplification of microsatellites, followed by fluorescent semi-automated detection of the amplification products in an ABI PRISM 377 DNA sequencer (PerkinElmer Life Sciences, Boston). Genomic DNA samples of each accession were used as templates in four parallel multiplex PCR mixtures, each of which included five to six primer pairs. Each primer pair included one oligonucleotide labeled with a fluorochrome (HEX, 6-FAM, and TET phosphoramidites). The microsatellites co-amplified in each reaction mixture and the fluorochrome used to identify the corresponding amplification product (in parentheses) were as follows: nga361 (TET), AthACS (TET), AthGAPAab (6-FAM), AthZFPG (6-FAM), nga1111 (HEX), and AthCHIB (HEX) in mixture 1; nga162 (6-FAM), nga1107 (TET), nga1145 (HEX), nga1139 (HEX), nga168 (6-FAM), and AthGENEA (TET) in mixture 2; nga6 (HEX), AthCTR1 (TET), AthPHYC (6-FAM), T27k12-Sp6 (6-FAM), and nga1126 (TET) in mixture 3; and MBK5 (HEX), nga12 (HEX), AM4 (TET), nga111 (6-FAM), and nga139 (TET) in mixture 4. The sequences of the oligonucleotides used were as described in Ponce et al. (1999).
Microsatellite lengths were determined using the GENESCAN 2.1 fragment analysis software (Applied Biosystems, Foster City, CA). The number of repeats for each allele was estimated by comparing the size of its amplification product with that of the Col-0 accession, which was determined by Bell and Ecker (1994).
Gene diversity (or expected heterozygosity, H) was estimated following Nei (1973) and Innan et al. (1997): n (1 − Σp2)/(n − 1), where n is the number of samples and p is the frequency of an allele. The Microsat 1.5d program (E. Minch, unpublished data; available at http://hpgl.stanford.edu/projects/microsat/) was used to obtain genetic distance measurements. This program allows calculation of the DAD parameter (Goldstein et al., 1996), among others, for microsatellite data. The distance matrices obtained were used to construct a consensus phylogenetic tree (Innan et al., 1997) with the NEIGHBOR program, included in the PHYLIP 3.5c package (Felsenstein, 1993). Trees were plotted with the Treeview program (Page, 1996).
QTL Analysis
We constructed a linkage map using 173 molecular markers (42, 24, 29, 38, and 40 markers, respectively, for chromosomes 1–5), all of which were already genotyped by previous authors in at least 90 of the RILs studied here (information available at http://nasc.nott.ac.uk/RI_data/full_markers. text). These markers covered 519.5 cM (more than 85% of the Arabidopsis genome) and were spaced at intervals ranging from 0.5 to 8 cM, their average distance being 3 cM. The progeny from two to four plants per RIL was tested, and the average percentage for each phenotypic trait was determined. The germination ratio data were transformed by arcsin transformation to improve normality of the data. To perform QTL analyses, mean values for each RIL were used.
The computer program MapQTL 4.0 (Van Ooijen and Maliepaard, 1995) was used to identify the QTL linked to the molecular markers by using first interval mapping (Lander and Botstein, 1989) and then MQM mapping (Jansen, 1994). After interval mapping analysis, different combinations of the markers linked to the identified QTL were tested as cofactors for MQM mapping. We refined QTL intervals in this way, selecting as cofactors those markers that maximized the variance explained by each QTL. Those QTL detected with a LOD score above the threshold of 2.4 were considered as major QTL, whereas those below 2.4 but above 1.5 LOD scores were considered as minor QTL. These threshold levels are assumed to be equivalent to P ≤ 0.001 and P ≤ 0.01 values, respectively, in single-marker analyses (Lander and Botstein, 1989). QTL likelihood plots were constructed from the LOD scores obtained, using the SigmaPlot 2000 program (v6.0, Statistical Products and Service Solutions, Chicago). One and two LOD support intervals were established as 90% and 95% confidence intervals, respectively (Van Ooijen, 1992). The MapQTL program was also used to obtain estimates of the additive effect and the percentage of variance explained by each QTL, as well as the total variance explained by all of the QTL affecting each of the traits were analyzed.
Measurement of Germination
To study salt tolerance in accessions, 100 seeds from each wild-type line were used. To search for loci involved in salt tolerance at germination, two to four progenies (100 seeds per progeny) from each RIL or 100 F2 seeds from each cross between salt-tolerant and -sensitive accessions were used. Water-suspended seeds were sown using a Pasteur pipette, at a density of 200 (RILs) and 100 (F2 and accessions) regularly spaced seeds per plate, in 150-mm petri dishes filled with 100 mL of agar medium supplemented with 250 mm NaCl. We considered as germinated those seeds whose radicle had emerged through the seed coat. Germination response was scored during the 4 weeks after sowing. For each RIL progeny, we determined the germination percentage and the number of days required to reach the 50% of the final germination (T50) on 250 mm NaCl.
Measurement of Growth
Salt tolerance at the vegetative growth stage was tested by sowing seeds from the accessions N938, N1302, N1038, and N9222 and three progenies from each RIL on both non-supplemented and 50 mm NaCl-supplemented agar medium. Ten and 15 stressed or non-stressed plants from each RIL progeny and the above-mentioned accessions, respectively, were collected 3 weeks after sowing. Fresh weight was determined immediately after harvest and dry weight after desiccation for 24 h at 50°C in an oven.
ACKNOWLEDGMENTS
We are grateful to J.M. Barrero, H. Candela, S. Jover, J.M. Pérez-Pérez, P. Robles, and two anonymous referees for comments on the manuscript, to the NASC for providing seeds of accessions, and to S. Gerber and J.M. Serrano for their expert technical assistance. We are especially grateful to C. Alonso-Blanco for his useful suggestions.
Footnotes
This research was supported by the Ministerio de Ciencia y Tecnología of Spain (grant no. BIO2000–1082). V.Q. and P.P. were fellows of the Conselleria de Cultura, Educació i Ciència of the Generalitat Valenciana.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.006536.
LITERATURE CITED
- Abel GH, Mackenzie AJ. Salt tolerance of soybean varieties (Glycine max L. Merrill) during germination and later growth. Crop Sci. 1963;3:159–161. [Google Scholar]
- Alonso-Blanco C, Blankestijn-De Vries H, Hanhart CJ, Koornneef M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Din El-Assal S, Coupland G, Koornneef M. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics. 1998;149:749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M. Naturally occurring variation in Arabidopsis: an underexploited resource for plants genetics. Trends Plant Sci. 2000;5:22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Pantoja O. Towards the production of salt-tolerant crops. In: Shadidi F, Kolodziejczyk P, Whitaker JR, Munguia AL, Fuller G, editors. Chemicals via Higher Plant Bioengineering. 464 of Advances in Experimental Medicine and Biology. New York: Kluwer Academic/Plenum Publishers; 1999. pp. 77–89. [DOI] [PubMed] [Google Scholar]
- Begum F, Karmoker JL, Fattah QA, Maniruzzaman AFM. The effect of salinity on germination and its correlation with K+, Na+, Cl− accumulation in germinating seedlings of Triticum aestivum L. cv. Akbar Plant Cell Physiol. 1992;33:1009–1014. [Google Scholar]
- Bell DJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol. 2000;124:1595–1604. doi: 10.1104/pp.124.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Clarke JH, Mithen R, Brown JKM, Dean C. QTL analysis of flowering time in Arabidopsis thaliana. Mol Gen Genet. 1995;248:555–564. doi: 10.1007/BF02191594. [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- El-Assal E-DS, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Yeo AR. Breeding for salinity resistance in crop plants: where next? Aust J Plant Physiol. 1995;22:875–884. [Google Scholar]
- Foolad MR. Comparison of salt tolerance during seed germination and vegetative growth in tomato by QTL mapping. Genome. 1999;42:727–734. [Google Scholar]
- Frary A, Clint N, Frary A, Grandillo S, Van der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB et al. A quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA. 2000;97:4718–4723. doi: 10.1073/pnas.97.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Zhivotovsky LA, Nayar K, Ruíz-Linares A, Cavalli-Sforza LL, Feldman MW. Statistical properties of the variation at linked microsatellite loci: implications for the history of human Y-chromosomes. Mol Biol Evol. 1996;13:1213–1218. doi: 10.1093/oxfordjournals.molbev.a025686. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds. Plant Physiol. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath DP, McLarney BK, Thomsahow MF. Regulation of Arabidopsis thaliana L. (Heynh) COR78 in response to low temperature. Plant Physiol. 1993;103:1047–1053. doi: 10.1104/pp.103.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugly S, McCourt P, Browse J, Patterson GW, Somerville C. A chilling sensitive mutant of Arabidopsis with altered steryl-ester metabolism. Plant Physiol. 1990;93:1053–1062. doi: 10.1104/pp.93.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI. An mRNA binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- Innan H, Terauchi R, Miyashita NT. Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics. 1997;146:1441–1452. doi: 10.1093/genetics/146.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC. Controlling the type I and type II errors in mapping quantitative trait loci. Genetics. 1994;138:871–881. doi: 10.1093/genetics/138.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DW, Smith SE, Dobrenz AK. Genetic and phenotypic relationships in response to NaCl at different developmental stages in alfalfa. Theor Appl Genet. 1992;83:833–838. doi: 10.1007/BF00226705. [DOI] [PubMed] [Google Scholar]
- Juenger T, Purugganan M, Mackay T. Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics. 2000;156:1379–1392. doi: 10.1093/genetics/156.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1982;61:385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- Kowalski SP, Lan T-H, Feldmann KA, Paterson AH. QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol Gen Genet. 1994;245:548–555. doi: 10.1007/BF00282217. [DOI] [PubMed] [Google Scholar]
- Koyama ML, Levesley A, Koebner RMD, Flowers TJ, Yeo AR. Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 2001;125:406–422. doi: 10.1104/pp.125.1.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz AR. Demonstration of new and additional population samples and mutant lines of the AIS-seed bank. Arabidopsis Inf Serv. 1978;15:118–139. [Google Scholar]
- Kumar D, Singh B, Singh RK. Salt tolerance in wheat varieties. Soc Adv Breed Res Asia Oceania J. 1983;15:71–83. [Google Scholar]
- Lan TJ, Paterson AH. Comparative mapping of quantitative trait loci sculpting the curd of Brassica oleracea. Genetics. 2000;155:1927–1954. doi: 10.1093/genetics/155.4.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Alvarez-Gil M, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K, Jones MGK. Plant Biotechnology in Agriculture. New York: John Wiley & Sons; 1989. [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kin CS, Zhu J-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is requires for salt tolerance. Proc Natl Acad Sci USA. 2000;97:3730–3740. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Mano Y, Takeda K. Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.) Euphytica. 1997;94:263–272. [Google Scholar]
- McCouch S, Doerge RW. QTL mapping in rice. Trends Genet. 1995;11:482–487. doi: 10.1016/s0168-9525(00)89157-x. [DOI] [PubMed] [Google Scholar]
- McCue KF, Hanson AD. Drought and salt tolerance: towards understanding and application. Trends Biotechnol. 1990;8:358–362. [Google Scholar]
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Shen B, Bohnert HJ. Salinity tolerance: mechanisms, models and the metabolic engineering of complex traits. In: Setlow JK, editor. Genetic Engineering. Vol. 20. New York: Plenum Press; 1999. pp. 153–176. [DOI] [PubMed] [Google Scholar]
- Ni BR, Bradford KJ. Germination and dormancy of abscisic acid and gibberellin-deficient mutant of tomato (Lycopersicon esculentum) seeds. Plant Physiol. 1993;101:607–617. doi: 10.1104/pp.101.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET. Differential expression of two related low temperature induced genes in Arabidopsis thaliana L. Heynh Plant Mol Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Ponce MR, Quesada V, Micol JL. Rapid discrimination of sequences flanking and within T-DNA insertions in the Arabidopsis genome. Plant J. 1998;14:497–501. doi: 10.1046/j.1365-313x.1998.00146.x. [DOI] [PubMed] [Google Scholar]
- Ponce MR, Robles P, Micol JL. High-throughput genetic mapping in Arabidopsis thaliana. Mol Gen Genet. 1999;261:408–415. doi: 10.1007/s004380050982. [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics. 2000;154:421–436. doi: 10.1093/genetics/154.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röbbelen G. The LAIBACH standard collection of natural races. Arabidopsis Inf Serv. 1965;2:36–47. [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 1999;20:67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- Szaboles I. The global problems of salt-affected soils. Acta Agron Hung. 1987;36:159–172. [Google Scholar]
- Taji T, Motoaki S, Yamaguchi-Shinozaki K, Kamada H, Giraudat J, Shinozaki K. Mapping od 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiol. 1999;40:119–123. doi: 10.1093/oxfordjournals.pcp.a029469. [DOI] [PubMed] [Google Scholar]
- Thorlby G, Veale E, Butcher K, Warren G. Map position of SFR genes in relation to other freezing related genes of Arabidopsis thaliana. Plant J. 1999;17:445–452. doi: 10.1046/j.1365-313x.1999.00395.x. [DOI] [PubMed] [Google Scholar]
- Todokoro S, Terauchi R, Kawano S. Microsatellite polymorphism in natural populations of Arabidopsis thaliana in Japan. Jpn J Genet. 1995;70:543–554. [Google Scholar]
- Tsugane K, Kobayashi K, Niwa Y, Ohba Y, Wada K, Kobayashi H. A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell. 1999;11:1195–1206. doi: 10.1105/tpc.11.7.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Schaar W, Alonso-Blanco C, Léon-Kloosterziel KM, Jansen RC, Van Ooijen JW, Koornneef M. QTL analysis of seed dormancy in Arabidopsis using recombinant inbred lines and MQM mapping. Heredity. 1997;79:190–200. doi: 10.1038/hdy.1997.142. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW, Maliepaard C. MapQTL 3.0: Software for the Calculation of QTL Positions on Genetics Maps. Wageningen, The Netherlands: CPRO-DLO; 1995. [Google Scholar]
- Werner JE, Finkelstein RR. Arabidopsis mutants with reduced response to NaCl and osmotic stress. Physiol Plant. 1995;93:659–666. [Google Scholar]
- Xin Z, Browse J. eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc Natl Acad Sci USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu J-K. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress-and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/TPC.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M. Genetic and molecular dissection of naturally occurring variation. Curr Opin Plant Biol. 2001;4:130–135. doi: 10.1016/s1369-5266(00)00148-5. [DOI] [PubMed] [Google Scholar]
- Zhang H-X, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001;19:765–768. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- Zhang H-X, Hodson JN, Williams JP, Blumwald E. Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA. 2001;98:12833–12836. doi: 10.1073/pnas.231476498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]